Abstract
Myocyte enhancer factor 2 (MEF2) is in the MADS (MCM1agamous-deficiens-serum response factor) family of transcription factors. Although MEF2 is known as a myogenic factor, the expression pattern of the MEF2 family of genes (MEF2A-D) in developing brain also suggests a role in neurogenesis. Here we show that transfection with MEF2C, the predominant form in mammalian cerebral cortex, induces a mixed neuronal/myogenic phenotype in undifferentiated P19 precursor cells. During retinoic acid-induced neurogenesis of these cells, a dominant negative form of MEF2 enhances apoptosis but does not affect cell division. The mitogen-activated protein kinase p38α activates MEF2C. Dominant negative p38α also enhances apoptotic death of differentiating neurons, but these cells can be rescued from apoptosis by coexpression of constitutively active MEF2C. These findings suggest that the p38α/MEF2 pathway prevents cell death during neuronal differentiation.
Myocyte enhancer factor 2 (MEF2) proteins are members of the MADS (MCM1-agamous-deficiens-serum response factor) family of transcription factors (1). Four members of the family (MEF2A, MEF2B, MEF2C, and MEF2D) have been reported in mouse and human (2–6), whereas one MEF2 homologue, D-MEF2, has been identified in Drosophila (7). Various mitogen-activated protein (MAP) kinases (p38α, p38β2, and big MAP kinase 1/extracellular signal-regulated kinase 5) phosphorylate and thereby activate the MEF2 family (8–11). These MAP kinase pathways lead to MEF2 modulation of gene expression (8–10). The MEF2 family of genes is highly expressed in cells of muscle lineage (4–6, 12). Several studies have suggested a role for MEF2 in myogenesis. In D-mef2 loss-of-function Drosophila, muscle cells do not differentiate (13). Mef2c-null mice are embryonic lethal because of malformation of the heart (14). A dominant negative form of MEF2 inhibits myotube formation in myoblastic cell lines (15). Additionally, the molecular mechanism of MEF2 action has been examined during myogenesis. MEF2 proteins physically interact with the basic helix–loop–helix (bHLH) myogenic transcription factors MyoD and myogenin to initiate muscle development (16, 17).
All MEF2 family members are highly expressed in neurons in the central nervous system (CNS) (18), especially MEF2C as we have shown (3). The level of MEF2 expression increases in differentiating neurons in the developing brain (3, 18–20), and we and others therefore have suggested the possible involvement of MEF2 in neuronal differentiation. Analogies between neurogenesis and myogenesis have been drawn in both Drosophila and vertebrates (21). In those models, bHLH transcription factors play critical roles in both myogenesis and neurogenesis. Several neuronal bHLH transcription factors have been identified during mammalian development (22). Ectopic overexpression of neuronal bHLH factors NeuroD (1)/BETA2, NeuroD2/KW8/NDRF, or NeuroD3/neurogenin1 in Xenopus causes neurogenic conversion of ectoderm (22). Additionally, physical and functional interaction between a neuronal bHLH transcription factor (Mash-1) and MEF2 proteins has been reported (23, 24). These findings suggested to us that the MEF2 family may play a role in neuronal differentiation in concert with neuronal bHLH transcription factors, just as the MEF2 family is involved in myogenesis along with myogenic bHLH transcription factors.
In this study, we examined the role of MEF2 in neuronal differentiation by using the P19 embryonal carcinoma cell line as a model system. P19 cells terminally differentiate into neuronal cells after retinoic acid treatment, and the process of neurogenesis in P19 cells is similar to that of the mammalian CNS (25, 26). Moreover, apoptotic cell death is observed in neuronally differentiating P19 cells, similar to that in the fetal brain, and common mechanisms have been suggested (27–31). Thus, P19 is thought to represent an excellent in vitro model for neuronal differentiation of the CNS in vivo. Here we show that transfection with MEF2C induces differentiation of P19 cells into a mixed neurogenic/myogenic phenotype. In contrast, a dominant negative form of MEF2 that inhibits MEF2 function enhances apoptotic cell death of differentiating P19 cells. Additionally, inhibition of p38α, a known activator of MEF2, also increases apoptosis in differentiating cells. Coexpression of constitutively active MEF2C reduces this apoptosis. These results suggest that the p38α/MEF2 cascade protects differentiating cells from death during neurogenesis.
Materials and Methods
Induction of Neuronal Differentiation in P19 Cells.
P19 cells were purchased from the American Type Culture Collection (CRL 1825) and maintained in α-MEM (Sigma), supplemented with 10% heat-inactivated FBS (Intergen, Purchase, NY). For neuronal differentiation, 1 × 106 P19 cells were cultured in a 10-cm diameter tissue culture dish with 300 pM 13-cis retinoic acid (Eastman Kodak) for 2 days. After trypsinization, the cells were again exposed to 300 pM retinoic acid and reseeded onto a bacterial grade Petri dish to allow the cells to aggregate, which is known to facilitate neuronal differentiation. After a 1-day incubation, cell aggregates were collected and dissociated with trypsin-EDTA. The dissociated cells were plated onto a tissue culture chamber slide (Nunc). The medium was changed the day after plating and every 2 days thereafter.
Gel Shift Assays.
Nuclear extracts of undifferentiated or retinoic acid-treated P19 cells were prepared as described (32). Protein concentrations were measured with a Micro BCA Protein Assay Reagent Kit (Pierce) using albumin as the standard. Nuclear extracts (5 μg/20 μl) were preincubated on ice for 10 min in a solution containing 20 mM Tris (pH 7.6), 10% glycerol, 1 mM DTT, 80 mM KCl, and 1 μg poly(dI-dC)⋅(dI-dC). 32P-end-labeled double-stranded oligonucleotide representing the MEF2 site (TGGGCTATAAATAGCCGC) of the brain-specific creatine kinase gene then was added and incubated at room temperature for 20 min. The binding mixture then was electrophoresed on a 6% nondenaturing acrylamide gel in 0.25 × 90 mM Tris/90 mM boric acid/2 mM EDTA, pH 8.3 for 1.5 h at 150 V. For supershift assays, antibodies against MEF2A (Santa Cruz Biotechnology), MEF2C (3), or MEF2D (generously provided by B. Kosofsky, Massachusetts General Hospital, Boston) were added to the preincubation binding mixtures.
Immunoblots.
Whole-cell lysates were prepared in RIPA buffer (1 × PBS/1% NP-40/0.5% sodium deoxycholate/0.1% SDS) containing 0.1 mg/ml PMSF and 1 mM sodium vanadate. Proteins in 50-μg aliquots were separated by SDS/PAGE and then transferred onto a nitrocellulose membrane (Amersham Life Science). Membranes were incubated overnight at 4°C with primary antibody to MEF2C (1:1,000), MEF2D (1:500), phospho p38 (1:1,000, New England Biolabs), or p38α (1:1,000, Santa Cruz Biotechnology). Horseradish peroxidase-linked anti-rabbit IgG (Vector) was used as the secondary antibody. Immunoblots were visualized with an enhanced chemiluminescence system (ECL, Amersham Pharmacia Biotech).
Plasmid Constructs.
The phosphoglycerate kinase gene promoter-driven expression vector (pGK) was kindly provided by M. W. McBurney (University of Ottawa, Canada). Human MEF2C cDNA was inserted between the BamHI and XhoI sites of pGK (termed pGK-MEF2C). Dominant negative MEF2 (amino acids 1–105) tagged with a flag sequence (pcDNAI-MEF2C 1–105 flag) and constitutively active MEF2C (pcDNAI-MEF2C 1–117/VP16) were the kind gifts of Eric N. Olson (Southwestern Medical Center, Dallas). MEF2C 1–105 flag acts as a MEF2 dominant negative by binding to the MEF2 site without producing activation because it lacks the transactivation domain (33). MEF2C 1–105 flag cDNA was ligated into the BamHI/XhoI sites of pGK (termed pGK-DN). We also engineered a mutation in this MEF2 dominant negative by changing the arginine residue at position 24 to a leucine (termed pGK-DNmt). This mutated MEF2 dominant negative was unable to bind to the MEF2 site and therefore served as control to rule out the possibility that the MEF2 dominant negative was affecting cell survival nonspecifically by binding to sites other than MEF2. An enhanced green fluorescent protein (GFP) vector (pEGFP-N1) was purchased from CLONTECH. Dominant negative expression vectors for p38α [pcDNA3-p38α(AF)] and p38β2 [pcDNA3-p38β2(AF)] were kindly provided by J. Han (The Scripps Research Institute, La Jolla, CA) (10).
Overexpression of MEF2C in P19 Cells.
A total of 2 × 105 P19 cells were plated in a 6-cm diameter dish 24 h before transfection. Then, 25 μg of the MEF2C expression vector (pGK-MEF2C) and 1 μg of the neomycin resistance gene expression vector (pSV neo) were cotransfected by calcium phosphate precipitation. The cells were washed 16 h posttransfection and cultured in MEM with 10% serum. After 24 h, the cells were trypsinized and seeded onto a tissue culture chamber slide. The cells were maintained in 200 μg/ml Geneticin for 5 days to select the transfected cells.
Establishment of P19 Cells Stably Expressing a Dominant Negative Form of MEF2.
Using the calcium phosphate precipitation method, 2 × 105 P19 cells were transfected with 24 μg of an empty expression vector (pGK), an expression vector encoding a dominant negative MEF2 (pGK-DN), or a mutated dominant negative MEF2 construct (pGK-DNmt), in addition to the neomycin resistance gene (pSV neo). The transfected cells were selected by exposure for 10 days to 200 μg/ml Geneticin. The selection medium was changed every 2 days. Stable clones expressing the MEF2 dominant negative and the mutated MEF2 dominant negative were selected with reverse transcriptase–PCR.
Immunocytochemistry.
Cultures were fixed with 3% paraformaldehyde at room temperature for 40 min. After washing three times with PBS, cells were permeabilized with 0.3% Triton X-100 for 5 min. The free aldehyde groups formed during the fixation were reduced by incubation with 1 mg/ml sodium borohydride three times for 5 min each. Cells then were washed three times in PBS. The fixed cells were incubated at 4°C overnight with primary mAbs to microtubule-associated protein 2 (MAP2, 1:500; Sigma, clone HM-2), neurofilament H (1:250; Sternberger Monoclonals, Baltimore, SMI311), myosin heavy chain (1:250; Developmental Studies Hybridoma Bank, Iowa City, MF20), glial fibrillary acidic protein (1:400; Sigma, clone G-A-5), or rabbit antiserum to MEF2C (1:250) (3). Cells then were washed three times in PBS containing 0.2% Tween 20, and rhodamine-conjugated anti-mouse IgG or fluorescein-conjugated anti-rabbit IgG (each at 1:100; Boehringer Mannheim) was added as the secondary antibody. After a 1-h incubation at room temperature, the cells were washed again and mounted. For Hu and nestin staining, cells were fixed with acid ethanol (95% ethanol/5% acetic acid) for 30 min at room temperature. The fixed cells then were washed three times with PBS and incubated with a mAb to Hu (1:200, gift of M. F. Marusich and J. A. Weston, University of Oregon, Eugene) or to nestin (1:20, Developmental Studies Hybridoma Bank, Rat-401). After overnight incubation at 4°C, the samples were further washed and incubated with anti-mouse Igs conjugated to horseradish peroxidase (1:100; Dako). A peroxidase reaction was performed by using 3,3′-diaminobenzidine tetrahydrochloride (Sigma). Stained preparations were examined under epifluorescence microscopy.
Apoptotic Assays.
Cells were incubated with the DNA dye Hoechst 33342 (1 μg/ml) for 5 min at 37°C to observe nuclear morphology. After washing with PBS, cells were fixed with acid ethanol (95% ethanol/5% acetic acid) for 10 min at room temperature. Samples then were washed with distilled water three times and mounted. Apoptotic nuclei were counted at ×400 magnification. Within several hours of dying by apoptosis, cells underwent secondary necrosis (because they were not phagocytosed in these cultures) and detached from the substrate (34). Hence, several hours after apoptotic cell death, dead cells were no longer present to be stained by Hoechst dye. This temporal separation made it possible to distinguish the number of cells recently undergoing apoptosis by using sequential Hoechst staining at different time points. Additionally, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assays were performed in a blinded fashion using an In Situ Cell Death Detection Kit tagged with tetramethyl-rhodamine (Roche Molecular Biochemicals) or an Apoptosis Detection System Kit tagged with fluorescein (Promega).
Cell Proliferation Assays.
To label proliferating cells, BrdUrd (1:1,000; Amersham Life Science) was added to cultures for 2 h at 37°C. After washing with PBS, cells were treated with acid ethanol (95% ethanol/5% acetic acid) for 30 min at room temperature and cellular DNA was denatured with 2 M HCl. After an additional wash in PBS, cells were incubated at 4°C overnight in rat monoclonal anti-BrdUrd antibody (1:10; Harlan Sera-Lab Limited, Belton Loughborough, U.K.) and mouse monoclonal anti-nestin antibody (11 μg/ml). After three washes in PBS, secondary antibodies were added: rhodamine red-X-conjugated anti-rat IgG (1:200) and biotin-SP-conjugated anti-mouse IgG (1:50; Jackson ImmunoResearch) followed by streptavidin-fluorescein (1:25; Amersham Life Science). Cells were examined under epifluorescence microscopy.
Transient Transfection During Differentiation of P19 Cells.
P19 cells (2 × 105) were seeded onto 6-well tissue culture plates and treated with 300 pM retinoic acid to induce neuronal differentiation. One day later, cells were transfected with 0.83 μg of a p38 dominant negative construct or vector alone [pcDNA3-p38α(AF), pcDNA3-p38β2(AF) or pcDNA3], 0.83 μg of a constitutively active MEF2C construct or vector alone (pcDNAI-MEF2C 1–117/VP16 or pcDNAI-Amp), plus 0.33 μg of a GFP construct to identify transfected cells (pEGFP-N1). A lipid-based transfection system was used (6 μl of TransFast, Promega). The next day cells were transferred to 3.5-cm bacterial dishes and treated with an additional 300 pM retinoic acid. TUNEL assays were performed on day 3 after initiating retinoic acid. More than 1,200 GFP-positive cells were scored for apoptosis from each culture plate under epifluorescence microscopy, and each experiment was replicated on three separate days.
Results
Retinoic Acid Induces MEF2 Protein Expression in P19 Cells.
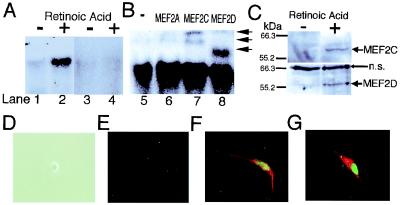
Treatment with retinoic acid induces neuronal differentiation of P19 cells (25, 26). We previously showed that MEF2, especially MEF2C, is expressed during neurogenesis in the rodent cerebral cortex (3). To examine whether MEF2 proteins are expressed during neurogenesis of P19 cells, gel shift assays were performed by using the MEF2 site as a probe. Only very faint binding activity was detected in undifferentiated P19 cells (Fig. 1A, lane 1). Binding activity increased to high levels 2 days after retinoic acid treatment (Fig. 1A, lane 2). Unlabeled MEF2 oligonucleotide abrogated the binding activity, indicating the specificity of the binding to the MEF2 site (Fig. 1A, lanes 3 and 4). Although anti-MEF2A antibody did not affect the formation of the complex (Fig. 1B, lanes 5 and 6), anti-MEFC and anti-MEF2D supershifted the bands (Fig. 1B, lanes 7 and 8), indicating the presence of MEE2C and MEF2D proteins in the complex. Immunoblotting revealed that the level of MEF2C and MEF2D proteins increased after retinoic acid treatment (Fig. 1C). These results indicate that retinoic acid treatment induces MEF2 site-binding activity by MEF2C and MEF2D, accompanied by an increase in MEF2C and MEF2D protein expression during neurogenesis of P19 cells.
Figure 1.
MEF2 binding activity, protein expression and transfection during neuronal differentiation of P19 cells. (A) Gel shift assays show that MEF2 binding activity increased during neuronal differentiation of P19 cells. A 32P-labeled MEF2 site oligonucleotide was incubated with nuclear extracts from undifferentiated P19 cells (lanes 1 and 3), or from P19 cells treated with retinoic acid for 2 days (lanes 2 and 4). Cold competition by unlabeled MEF2 site oligonucleotides (lanes 3 and 4). (B) Antibody to MEF2A (lane 6), MEF2C (lane 7), or MEF2D (lane 8) was added to the binding mixture for supershift assays. Anti-MEF2C yielded two supershifted bands, representing one or more DNA complexes containing MEF2C, whereas anti-MEF2D produced a single supershifted complex (arrows) (3). (C) Immunoblots revealed that protein expression of MEF2C and MEF2D was induced during neuronal differentiation of P19 cells. Whole-cell lysates from undifferentiated P19 cells or P19 cells treated with retinoic acid for 2 days were used for these immunoblots (n.s., nonspecific bands). (D–G) Overexpression of MEF2C induced a mixed neurogenic/myogenic phenotype. Undifferentiated P19 cells did not display immunoreactivity for MEF2C or neurofilament (D, phase contrast image; E, immunocytochemistry). Undifferentiated P19 cells were transfected with an expression vector for MEF2C. Immunoreactivity for MEF2C is represented in green (E–G), neurofilament in red (E and F), and myosin heavy chain in red (G). Similar findings were observed in 16 experiments in which more than 200 cells were scored.
Overexpression of MEF2C Transforms P19 Cells into a Mixed Neurogenic/Myogenic Phenotype Expressing Neurofilament and Myosin Heavy Chain.
P19 cells will develop into myogenic rather than neurogenic cells if exposed to DMSO instead of retinoic acid (25). Therefore, to examine the potential role of MEF2C in the differentiation process, we overexpressed MEF2C in undifferentiated P19 cells in the absence of retinoic acid or DMSO. Undifferentiated P19 cells did not show immunoreactivity for MEF2C or neurofilament (Fig. 1E). MEF2C-transfected cells manifest MEF2C protein in the nucleus, as determined by specific antibody labeling, and were neuronal in character because they labeled with antineurofilament. In fact, all transfected cells (from more than 200 such cells scored) expressed MEF2C, stained with antineurofilament, and extended two neuronal-like processes, producing a bipolar appearance (Fig. 1F). None of the MEF2C-transfected cells stained positively for glial fibrillary acidic protein, indicating that they did not manifest an astrocytic phenotype.
Previously, it was reported that overexpression of MEF2A initiated the myogenic phenotype in the 10T1/2 fibroblast cell line (16), but Olson and colleagues (17) found that other factors (MyoD or myogenin) were also necessary to activate the myogenic program. To test whether MEF2C induces myogenic features in P19 cells, we stained cells transfected with MEF2C with anti-myosin heavy chain antibody and found that all cells expressing MEF2C (more than 200 counted) were also positive for myosin heavy chain label (Fig. 1G). Taken together, these results indicate that transfection of undifferentiated P19 cells with MEF2C induces a bipolar cell phenotype that expresses both neuronal (neurofilament) and myogenic (myosin heavy chain) markers.
P19 Clones Stably Expressing a Dominant Negative Form of MEF2.
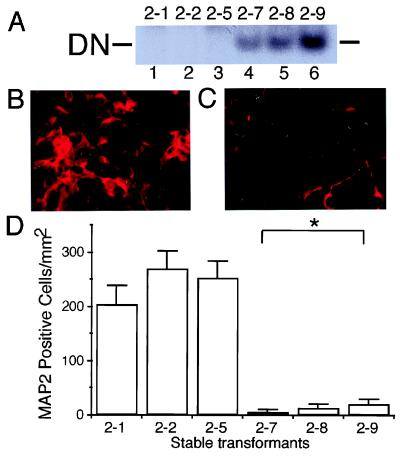
To investigate the role of endogenous MEF2 proteins in retinoic acid-induced neuronal differentiation of P19, we used a dominant negative form of MEF2. MEF2 proteins are functionally divided into two regions. The N-terminal region [containing MADS (MCM1-agamous-deficiens-serum response factor) and MEF2 domains] is responsible for binding to the MEF2 site, whereas the C-terminal region is necessary for transcriptional activity (5, 33). Because the MADS and MEF2 domains alone lack transcriptional activity, the N-terminal region of MEF2 acts as a dominant negative (5). Dominant negative MEF2 has been shown to inhibit myotube formation in myoblastic cell lines (15). In the present study, we established stable transformants of P19 cells expressing an N-terminal (amino acids 1–105) MEF2C dominant negative. We obtained three “vector-alone” transfected control clones (designated clones 2–1, 2–2, and 2–5) and three clones expressing the MEF2 dominant negative (designated clones 2–7, 2–8, and 2–9). As a further control, two additional clones (clones 2–16 and 2–21) were produced that expressed a mutated form of the MEF2 dominant negative construct (see Materials and Methods). Expression of the MEF2 dominant negative was monitored with gel shift assays. Binding activity of the MEF2 dominant negative was detected for clones 2–7, 2–8, and 2–9, but not for control clones 2–1, 2–2, 2–5, 2–16, or 2–21 (Fig. 2A and data not shown). All transformants were morphologically indistinguishable from parent P19 cells.
Figure 2.
Inhibition of MEF2 function decreases the number of neuronal (MAP2-positive) P19 cells after retinoic acid treatment. (A) Undifferentiated P19 cells were stably transfected with empty vector (clones 2–1, 2–2, and 2–5 in lanes 1, 2, and 3, respectively) or MEF2 dominant negative (clones 2–7, 2–8, and 2–9 in lanes 4, 5, and 6, respectively). Expression of the MEF2 dominant negative was demonstrated in a gel shift assay using a radiolabeled MEF2 site oligonucleotide and nuclear extracts of each clone. DN: binding complex of the MEF2 site and dominant negative MEF2 protein. (B and C) Cultures from control (B, clone 2–1) and MEF2 dominant negative (C, clone 2–7) transformants were treated with retinoic acid for 7 days to induce neurogenesis, and neuronal differentiation then was evaluated by immunocytochemistry with anti-MAP2. (D) Control cultures (clones 2–1, 2–2, and 2–5) and MEF2 dominant negative cultures (clones 2–7, 2–8, and 2–9) were treated with retinoic acid and scored for the number of MAP2-positive cells (n = 6 experiments; *, P < 0.0001 by ANOVA and post hoc comparison).
Inhibition of MEF2 Function Diminishes the Number of Neuronal Cells.
Neuronal differentiation occurs via multiple sequential steps (35). Nerve cells differentiate from unipotent neuronal precursors, which arise from multipotent precursor cells. To begin to monitor the effect of the MEF2 dominant negative on these steps of neuronal differentiation, we first examined the appearance of differentiated neurons by using antibodies to neuronal markers, such as neurofilament and MAP2. Stable transformants expressing a dominant negative form of MEF2 to inhibit MEF2 function were treated with retinoic acid to induce neuronal differentiation. After 7 days, cells were fixed and stained with an antibody to MAP2. Many MAP2-positive cells appeared in the control cultures (Fig. 2B), whereas the number of MAP2-positive cells was dramatically reduced in the MEF2 dominant negative cultures (Fig. 2C). The number of MAP2-positive cells in the MEF2 dominant negative cultures was statistically smaller than that of the control groups (Fig. 2D and data not shown).
Inhibition of MEF2 Function Reduces the Number of Neuronal Precursor Cells.
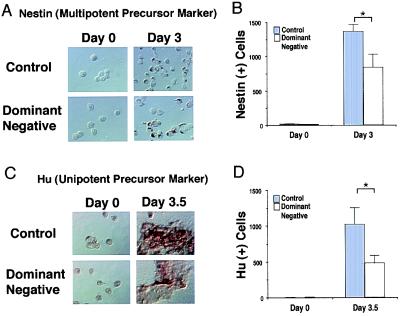
Next, we investigated whether the decrease in terminally differentiated neurons after inhibition of MEF2 function was caused by a decrease in the number of precursor cells (multipotent precursor or unipotent precursor cells). The appearance of multipotent neuronal precursors (nestin-positive cells) and unipotent precursors (Hu-positive cells) was monitored in control (clone 2–1) and MEF2 dominant negative cultures (clone 2–7). Nestin-positive and Hu-positive cells were counted before and after retinoic acid treatment for 3.0 and 3.5 days, respectively. At these time points, multipotent precursor cells (3.0 days) followed by unipotent precursor cells (3.5 days) appear in these cultures before the neuronally differentiated state (25). Hu-positive and nestin-positive cells were induced after retinoic acid treatment in both control (clone 2–1) and MEF2 dominant negative cultures (clone 2–7; Fig. 3 A and C). However, the number of Hu-positive and nestin-positive cells in the MEF2 dominant negative cultures was significantly smaller than that of the control cultures (Fig. 3 B and D). These results suggest that interference with MEF2 activity reduces the number of multipotent precursor and unipotent neuronal precursor cells.
Figure 3.
Inhibition of MEF2 function decreases the number of multipotent and unipotent precursor cells. Control cultures (clone 2–1) and MEF2 dominant negative cultures (clone 2–7) were treated with retinoic acid for 3.0 or 3.5 days. (A and C) Cells incubated with anti-nestin to label multipotent precursor cells (A) or anti-Hu to label unipotent precursor cells (C). Labeled cells were visualized with peroxidase. (B and D) The number of nestin-positive cells (B) and Hu-positive cells (D) in 40 randomly selected fields was scored in a blinded fashion. Values are mean ± SD from at least three independent experiments (*, P < 0.02 by Student's t test).
Inhibition of MEF2 Function Enhances Apoptotic Cell Death During Neuronal Differentiation.
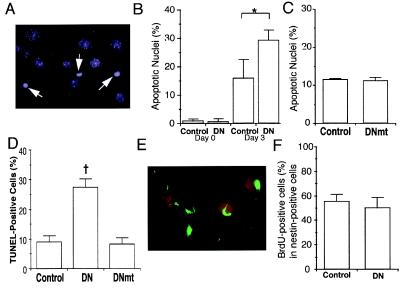
Apoptotic cell death of differentiating cells is found widely in the developing fetal brain (29). Similarly, apoptosis also is observed during the course of neuronal differentiation of P19 cells (27, 28). We therefore hypothesized that MEF2 might be essential for survival during development, and that interference with MEF2 activity by the dominant negative might enhance apoptotic death of differentiating cells, leading to a reduction in the number of multipotent or unipotent precursor cells. To test this premise, we examined nuclear morphology with the DNA dye Hoechst 33342 (Fig. 4A) to score the number of apoptotic cells after 3 days of retinoic acid treatment, at which time multipotent precursors were detected. Before the addition of retinoic acid, fewer than 1% of the cells manifest apoptotic nuclei in control or MEF2 dominant negative cultures. After 3 days of retinoic acid treatment, a significant number of cells displayed apoptotic nuclei in the controls (transfected with either empty vector or the mutated form of the MEF2 dominant negative) (Fig. 4 B and C). However, apoptosis was increased in the MEF2 dominant negative cultures (Fig. 4B). These findings were confirmed by another apoptosis assay using the TUNEL technique (Fig. 4D). Additionally, the number of apoptotic cells was increased in the MEF2 dominant negative cultures after 3.5 days of retinoic acid treatment, when unipotent precursor cells predominate. These results suggest that interference with MEF2 activity increases apoptotic cell death during neuronal differentiation of P19 cells. Moreover, these results are consistent with the notion that MEF2 transcriptional activity is essential for the prevention of cell death during neuronal development.
Figure 4.
Effects of inhibition of MEF2 function during neuronal differentiation of P19 cells. Control cultures (clone 2–1), MEF2 dominant negative cultures (clone 2–7, labeled DN), and mutated MEF2 dominant negative cultures (clone 2–16, labeled DNmt) were treated with retinoic acid for 3 days. (A) Representative apoptotic cells with condensed nuclei from a MEF2 dominant negative clone treated with retinoic acid and stained with Hoechst dye to detect apoptotic morphology (white arrows). (B) Percentage of apoptotic cells in control or MEF2 dominant negative cultures before and after retinoic acid treatment. (C) Similar percentage of apoptotic cells in control or mutated MEF2 dominant negative cultures after 3 days of retinoic acid. (D) Apoptosis in control, dominant negative, or mutated dominant negative cultures treated with retinoic acid for 3 days scored by the TUNEL technique. (E and F) Lack of effect of MEF2 dominant negative on multipotent precursor cell proliferation. Control cultures and MEF2 dominant negative cultures were treated with retinoic acid for 3 days. BrdUrd then was added to visualize proliferating cells. (E) Dividing multipotent precursor cells detected by double staining with anti-BrdUrd antibody (red) and anti-nestin antibody (green) in retinoic acid-treated control cells. (F) Comparison of BrdUrd incorporation into multipotent (nestin-positive) precursor cells in control and MEF2 dominant negative cultures. Values are mean ± SD from at least three independent experiments (*, P < 0.05 by Student's t test; †, P < 0.001 by ANOVA and post hoc comparison).
Inhibition of MEF2 Function Does Not Affect Cell Division of Multipotent Precursor Cells.
Multipotent precursor cells are known to proliferate. The proliferation of multipotent precursors leads to an expansion of the cell population that eventually can differentiate into neurons. We therefore next determined whether dominant negative MEF2 affects the proliferation of multipotent precursor cells. Control and MEF2 dominant negative cultures were treated with retinoic acid for 3 days, and then BrdUrd was added to detect dividing cells. BrdUrd-positive cells and multipotent precursor cells were identified by double labeling with anti-BrdUrd and anti-nestin antibodies, respectively (Fig. 4E). The percentage of cells positive for both BrdUrd and nestin (proliferating, multipotent precursor cells) was similar in the control and MEF2 dominant negative cultures (Fig. 4F). This finding suggests that MEF2 transcriptional activity has no significant effect on cell division of multipotent precursor cells.
Inhibition of p38 MAP Kinase Increases Apoptosis During Neuronal Differentiation.
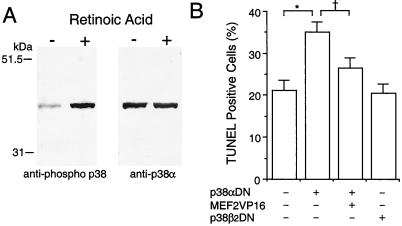
Interference with MEF2 function enhanced apoptosis in differentiating P19 cells, suggesting that MEF2 transcriptional activity is necessary for the prevention of cell death during neuronal development. Two members of the p38 MAP kinase family, p38α and p38β2, are known to activate MEF2 via phosphorylation of Ser/Thr residues (8, 10, 11). Moreover, the p38 MAP kinase family has been reported to play an essential role in life or death decisions in several cell types (36). Thus we hypothesized that the p38/MEF2 pathway prevents apoptosis in differentiating P19 cells. To begin to test this possibility, we examined activation of p38 family members by immunoblotting with an anti-phospho-pan p38 antibody, which recognizes all activated/phosphorylated p38 MAP kinases. One band was strongly induced after retinoic acid treatment, and the mobility of this band was the same as p38α (Fig. 5A). Total (phosphorylated and unphosphorylated) p38α protein appeared to be present at similar levels before and after stimulation with retinoic acid (Fig. 5A Right). In contrast, p38β2 was undetectable with two different antibodies, although these antibodies clearly reacted with recombinant p38β2 protein under the same conditions in our experiments (data not shown). These results indicate that p38α is activated/phosphorylated during the induction of neurogenesis. Additionally, we found that transfection of dominant negative p38α, but not dominant negative p38β2, enhanced apoptotic cell death in differentiating cells. Coexpression of constitutively active MEF2C significantly rescued these differentiating cells from apoptosis (Fig. 5B). These findings are consistent with the notion that the p38α/MEF2 cascade plays a role in preventing apoptotic cell death during neuronal differentiation.
Figure 5.
Involvement of the p38α/MEF2 pathway in preventing apoptosis during neuronal differentiation of P19 cells. (A) p38α was phosphorylated during induction of neuronal differentiation by retinoic acid. Anti-phospho p38 was used to detect activated/phosphorylated p38 family members on immunoblots during induction of neuronal differentiation. The same membrane then was stripped and reblotted with a p38α-specific antibody that labeled the same band. (B) Dominant negative p38α (p38αDN) enhanced apoptosis during neuronal differentiation. Constitutively active MEF2C (MEF2VP16) significantly rescued the differentiating cells from apoptosis. Dominant negative p38β2 (p38β2DN) had no effect on apoptosis compared with control (expression vector only). After treatment with retinoic acid for 1 day, cells were transfected with the indicated expression vector(s) along with a GFP expression vector to identify the transfected cells. The number of transfected apoptotic cells was determined in a blinded fashion by TUNEL assay on day 3 of retinoic acid treatment. More than 1,200 GFP-positive cells were scored in each culture. Mean ± SD are shown from three experiments (*, P < 0.001; †, P < 0.01 by ANOVA and post hoc comparison).
Discussion
Neuronal differentiation in the mammalian CNS is driven by multiple events. Clonal analysis in vivo and in vitro has shown that neuronal differentiation is composed of at least two steps (35): (i) Multipotent precursor cells give rise to unipotent precursors, and, in turn, (ii) neurons differentiate from these unipotent precursors. Both proliferation and cell death are essential components of neuronal differentiation. Multipotent precursor cells proliferate to expand their population, but up to 70% of the differentiating cells die by apoptosis in the developing cerebral cortex (29). In the present study, we show that MEF2 protein expression and binding activity, similar to that in the developing mammalian cerebral cortex (3), are induced during neuronal differentiation of P19 cells. Transfection of undifferentiated P19 cells with MEF2C, the predominant form in the developing cerebral cortex, induced a mixed neurogenic/myogenic phenotype, implying that additional factors in conjunction with MEF2 are necessary to determine ultimate cell lineage. On the other hand, if MEF2 activity was inhibited with dominant negative MEF2 during retinoic acid-induced neuronal differentiation, the number of multipotent precursor, unipotent precursor, and neuronal cells in the cultures was reduced. This decrease in cell number resulted from enhanced apoptotic cell death rather than from inhibition of multipotent precursor cell division. Additionally, inhibition of p38α MAP kinase, an event upstream from MEF2 activation, also increased apoptosis in differentiating cells. However, differentiating cells could be rescued from apoptosis because of transfection of a dominant negative p38α construct by coexpression of a constitutively active form of MEF2C, proving the specificity of MEF2C in these events. These findings suggest that the p38α/MEF2 pathway plays an important role in preventing apoptotic cell death during neuronal differentiation. Although P19 is an embryonal carcinoma cell line, the process of retinoic acid-induced maturation in these cells follows a process very similar to that of neuronal differentiation in the mammalian CNS in vivo (25, 26). Two important and distinct advantages of using P19 cells over primary brain cells in these studies are (i) the ability to produce multiple, stably transformed clonal cell lines that express consistent levels of MEF2 dominant negative or control constructs, and (ii) the ability to test these constructs on a uniform population of precursor cells that have been synchronized by retinoic acid treatment. Additionally, the characteristics of retinoic acid-induced P19 neurons are very similar to those of native CNS neurons (25, 26). Therefore, the findings described here using P19 cells may be similar to mechanisms for cell death/survival in differentiating neurons in the mammalian CNS.
The MEF2 family of transcription factors, especially MEF2C, is known to be highly expressed in developing cerebral cortex, where death and survival are critical events (3, 18). Although p38 MAP kinase has been reported to be an essential regulator of cell death/survival (36), heretofore it was not clear which of its substrates participate in signaling pathways that influence decisions on cell fate. Recently, the p38/MEF2 pathway was shown to be critical for Ca2+-mediated survival of cerebellar granule cells developing in culture (37), indicating that this pathway is indeed important in primary neurons. In the present study, we report that the p38α/MEF2 pathway prevents apoptotic cell death during neuronal differentiation of P19 cells. Future studies of the p38/MEF2 pathway may provide new approaches to prevent neuronal cell death in disease states involving alterations in neuronal Ca2+ levels or during abnormal CNS development.
Acknowledgments
We thank Dr. B. Kosofsky for providing anti-MEF2D antibody, Drs. M. F. Marusich and J. A. Weston for anti-Hu antibody, Dr. J. Han for p38 constructs, and Dr. E. N. Olson for the MEF2-related expression vectors. mAbs to nestin and myosin heavy chain were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of National Institute of Child Health and Human Development/National Institutes of Health and maintained by the University of Iowa, Department of Biological Science. This work was supported in part by National Institutes of Health Grant P01 HD29587 to S.A.L. and research fellowships to S.-i.O. from the American Heart Association, Massachusetts Affiliate, Inc. and the American Parkinson Disease Association, Inc.
Abbreviations
- bHLH
basic helix–loop–helix
- CNS
central nervous system
- GFP
green fluorescent protein
- MAP
mitogen-activated protein
- MAP2
microtubule-associated protein 2
- MEF2
myocyte enhancer factor 2
- pGK
phosphoglycerate kinase
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130502697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130502697
References
- 1.Treisman R. Nature (London) 1995;376:468–469. doi: 10.1038/376468a0. [DOI] [PubMed] [Google Scholar]
- 2.Pollock R, Triesman R. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 3.Leifer D, Krainc D, Yu Y-T, McDermott J, Breitbart R E, Heng J, Neve R L, Kosofsky B, Nadal-Ginard B, Lipton S A. Proc Natl Acad Sci USA. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin J F, Schwarz J J, Olson E N. Proc Natl Acad Sci USA. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin J F, Miano J M, Hustad C M, Copeland N G, Jenkins N A, Olson E N. Mol Cell Biol. 1994;14:1647–1656. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molkentin J D, Firulli A B, Black B L, Martin J F, Hustad C M, Copeland N, Jenkins N, Lyons G, Olson E N. Mol Cell Biol. 1996;16:3814–3824. doi: 10.1128/mcb.16.7.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lilly B, Galewsky S, Firulli A B, Schulz R A, Olson E N. Proc Natl Acad Sci USA. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Nature (London) 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 9.Kato Y, Kravchenko V V, Tapping R I, Han J, Ulevitch R J, Lee J-D. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao M, New L, Kravchenko V V, Kato Y, Gram H, Padova F D, Olson E N, Ulevitch R J, Han J. Mol Cell Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S-H, Galanis A, Sharrocks A D. Mol Cell Biol. 1999;19:4028–4038. doi: 10.1128/mcb.19.6.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDermott J C, Cardoso M C, Yu Y-T, Andres V, Leifer D, Krainc D, Lipton S A, Nadal-Ginard B. Mol Cell Biol. 1993;13:2564–2577. doi: 10.1128/mcb.13.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lilly B, Zhao B, Ranganayakulu G, Paterson B M, Schulz R A, Olson E N. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 14.Lin Q, Schwartz J, Bucana C, Olson E N. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ornatsky O I, Andreucci J J, Mcdermott J C. J Biol Chem. 1997;272:33271–33278. doi: 10.1074/jbc.272.52.33271. [DOI] [PubMed] [Google Scholar]
- 16.Kaushal S, Schneider J W, Nadal-Ginard B, Mahdavi V. Science. 1994;266:1236–1240. doi: 10.1126/science.7973707. [DOI] [PubMed] [Google Scholar]
- 17.Molkentin J D, Black B L, Martin J F, Olson E N. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 18.Lyons G E, Micales B K, Schwarz J, Martin J F, Olson E N. J Neurosci. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leifer D, Golden J, Kowall N W. Neuroscience. 1994;63:1067–1079. doi: 10.1016/0306-4522(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 20.Lin X, Shah S, Bulleit R F. Mol Brain Res. 1996;42:307–316. doi: 10.1016/s0169-328x(96)00135-0. [DOI] [PubMed] [Google Scholar]
- 21.Jan Y N, Jan L Y. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y, Nadal-Ginard B, Mahdavi V, Izumo S. Mol Cell Biol. 1997;17:2745–2755. doi: 10.1128/mcb.17.5.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao Z, Nadal-Ginard B. J Biol Chem. 1996;271:14371–14375. doi: 10.1074/jbc.271.24.14371. [DOI] [PubMed] [Google Scholar]
- 24.Black B L, Ligon K L, Zhang Y, Olson E N. J Biol Chem. 1996;271:26659–26663. doi: 10.1074/jbc.271.43.26659. [DOI] [PubMed] [Google Scholar]
- 25.McBurney M W. Int J Dev Biol. 1993;37:135–140. [PubMed] [Google Scholar]
- 26.Bain G, Ray W J, Yao M, Gottlieb D I. BioEssays. 1994;16:343–348. doi: 10.1002/bies.950160509. [DOI] [PubMed] [Google Scholar]
- 27.Slack R S, Skerjanc I S, Lach B, Craig J, Jardine K, McBurney M W. J Cell Biol. 1995;129:779–788. doi: 10.1083/jcb.129.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mukasa T, Khoroku Y, Tsukahara T, Momoi M Y, Kimura I, Momoi T. Biochem Biophys Res Commun. 1997;232:192–197. doi: 10.1006/bbrc.1997.6234. [DOI] [PubMed] [Google Scholar]
- 29.Blaschke A J, Stanley K, Chun J. Development (Cambridge, UK) 1996;122:1165–1174. doi: 10.1242/dev.122.4.1165. [DOI] [PubMed] [Google Scholar]
- 30.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Nature (London) 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 31.Kuida K, Zheng T S, Na S, Kuan C-Y, Yang D, Karasuyama H, Rakic P, Flavell R A. Nature (London) 1996;384:368–372. doi: 10.1038/384368a0. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto S-i, Sherman K, Lipton S A. Brain Res Mol Brain Res. 1999;74:44–54. doi: 10.1016/s0169-328x(99)00250-8. [DOI] [PubMed] [Google Scholar]
- 33.Molkentin J D, Black B L, Martin J F, Olson E N. Mol Cell Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonfoco E, Krainc D, Ankarcrona M, Nicotera P, Lipton S A. Proc Natl Acad Sci USA. 1995;92:7162–7166. doi: 10.1073/pnas.92.16.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stemple D L, Mahanthappa N K. Neuron. 1997;18:1–4. doi: 10.1016/s0896-6273(01)80018-0. [DOI] [PubMed] [Google Scholar]
- 36.New L, Han J. Trends Cardiovasc Med. 1998;8:220–229. doi: 10.1016/s1050-1738(98)00012-7. [DOI] [PubMed] [Google Scholar]
- 37.Mao Z, Bonni A, Xia F, Nadal-Vincens M, Greenberg M E. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]