Abstract
What do functionally important DNA sites, those scrutinized and shaped by natural selection, tell us about the place of humans in evolution? Here we compare ≈90 kb of coding DNA nucleotide sequence from 97 human genes to their sequenced chimpanzee counterparts and to available sequenced gorilla, orangutan, and Old World monkey counterparts, and, on a more limited basis, to mouse. The nonsynonymous changes (functionally important), like synonymous changes (functionally much less important), show chimpanzees and humans to be most closely related, sharing 99.4% identity at nonsynonymous sites and 98.4% at synonymous sites. On a time scale, the coding DNA divergencies separate the human–chimpanzee clade from the gorilla clade at between 6 and 7 million years ago and place the most recent common ancestor of humans and chimpanzees at between 5 and 6 million years ago. The evolutionary rate of coding DNA in the catarrhine clade (Old World monkey and ape, including human) is much slower than in the lineage to mouse. Among the genes examined, 30 show evidence of positive selection during descent of catarrhines. Nonsynonymous substitutions by themselves, in this subset of positively selected genes, group humans and chimpanzees closest to each other and have chimpanzees diverge about as much from the common human–chimpanzee ancestor as humans do. This functional DNA evidence supports two previously offered taxonomic proposals: family Hominidae should include all extant apes; and genus Homo should include three extant species and two subgenera, Homo (Homo) sapiens (humankind), Homo (Pan) troglodytes (common chimpanzee), and Homo (Pan) paniscus (bonobo chimpanzee).
As we have no record of the lines of descent, the lines can be discovered only by observing the degrees of resemblance between the beings which are to be classed. For this object numerous points of resemblance are of much more importance than the amount of similarity or dissimilarity in a few points.
Charles Darwin (1)
The most direct, but unfortunately not the most useful, approach to the phylogeny of recent animals is through their genetics. The stream of heredity makes phylogeny; in a sense, it is phylogeny. Complete genetic analysis would provide the most priceless data for the mapping of this stream.
George Gaylord Simpson (2)
What was not at all feasible in 1945 (2), mapping the stream of heredity that makes phylogeny, is now entirely feasible because of the emerging wealth of genomic DNA data. The nearly finalized complete genomic DNA sequence representing humans and the mounting DNA sequence data for other primates (now growing most rapidly for chimpanzees) are making possible a comprehensive genomewide genetic analysis of the place of humans in evolution. The results obtained so far show that, genetically, humans share much in common with other primates and are highly similar to their closest living relatives, the common and bonobo chimpanzees. This challenges traditional taxonomic classifications that have the two chimpanzee species closest to gorillas, place these three African ape species along with orangutans within the family Pongidae, and have humans as the only extant members of the family Hominidae.
These traditional classifications, as in Simpson (2), Martin (3), and Fleagle (4), use the anthropocentric concept of grades to subdivide the order Primates into groups that form a series progressing from primitive to advanced as estimated by such human-important features as brain capacity and mental abilities. The grade concept traces back to Aristotle's “Great Chain of Being,” in which animals are arranged “in a single graded scala naturae according to their degree of 'perfection'” (see ref. 5, page 58). Simpson (6) excluded humans from being embraced by the term apes and rationalized having a large taxonomic separation between humans and apes by claiming that the lineage ascending to humans diverged markedly from the ancestral state and entered a new “adaptive and structural–functional” zone called the “hominid” zone, whereas the lineages to apes were conservative and remained in the “pongid” zone. This concept of greatly different “hominid” and “pongid” zones has perpetuated the widespread continuing use of the term “hominids” to refer solely to humans and those fossils judged to be more closely related to humans than to any other living primates.
The accumulating DNA evidence provides an objective nonanthropocentric view of the place of humans in evolution. We humans appear as only slightly remodeled chimpanzee-like apes. This is apparent when the DNA evidence is translated into a phylogenetic classification based on principles first envisioned by Darwin (1, 7) and elaborated on by Hennig (8). The paramount principle is that each taxon should represent a clade in which all species in the clade share a more recent common ancestor (mrca) with one another than with any species from any other taxon. The second basic principle, a corollary of the first, is that the hierarchical groupings of lower ranked taxa into higher-ranked taxa (e.g., species into genera, genera into families) should describe the degrees of phylogenetic relationships among taxa, i.e., among clades. Hennig (8) also proposed that the age of origin of a clade could provide an objective measure for assigning a rank to the taxon representing that clade. Among a group of organisms, say primates or, more broadly, mammals, taxa assigned the same rank would represent clades of about the same absolute age, thus being evolutionarily equivalent with respect to an objective yardstick. A classification of primates proposed by Goodman et al. (9) used both DNA and fossil evidence to determine the ages of the clades. This classification places all apes, including humans, in the family Hominidae, and within Hominidae places common and bonobo chimpanzees with humans in the genus Homo (Table 1). The DNA that was analyzed (9) was noncoding, i.e., it did not code for proteins. Most noncoding DNA is not closely scrutinized and shaped by natural selection, and thus on average evolves more rapidly than the DNA that codes for proteins (coding DNA). Although interspecies comparisons based on typical noncoding DNA have revealed degrees of phylogenetic kinship that exist among primates, these comparisons have left open the question as to whether during evolution the changes in functionally important characters show chimpanzees to be closest to gorillas (the traditional view) or, alternatively, closest to humans.
Table 1. Time-based phylogenetic classification of extant superfamily Cercopithecoidea.
| Superfamily Cercopithecoidea (25 Ma) |
| Family Cercopithecidae (OWM clade) |
| Family Hominidae (Ape clade) |
| Subfamily Homininae (18 Ma) |
| Tribe Hylobatini |
| Subtribe Hylobatina (8 Ma) |
| Symphalangus: siamang |
| Hylobates: gibbon |
| Tribe Hominini (14 Ma) |
| Subtribe Pongina |
| Pongo: orangutans |
| Subtribe Hominina (7 Ma) |
| Gorilla: gorilla |
| Homo (6 Ma) |
| Homo (Pan) (3 Ma) |
| Homo (Pan) troglodytes: common chimpanzee |
| Homo (Pan) paniscus: bonobo chimpanzee |
| Homo (Homo) |
| †Homo (Homo) ramidus (Ardipithecus ramidus, 4.5 Ma) |
| †Homo (Homo) afarensis (Australopithecus afarensis, 4.2 Ma) |
| †Homo (Homo) robustus (Paranthropus robustus, 1.9 Ma) |
| †Homo (Homo) erectus (H. erectus, 1.8 Ma) |
| †Homo (Homo) neanderthalensis (H. neanderthalensis, 0.3 Ma) |
| Homo (Homo) sapiens (H. sapiens, 0.15 Ma) |
| Ranks of the taxa follow the scheme in refs. 9 and 10. All extant hominid genera are included. However, because the gibbon-siamang clade is poorly represented by DNA coding sequence data, this clade is not sampled in our present study. Representative fossil Homo species, their traditional generic names, and times of origin are listed as they appear in ref. 11. Dates other than those listed for fossils refer to the approximate time of origin of the taxon as treated as a crown group (the mrca and its descendants). The name Cercopithecoidea for the superfamily that includes OWM and apes has taxonomic priority over the name Hominoidea (12) |
, Extinct species
DNA that codes for proteins provides a rich source of functionally important characters with which to reexamine the place of humans in primate evolution. Here, taking advantage of the primate DNA sequence data accumulating in the public genomic databases, we compare ≈90 kb of coding DNA nucleotide sequence from 97 human genes to their sequenced chimpanzee counterparts and to available sequenced gorilla, orangutan, and Old World monkey (OWM) counterparts, and, on a more limited basis, to mouse. We subdivide the nucleotide substitutions that occur in coding DNA during evolution into nonsynonymous substitutions (amino acid-changing, thus functionally important) and synonymous substitutions (amino acid-unchanging, thus functionally much less important). Using each of the two categories of substitutions and other partitions of the coding sequence data, we found an evolutionary tree that measured the degrees of relationships among its six terminal branches. Then, in units of millions of years ago (Ma), we estimated both the dates of the branch points in the tree and, on the branches, rates of nonsynonymous and synonymous substitutions. Among the 97 genes, we identified those genes and their nonsynonymous substitutions that provide evidence of having evolved under the force of positive natural selection. In that our results support the classification shown in Table 1, we discuss the implications of enlarging the genus Homo to include chimpanzees.
Materials and Methods
Data Sources. Sequences are primarily from GenBank. The genes representing chimpanzees are, in most cases, from the species Homo (Pan) troglodytes, but in a few cases are from Homo (Pan) paniscus. If a gene appeared from its sequence to be nonfunctional, i.e., a pseudogene, it was discarded. In choosing the nonhuman genes to compare with a human gene and to one another, we also discarded any suspected of being paralogously related to the human gene, i.e., in this case, suspected of being related by a last common gene ancestor that duplicated long before the most recent common species ancestor. Our aim was to compare functional coding sequences that are orthologously related; i.e., each interspecies pair traces back to a single last common gene ancestor that existed in the most recent common species ancestor. However, without transcriptional data on many of the loci it is possible that some pseudogenes and/or paralogs were inadvertently compared. Our dataset of inferred orthologous functional coding sequences encompasses 97 loci for both humans and chimpanzees, and, among the 97, 67 were available for gorilla (Gorilla gorilla), 69 for orangutan (Pongo pygmaeus), 58 for at least one OWM, and 49 for mouse (Mus musculus; chosen because they were represented by at least four primate taxa). Sequences were aligned with the clustal algorithm as implemented in MACVECTOR 7.0 (Accelrys, Burlington, MA) and verified by eye. Putative orthologous sequences were first aligned on a gene-by-gene basis and subsequently concatenated for further analysis into a single coding “supergene” alignment that represented 93,045 nucleotide positions including indels (insertions/deletions). Human RefSeq numbers for each individual locus examined and GenBank accession numbers for nonhuman sequences can be found in Table 6, which is published as supporting information on the PNAS web, site www.pnas.org, and at www.genetics.wayne.edu/lgross/primates.htm. Previously unpublished sequences from the cytochrome c locus (CYCS) were obtained by using standard PCR-based procedures and f luorescence-based automated sequencing protocols. Primer sequences and cycling conditions are presented in Supporting Text, which is published as supporting information on the PNAS web site.
Phylogenetic Analyses. Phylogenetic trees were inferred for the concatenated supergene alignment of all six taxa (human, chimpanzee, gorilla, orangutan, OWM, and mouse) and all 97 protein-coding loci. This full dataset does not include all six taxa for each locus. To remove any possible artifactual effects from missing genes in some of the taxa, phylogenetic trees were also constructed for restricted datasets in which each taxon in each such dataset was represented by all loci. Phylogenies were inferred by maximum parsimony (MP) and maximum likelihood (ML) analyses using the program PAUP* (13). The algorithm called Deltran was used to reconstruct ancestral sequences at the internodes of the MP tree. For likelihood analyses the model of sequence evolution was chosen by hierarchical log-likelihood ratio tests using the program MODELTEST, Version 3.06 (14).
Sequence Divergence. We estimated pairwise nucleotide and amino acid sequence distances between extant taxa and between ancestral and descendent nodes in the phylogenetic trees that were constructed. The programs PAUP* and MEGA2 calculated for each pairwise comparison the observed distance between the two aligned sequences as the percent of nucleotide positions (or amino acid positions) with differing nucleotides (or amino acids). These observed distances were calculated with any gaps due to indels excluded from the count of differences. The observed distances were also calculated with each alignment position in an indel counted as a sequence difference. PAUP* calculated the ML distances between each pair of sequences. The ML distances augment the observed distances by including in the calculations presumed numbers of superimposed nucleotide substitutions.
We also calculated nonsynonymous substitutions per nonsynonymous site (Ka) and synonymous substitutions per synonymous site (Ks) to determine, respectively, the rate of amino acid changing and amino acid unchanging nucleotide substitutions (15, 16). An elevation of nonsynonymous substitution rate greater than the synonymous substitution rate indicates positive selection for advantageous mutations, whereas a nonsynonymous rate much less than the synonymous rate indicates purifying selection to prevent the spread of detrimental mutations (17). Ka/Ks was estimated in the programs MEGA2 (18) and FENS (19) for concatenated data and for individual genes. We first calculated pairwise Ka/Ks ratios between extant taxa. If a gene had Ka/Ks > 1 for any taxon pair, then on the phylogenetic tree for the six taxa (see next section) we estimated Ka/Ks for each individual branch of that tree. This allowed us to identify where on the phylogenetic tree the genes had evolved under the force of positive selection.
There are six taxa and 10 branches in the dataset (Fig. 1). Six of the 10 branches are the terminal branches to human (A), chimpanzee (B), gorilla (C), orangutan (D), OWM (E), and mouse (F). The remaining four branches are the human–chimpanzee stem (G), the African ape stem (H), the ape stem (I) from the mrca of Pongo and the three extant African hominids to the mrca of the catarrhines (because gibbons/ siamangs are not included in the study, the mrca node of all living apes is not estimated), and the long stem (J) from the mrca of catarrhines to the mrca of primates and rodents.
Fig. 1.
The optimal MP and ML tree topology when all sites are included. Additionally, this topology represents the most parsimonious tree under a variety of data partitions including only first, second, and third positions. Branch lengths for branches A–J are in percent value and are Ka, Ks, and ML 1 and 2 distances. ML 1 distances are from the six datasets (Materials and Methods). Model parameters varied for different branches; however, the HKY+Γ model was always chosen as described in the text. ML 2 branch lengths are from the 90-kb dataset (Table 2) and were obtained by using the HKY+Γ model; α shape parameter = 0.3508. The names for the branches are: A, human terminal; B, chimpanzee terminal; C, gorilla terminal; D, orangutan terminal; E, OWM terminal; F, mouse terminal; G, human–chimpanzee stem; H, African ape stem; I, ape stem; and J, catarrhine stem extended to the primate–rodent mrca. Branches are not drawn to scale. Dates for the catarrhine mrca, African ape mrca, and human–chimpanzee mrca are averages and standard deviations of estimates obtained by global and local molecular clock models (see text and Table 4). Tree scores, bootstrap support values, and data partitions are shown in Table 2. *, Equally partitioned ML distances on branches F and J.
All reconstructions for individual branches used human and chimpanzee sequences, as well as only those other sequences required to estimate the branch. Thus, to reconstruct branch lengths for branches A and B we used those loci that were always represented in human, chimpanzee, and gorilla (n = 67); to estimate lengths for branches C and G we used those loci represented in human, chimpanzee, gorilla, and orangutan (n = 58); for branch D we used the loci available for human, chimpanzee, orangutan, and OWM (n = 50); branches E, F, and J used available loci from human, chimpanzee, OWM, and mouse (n = 36); for branch H we used loci available from human, chimpanzee, gorilla, orangutan, and OWM (n = 42); and to estimate the length of branch I we used those loci available for human, chimpanzee, orangutan, OWM, and mouse (n = 31). With each of these six datasets we calculated for the relevant branches Ka, Ks, and ML 1 distances.
To calculate divergence dates we used the model of a global clock for pairwise distances, and a local clock model for branch lengths (20–22). One series of global and local clock calculations used a preassigned divergence date of 25 Ma for the mrca of OWMs and apes (fossil evidence for this date is discussed in refs. 9 and 23–26). Another series used a preassigned date of 14 Ma for the mrca of Pongo and the African apes (9, 27).
Results and Discussion
Sequence Divergence and Similarity. The pairwise nucleotide and amino acid differences among the six taxa are given in Table 2. Our results are in general agreement with previous DNA studies (28–35). Humans and chimpanzees are >99.1% identical at the coding sequence level by both the measures of observed distance and ML augmented distance. In terms of observed distances, humans differ from chimpanzees by 0.9% and each differs from gorillas by 1.0%. Orangutans are slightly more than 2% divergent from each of the African hominids, whereas OWMs are slightly less than 4% divergent from the apes. The mouse is ≈20% divergent from these primates when using observed distances, although this value doubles when ML augmented distances are used.
Table 2. DNA and amino acid divergence.
| Taxon pair | Loci | bp | % distance | ML distance, % | % distance with indels | Ka, % | Ks, % | AA % distance |
|---|---|---|---|---|---|---|---|---|
| Human vs. chimpanzee | 97 | 92,451 | 0.87 | 0.90 | 1.14 | 0.58 | 1.63 | 1.19 |
| Human vs. gorilla | 67 | 57,861 | 1.04 | 1.07 | 1.33 | 0.74 | 1.76 | 1.53 |
| Human vs. orangutan | 68 | 57,935 | 2.18 | 2.32 | 2.52 | 1.59 | 3.68 | 3.15 |
| Human vs. OWM | 57 | 45,965 | 3.76 | 4.21 | 3.98 | 3.01 | 5.88 | 5.84 |
| Human vs. mouse | 49 | 38,778 | 20.58 | 42.05 | 21.78 | 15.81 | 51.62 | 24.59 |
| Chimpanzee vs. gorilla | 67 | 57,716 | 0.99 | 1.01 | 1.45 | 0.69 | 1.69 | 1.42 |
| Chimpanzee vs. orangutan | 68 | 57,878 | 2.14 | 2.27 | 2.58 | 1.55 | 3.64 | 3.09 |
| Chimpanzee vs. OWM | 57 | 45,963 | 3.76 | 4.20 | 4.01 | 3.05 | 5.79 | 5.90 |
| Chimpanzee vs. mouse | 49 | 38,758 | 20.57 | 42.01 | 21.64 | 15.78 | 51.64 | 24.56 |
| Gorilla vs. orangutan | 58 | 48,436 | 2.25 | 2.40 | 2.44 | 1.69 | 3.62 | 3.35 |
| Gorilla vs. OWM | 44 | 32,253 | 3.99 | 4.47 | 4.14 | 3.35 | 5.88 | 6.46 |
| Gorilla vs. mouse | 45 | 34,782 | 19.83 | 39.28 | 21.35 | 14.76 | 50.73 | 23.29 |
| Orangutan vs. OWM | 50 | 37,402 | 3.83 | 4.30 | 3.99 | 3.16 | 5.81 | 6.14 |
| Orangutan vs. mouse | 44 | 34,799 | 20.17 | 40.60 | 21.65 | 15.27 | 51.01 | 23.90 |
| OWM vs. mouse | 36 | 24,711 | 21.62 | 45.67 | 23.03 | 18.01 | 50.78 | 28.12 |
Shown are the number of genes (Loci) and base pairs (bp) compared between pairs of taxa. DNA percent divergence was measured three different ways with all sites. % distance = observed percent difference; ML distance = ML (HKY+Γ) augmented distance; % distance with indels counts every alignment position within an indel as equivalent to a nucleotide substitution. We also translated proteins into amino acids and calculated uncorrected percent divergence values (AA % distance)
A recent study has proposed that when all alignment positions in indels are counted as if a nucleotide substitution had occurred at each of these indel positions, humans and chimpanzees are only 95% similar at the DNA level (36). We analyzed our data in a similar manner and found that the total coding divergence between humans and chimpanzees increased from 0.9% to 1.14%. Thus, in the >90,000 coding bases examined between humans and chimpanzees, the percent of sequence difference due to indels is much less within coding regions than for average genomic DNA. Humans and chimpanzees are more similar to each other from estimates made from protein-encoding DNA than from DNA samples in which noncoding DNA predominates (36, 37). Clearly, any indel that alters the reading frame of a gene will result in that gene coding for a completely different set of amino acid residues from the indel to the end of the coding sequence. Such mutations are very likely to be detrimental and selected against by purifying selection.
Nonsynonymous change is less common than synonymous change. Human and chimpanzee divergence is <0.6% at the nonsynonymous level but 1.6% at the synonymous level (Table 2). This result suggests that when all sites are considered the purifying form of natural selection acts on nonsynonymous sites but not on (or not nearly as much on) synonymous sites. Additionally, the amount of nonsynonymous change was only slightly larger on the terminal human branch than on the terminal chimpanzee branch.
Phylogenetic Results. Fig. 1 shows the phylogenetic relationship among the study taxa as inferred by either MP or ML analysis of the 97 loci combined dataset. The tree topology shown in Fig. 1 for all codon positions is also most parsimonious for partitioned datasets consisting of first codon position only, second codon position only, third codon position only, and first and second codon positions only. These partitions were made because of all possible substitutions, nearly all (96%) first and all (100%) second codon position substitutions are nonsynonymous, whereas substitutions at third codon positions are usually (70%) synonymous (38). As judged by the bootstrap procedure (39), the order of branching among the hominid branches is strongly supported (Table 3).
Table 3. Tree support and lengths.
| Clade that includes | First | Second | Third | First + second | All |
|---|---|---|---|---|---|
| Human and chimpanzee | 88 | 99 | 100 | 98 | 100 |
| Human, chimpanzee, and gorilla | 100 | 100 | 100 | 100 | 100 |
| Human, chimpanzee, gorilla, and orangutan | 100 | 100 | 100 | 100 | 100 |
| Parsimony length | 2,949 | 2,363 | 5,753 | 5,312 | 11,065 |
| GTR - In L | 56,777.9 | 54,676.8 | 64,890.7 | 112,140.2 | 179,335.4 |
| HKY+Γ - In L | 56,735 | 55,178.4 | 64,571.6 | 111,960 | 177,330.4 |
| Γ shape parameter (α) | — | — | — | — | 0.3508 |
Branch and bound bootstrap support values (500 replicates) for codon position partitions (first, second, third, first + second, and all positions) on the tree presented in Fig. 1. Also indicated are the parsimony tree lengths and likelihood scores for this topology, using the most general model of sequence evolution (GTR) for data partitions and the best fit HKY+Γ model chosen by likelihood ratio test for the data set in which all sites were included. Parsimony scores are for the concatenated data set of 97 protein-coding loci (93 kb). Indels were treated as missing data for the purpose of bootstrap analyses. The bootstrap values for all sites are equivalent whether parsimony or likelihood is used. Tree lengths were computed in paup* (V.4.Ob 10)
The MP and ML phylogenetic branching pattern sister-groups humans and chimpanzees, then joins gorillas, next orangutans, and then OWMs. The groupings of a presumed monophyletic “Pongidae” or a chimpanzee–gorilla clade are much less well supported. For example, if one examines the complete dataset, the number of parsimony steps for the tree shown in Fig. 1 is 11,065. Contrast this with the number of steps in a tree that has a chimpanzee–gorilla clade (11,105 steps), a human–gorilla clade (11,109 steps), or a monophyletic “pongid” clade (11,331 steps). Clearly, for either of these topologies to be supported one would have to assume an inordinate amount of homoplastic change among taxa that are on the whole quite similar (Table 2). Further, a statistical test (Kishino–Hasegawa) used in phylogenetic studies to assess topological support showed that the human–chimpanzee grouping was statistically better supported (P < 0.0001 for MP and P < 0.05 for ML) than the alternative topologies.
Of the 97 genes examined, there were 60 loci that were represented by human, chimpanzee, gorilla, and at least one outgroup. With each of these 60 loci we examined the four possible tree patterns among the three African hominid taxa. Twenty-six loci did not resolve the relationships among humans, chimpanzees, and gorillas, 22 supported a human–chimpanzee grouping, 5 supported a chimpanzee–gorilla grouping, and 7 supported a human–gorilla grouping. If there were a true trichotomy separating the humans, chimpanzees, and gorillas, the chimpanzee–gorilla grouping and the human–gorilla grouping would each be about as well supported as the human–chimpanzee grouping. Instead, the human–chimpanzee grouping received three to four times more support than did either of the two other dichotomous arrangements. Moreover, the grouping of chimpanzees and gorillas to the exclusion of humans is the most poorly supported arrangement. The phylogenetic groupings supported by the individual genes are presented in Table 6.
Divergence Dates. The branch lengths on the phylogenetic tree in Fig. 1 show that from the catarrhine mrca to the present about the same amount of change accumulated in descent to each of the five catarrhines represented in our data. Branch-points among these catarrhines could be dated by global clock calculations (which assume constancy of rates), as well as by local clock calculations (which adjust for rate variability). The results of the global and local molecular clock analyses estimate that humans shared a mrca with chimpanzees between 4.0 and 7.0 Ma; that gorillas shared a mrca with humans and chimpanzees between 5.2 and 7.4 Ma; that orangutans shared a mrca with humans, chimpanzees, and gorillas between 12.2 and 15.6 Ma; and that OWMs shared a mrca with apes between 22.4 and 28.1 Ma (Table 4). Global clock dates for the rodent–primate divergence ranged from 127 to 253 Ma and greatly exceed the 80 Ma date commonly cited in the literature and supported by clock calculations constrained by the fossil evidence (40, 41). This discrepancy between a supposed global molecular clock and fossil evidence supports findings that indicate that the rate of molecular evolution is faster in rodents than in catarrhine primates (42, 43).
Table 4. Divergence dates.
| mrca | Nonsynonymous | Synonymous | All sites ML 1 | All sites ML 2 |
|---|---|---|---|---|
| Human (H)-chimpanzee (C) | 4.3, 4.0, 5.0, 4.6 | 4.9, 4.8, 6.3, 7.0 | 4.6, 4.9, —, — | 4.9, 5.3, 5.4, 5.2 |
| H-C-gorilla (G) | 6.0, 5.2, 6.2, 5.9 | 7.0, 6.4, 6.6, 7.4 | 6.3, 6.3, —, — | 6.4, 6.5, 6.2, 6.1 |
| H-C-G-orangutan (O) | 14, 12.2, 14, 12.8 | 14, 12.9, 14, 15.6 | 14, 14.2, —, — | 14, 14.2, 14, 13.6 |
| H-C-G-O-OWM | 28.1, 25, 27.3, 25 | 26.3, 25, 22.4, 25 | 24.3, 25, —, — | 24.4, 25, 25.8, 25 |
Local molecular dock dates were calculated from branch lengths (Fig. 1); global molecular clock dates were estimated from pairwise distances (Table 2). Reference dates are in bold. Clock dates are presented as follows using the H-C synonymous mrca as an example: 4.9 = local with a 14-Ma divergence assumed for the mrca of H, C, G, and O; 4.8 = local with a 25-Ma divergence assumed for the mrca of H, C, G, O, and OWM; 6.3 = global with a 14-Ma divergence assumed for mrca of H, C, G, and O; 7.0 = global with a 25-Ma divergence assumed for the mrca of H, C, G, O, and OWM. Two ML distances (1 and 2) used all codon positions. No global clocks were calculated for the ML 1 class because datasets used to infer branch lengths in some cases used different genes and/or taxa (see Materials and Methods)
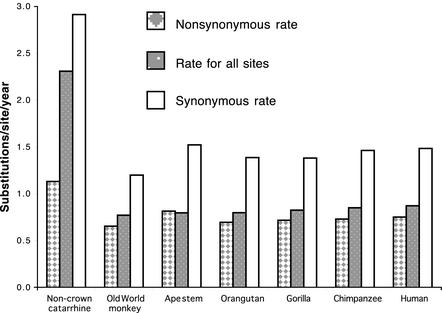
Rates of Evolution. The average rate for human and chimpanzee noncoding DNA has been reported as 1.1 × 10-9 substitutions per site per year (21, 22), and more recently as 0.99 × 10-9 substitutions per site per year (44). Coding DNA, because of its high proportion of nonsynonymous sites, would be expected to evolve at a slower rate. Our results confirm this expectation. Human and chimpanzee coding DNA evolved at an average rate of 0.86 × 10-9 substitutions per site per year. Moreover, as illustrated in Fig. 2, the catarrhine primate lineages sampled show a much slower rate than the non-crown catarrhines (branches F and J). This is the case for both nonsynonymous and synonymous rates, as well as for all coding sites. This finding is consistent with the hypothesis that the rate of occurrence of mutations decreased in those primates that have improved mechanisms for both preventing DNA damage and repairing damaged DNA, and that have life history strategies that favor prolonged periods of development.
Fig. 2.
Rates of nucleotide substitution from the catarrhine mrca to present day OWMs, the catarrhine mrca to the ape mrca, the ape mrca to each of the four present day apes, and the catarrhine mrca to mouse. Rates, as number of substitution per site per year × 10-9, are for nonsynonymous sites, all sites (ML 1), and synonymous sites. Rates were calculated by using the branch lengths and dates shown in Fig. 1, and assigning the date of 80 Ma to the rodent–primate mrca.
Positively Selected Genes. Thirty of the 97 genes analyzed have Ka values greater than Ks values on at least one of the eight catarrhine branches (Table 5). That a substantial fraction of these gene loci show evidence of positive selection at one or more times during catarrhine descent is consistent with other studies that point to natural selection as a powerful force in shaping the coding sequences of eukaryotic genomes (45–48).
Table 5. Genes showing elevated Ka and the branch on which the elevation occurs.
| Gene | Human | Chimpanzee | Gorilla | Orangutan | OWM | N | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| symbol | RefSeq | A | B | G | C | H | D | I | E | node | Biological process |
| AHSG | NM_001622 | X | X | 2 | Ossification | ||||||
| ANG | NM_001145 | X | X | X | 3 | RNA catabolism | |||||
| APOE | NM_000041 | X | 1 | Cholesterol metabolism | |||||||
| BRCA1 | NM_007294 | X | X | 2 | DNA repair | ||||||
| COX4I1 | NM_001861 | X | X | X | X | 4 | Energy pathways | ||||
| COX7C | NM_001867 | X | X | X | 3 | Energy pathways | |||||
| COX8L | NM_004074 | X | X | X | X | 4 | Energy pathways | ||||
| DAF | NM_000574 | X | X | X | X | X | 5 | [Complement control] | |||
| DAZ | NM_004081 | X | X | 2 | Spermatogenesis | ||||||
| DRD4 | NM_000797 | X | X | 2 | Dopamine receptor | ||||||
| FPRL2 | NM_002030 | X | X | 2 | G protein-coupled receptor | ||||||
| HBA1 | NM_000558 | X | X | X | 3 | Oxygen transport | |||||
| ICAM1 | NM_000201 | X | X | 2 | Cell-cell adhesion | ||||||
| IL3 | NM_000588 | X | X | X | X | 4 | Cell proliferation | ||||
| IL8RA | NM_000634 | X | X | X | 3 | G protein-coupled receptor | |||||
| IL8RB | NM_001557 | X | X | X | X | X | 5 | G protein-coupled receptor | |||
| IL16 | NM_004513 | X | 1 | Immune response | |||||||
| LEP | NM_000230 | X | X | 2 | Energy reserve metabolism | ||||||
| LYZ | NM_000239 | X | X | X | X | 4 | Inflammatory response | ||||
| NR0B1 | NM_000475 | X | X | X | X | 4 | Sex determination | ||||
| OR1E1 | NM_003553 | X | X | 2 | [Olfactory receptor] | ||||||
| OR1G1 | NM_003555 | X | X | 2 | G protein-coupled receptor | ||||||
| PRM1 | NM_002761 | X | X | X | X | X | X | 6 | Spermatogenesis | ||
| PRM2 | NM_002762 | X | X | X | X | X | 5 | Spermatogenesis | |||
| RHAG | NM_000324 | X | X | X | X | X | X | X | 7 | Protein complex assembly | |
| RNASE1 | NM_002933 | X | X | X | 3 | (Pancreatic ribonuclease) | |||||
| RNASE3 | NM_002935 | X | X | X | X | X | 5 | RNA catabolism | |||
| SP100 | NM_003113 | X | X | 2 | Regulation of transcription | ||||||
| SRY | NM_003140 | X | X | X | X | 4 | Male sex determination | ||||
| ZNF80 | NM_007136 | X | X | 2 | Regulation of transcription | ||||||
| N genes | 30 | 14 | 14 | 11 | 13 | 10 | 6 | 14 | 14 | ||
| Total Ka | 0.68% | 0.62% | 0.44% | 0.79% | 1.39% | 1.37% | 1.96% | 2.96% | |||
| Total Ks | 0.15% | 0.21% | 0.19% | 0.32% | 0.60% | 0.69% | 1.05% | 1.75% | |||
| Ka/Ks | 4.5 | 2.9 | 2.3 | 2.5 | 2.3 | 2 | 1.9 | 1.7 | |||
| P < % | 0.1 | 0.1 | 5 | 0.5 | 0.1 | 5 | 0.5 | 0.1 |
Ka/Ks was calculated on eight branches (Fig. 1). X, branches where Ka > Ks. Ka/Ks and associated P values are provided for concatenated alignments comprised of the all elevated genes on a particular branch. A representative biological process (or molecular function) is listed for genes as provided by the 04/03 build of the Gene Ontology (GO) database. If no GO information was available, the LocusLink [gene product] is listed. N node, total number of branches on which the elevation occurred; N genes, number of genes that were elevated on a given branch
These 30 loci when concatenated have an alignment with 24,645 nucleotide positions that yields by both MP and ML analyses the tree topology shown in Fig. 1. This 24.6-kb dataset supports the sister grouping of humans and chimpanzees by a bootstrap value of 100%, and also supports the human–chimpanzee–gorilla clade and the human–chimpanzee–gorilla–orangutan clade by bootstrap values of 100%. This result holds if the mostly synonymous third codon positions are removed or if only the most functionally important class of sites, second codon positions, is retained.
Table 5 lists the genes with elevated Ka compared with Ks and, for each of these genes, the branches that show the elevated Ka. Some genes show elevations of Ka throughout the tree, whereas others show only localized elevation. For example, RHAG shows an elevation on seven of the eight examined branches, whereas APOE shows an elevation on one branch only, the terminal gorilla branch.
The concatenations of the genes that show elevated Ka always support a human–chimpanzee grouping to the exclusion of gorilla. These are the concatenations for the human-terminal branch (n = 14 genes), the chimpanzee-terminal branch (n = 14), the human–chimpanzee stem (n = 11), the gorilla-terminal branch (n = 13), the African hominid stem (n = 10), the orangutan-terminal branch (n = 6), the ape stem (n = 14), and the OWM-terminal branch (n = 14). The Ka/Ks value for the concatenation of the 14 positively selected genes on the human-terminal branch is 4.5, a statistically significant elevation (P < 0.001). Ka significantly exceeds Ks on all other branches as well (Table 5).
The concatenation of eleven genes that show elevated Ka/Ks on the human–chimpanzee stem was analyzed more thoroughly. Most notably, a MP analysis that included only second codon positions recovered the human–chimpanzee grouping (bootstrap value = 98%), which was joined by the gorilla (100%) and then the orangutan (96%), as has been shown with other analytical permutations in this study. Thus, humans and chimpanzees group with one another to the exclusion of other extant primates when using sites where every nucleotide substitution causes an amino acid replacement and when using just those nucleotide substitutions in genes that show evidence of positive selection on the stem that groups humans and chimpanzees. Moreover, the terminal chimpanzee branch's 14 genes with elevated Ka and Ka/Ks values accumulates about as much Ka change (0.62%) as the terminal human branch's 14 positively selected sequences (0.68%).
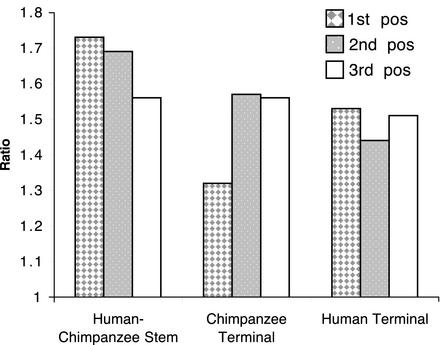
Selection in Substitutions That Group Humans and Chimpanzees. To further examine the role of natural selection in shaping the course of coding sequence evolution, we used a MP tree constructed for 45 loci representing, with a minimum amount of missing data, the six terminal branches and four stems. We focused on those nucleotide sites where nucleotide changes occurred on the human–chimpanzee stem, the human-terminal branch, and the chimpanzee-terminal branch. We counted the number of nucleotide substitutions for each of these three branches at each first, second, and third codon positions throughout the alignment and, at those particular alignment sites, the number of substitutions throughout the full tree. We then divided the number for the full tree by the number for that branch. These ratios are presented in Fig. 3. Although third-position ratios do not vary appreciably among the three branches, first- and, to a lesser extent, second-position ratios are higher on the stem than on the terminal branches. In addition, on the stem the first- and second-position ratios are higher than third-position ratios. Of the derived first-position changes, 86–94% are nonsynonymous; of second-position changes, 100% are nonsynonymous; of third-position changes, only 0–8% are nonsynonymous. It is of significance that the majority of first- and second-position nonsynonymous changes occur in genes that are positively selected during descent of catarrhines, with this number reaching 100% for nonsynonymous changes at third positions.
Fig. 3.
Variability at first, second, and third codon position sites showing nucleotide changes on the human–chimpanzee stem, the chimpanzee-terminal branch, and the human-terminal branch. Such variability is estimated for each of the three branches by the ratio: number of changes throughout the full tree at those sites showing nucleotide changes on that branch/number of nucleotide changes on that branch. The dataset used to construct the MP tree for the six extant taxa consisted of concatenated coding sequences from 45 genes. Chimpanzee- and human-terminal branches were represented by all 45 genes. Each of the remaining taxa had a minimum of missing sequence data.
The difference in ratios seen in first- to third-codon position ratios in stem vs. terminal human and chimpanzee branches may be explained in part by reference to the covarion hypothesis, originally proposed by Fitch and Markowitz (49) and since elaborated by others (50–52). This theory has for a protein a subset of amino acid residues that are much freer to vary (the so-called “covarions”) than the remaining residues. Even though the theory's authors suggest that at these covariable residues the variation among descendant lineages is due to selectively neutral amino acid replacements, the authors also suggest that these amino acid replacements impose new functional constraints on the protein. We would amend these suggestions by proposing that in the subset of residues that are freer to vary, a fraction of the replacements that occurred were favored by natural selection.
An example of this is observed for nonsynonymous changes that occur in positively selected genes encoding proteins that function as cell receptors. Such genes encode a class of molecules that interface with the cell surface's outside environment, making these genes likely candidates for experiencing strong selective pressures for change. Four of the receptor-encoding genes (DRD4, FPRL2, IL8RA, and IL8RB) account for 58% of the 45 loci's nonsynonymous changes (14 of 24 substitutions) on the human–chimpanzee stem but only 6% (3 of 49) and 11% (7 of 62) on the chimpanzee- and human-terminal branches, respectively. Given the spans of time on these branches (≈1.2 Ma on the stem vs. 5.1 Ma on each terminal), the rate of nonsynonymous change for these four genes decreases by ≈12-fold on the human- and chimpanzee-terminal branches compared with the stem. This finding suggests that natural selection, first in its positive form, spread beneficial amino acid replacements through the human–chimpanzee stem, then, in its purifying form, acted on the changed proteins to reduce their rate of further changes during descent of human- and chimpanzee-terminal branches.
Classification of Humans. Our results with coding DNA provide dates for branch-points in humankind's ape ancestry that agree with the dates found by using noncoding DNA (9, 22). Thus, the coding DNA results support the position of humans in the age-related classification shown in Table 1. In these previous studies, the principle of rank equivalence with other primate clades of the same age required grouping the chimpanzee clade with the human clade within the same genus. An age of <6 Ma for the mrca of Homo's two subgenera, Homo (Pan) and Homo (Homo), is well within the range of ages found in other mammals for intrageneric divergencies (11, 53–56). To have rank equivalence, any fossil species that shares a mrca with humans to the exclusion of chimpanzees should be in the subgenus Homo (Homo). For example, A. afarensis should be called H. (Homo) afarensis (Table 1).
Simpson (1963) provided a precedent for the very close taxonomic grouping of humans and chimpanzees (6). On the basis of his broad knowledge of mammalian systematics, he eliminated the genus Gorilla, grouped gorillas and chimpanzees together in the genus Pan, and grouped Pan and Pongo (the orangutan's genus) into the subfamily Ponginae, which along with the gibbon subfamily Hylobatinae constituted the family Pongidae (6). This taxonomic arrangement captured the cladistic relationships of the living nonhuman apes to one another but not to humans. As already noted, the traditional view advocated by Simpson and others treats humans as outside the ape clade and has the lineage to humans diverge radically from the supposed ancestral pongid state (2–4, 6, 57). In contrast to this traditional view, the results we obtained by using a sample of functionally important DNA depict humans to be as conservative as chimpanzees, gorillas, and orangutans. We argue that if it is valid from the standpoint of mammalian systematics to place gorillas and chimpanzees in the same genus, it is even more valid in light of findings such as 99.4% identity between humans and chimpanzees at nonsynonymous DNA sites to place these two closely related genetic relatives in the same genus.
The evidence both from cladistic analyses and from simply measuring degrees of genetic correspondence call for grouping chimpanzees and humans together as sister subgenera of the same genus and justify believing that chimpanzees can provide insights into distinctive features of humankind's own evolutionary origins. Chimpanzees use tools, have material cultures, are ecological generalists, and are highly social (58–63). Their anatomical inability to produce most of the sounds of human speech long obscured the fact that chimpanzees are also capable of understanding and using rudimentary forms of language, as shown by recent studies on communication via sign language and lexigrams (64–66).
It is of course entirely possible that once the genetic underpinnings of “human-important” phenotypic features are uncovered, these particular underpinnings will be seen to have diverged more in the terminal human lineage than in the terminal chimpanzee lineage. But it might also be speculated with regard to the genetic underpinnings of “chimpanzee-important” phenotypic features that those particular underpinnings will be seen to diverge more in the terminal chimpanzee than in the terminal human lineage.
Looking to the future, once the DNA sequences of complete genomes from chimpanzees, gorillas, orangutans, and some other primates are known, it will be relatively straightforward to identify among the 20,000–30,000 or more genes of each genome those coding sequences that evolved under the force of positive selection. Eventually it should also be possible to similarly identify the positively selected changes in cis-acting regulatory DNA elements. As such molecular genetic data are integrated with organismal phenotypic data, humans will continue to gain a much better understanding of their place in evolution.
Supplementary Material
Acknowledgments
We thank Jeffrey Doan, Allon Goldberg, and Timothy Schmidt for valuable comments on previous versions of this manuscript, and Mark Weiss, Richard Tashian, and Jerry Slightom for insightful discussion. This work could not have been completed without the efforts of many students, technicians, postdoctoral scientists, and principal investigators who collected much of the original sequence data used in the analyses. RNA from Trachypithecus cristatus was provided by the Center for Research in Endangered Species (CRES) at the San Diego Zoo. This work was supported by National Science Foundation Grants 0118696 and 9910679 and National Institutes of Health Grants DK56927 and GM65580.
This contribution is part of a special series of Inaugural Articles by members of the National Academy of Sciences elected on April 30, 2002.
Abbreviations: Ma, million years ago; ML maximum likelihood; MP, maximum parsimony; mrca, most recent common ancestor; OWM, Old World monkey.
Data deposition: The sequences for cytochrome c (CYCS) have been deposited in the GenBank database (accession nos. AY268592–AY268594).
References
- 1.Darwin, C. (1871) The Descent of Man, and Selection in Relation to Sex (Murray, London).
- 2.Simpson, G. G. (1945) The Principles of Classification and a Classification of Mammals (American Museum of Natural History, New York).
- 3.Martin, R. D. & Martin, A.-E. (1990) Primate Origins and Evolution: A Phylogenetic Reconstruction (Chapman & Hall, London).
- 4.Fleagle, J. G. (1999) Primate Adaptation and Evolution (Academic, San Diego).
- 5.Lovejoy, A. (1936) The Great Chain of Being: A Study of the History of an Idea (Harvard Univ. Press, Cambridge, MA).
- 6.Simpson, G. G. (1963) in Classification and Human Evolution, ed. Washburn, S. L. (Wenner-Gren Foundation for Anthropological Research, New York), pp. 1-31.
- 7.Darwin, C. (1859) On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life (Murray, London); reprinted (1950) (Watts & Co., London).
- 8.Hennig, W. (1966) Phylogenetic Systematics (Univ. of Illinois Press, Urbana).
- 9.Goodman, M., Porter, C. A., Czelusniak, J., Page, S. L., Schneider, H., Shoshani, J., Gunnell, G. & Groves, C. P. (1998) Mol. Phylogenet. Evol. 9, 585-598. [DOI] [PubMed] [Google Scholar]
- 10.Goodman, M., Page, S. L., Meireles, C. M. & Czeluzniak, J. (1999) in Evolutionary Theory and Processes: Modern Perspectives. Papers in Honour of Eviater Nevo, ed. Wasser, S. P. (Kluwer, Dordrecht, The Netherlands), pp. 193-211.
- 11.Wood, B. & Richmond, B. G. (2000) J. Anat. 197, 19-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna, M. C., Bell, S. K. & Simpson, G. G. (1997) Classification of Mammals Above the Species Level (Columbia Univ. Press, New York).
- 13.Swofford, D. L. (2000) paup*, Phylogenetic Analysis Using Parsimony (*And Other Methods) (Sinauer, Sunderland, MA), Version 4.0b10.
- 14.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 15.Li, W.-H. (1993) J. Mol. Evol. 36, 96-99. [DOI] [PubMed] [Google Scholar]
- 16.Pamilo, P. & Bianchi, N. O. (1993) Mol. Biol. Evol. 10, 271-281. [DOI] [PubMed] [Google Scholar]
- 17.Czelusniak, J., Goodman, M., Hewett-Emmett, D., Weiss, M. L., Venta, P. J. & Tashian, R. E. (1982) Nature 298, 297-300. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 19.de Koning, A. P. J., Palumbo, M., Messier, W. & Stewart, C.-B. (1998) fens, Facilitated Estimates of Nucleotide Substitution (State Univ. of New York, Albany), Version 1.0.
- 20.Goodman, M. (1986) in Evolutionary Perspectives and the New Genetics (Liss, New York), pp. 121-132.
- 21.Bailey, W. J., Fitch, D. H., Tagle, D. A., Czelusniak, J., Slightom, J. L. & Goodman, M. (1991) Mol. Biol. Evol. 8, 155-184. [DOI] [PubMed] [Google Scholar]
- 22.Bailey, W., Hayasaka, K., Skinner, C., Kehoe, S., Sieu, L., Slightom, J. & Goodman, M. (1992) Mol. Phylogenet. Evol. 1, 97-135. [DOI] [PubMed] [Google Scholar]
- 23.Pilbeam, D. (1996) Mol. Phylogenet. Evol. 5, 155-168. [DOI] [PubMed] [Google Scholar]
- 24.Gebo, D. L., MacLatchy, L., Kityo, R., Deino, A., Kingston, J. & Pilbeam, D. (1997) Science 276, 401-404. [DOI] [PubMed] [Google Scholar]
- 25.Harrison, T. & Gu, Y. (1999) J. Hum. Evol. 37, 225-277. [DOI] [PubMed] [Google Scholar]
- 26.Rossie, J. B., Simons, E. L., Gauld, S. C. & Rasmussen, D. T. (2002) Proc. Natl. Acad. Sci. USA 99, 8454-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kappelman, J., Kelley, J., Pilbeam, D., Sheikh, K. A., Ward, S., Anwar, M., Barry, J. C., Brown, B., Hake, P., Johnson, N. M., et al. (1991) J. Hum. Evol. 21, 61-73. [Google Scholar]
- 28.Miyamoto, M. M., Slightom, J. L. & Goodman, M. (1987) Science 238, 369-373. [DOI] [PubMed] [Google Scholar]
- 29.Sibley, C. G. & Ahlquist, J. E. (1987) J. Mol. Evol. 26, 99-121. [DOI] [PubMed] [Google Scholar]
- 30.Goodman, M., Tagle, D. A., Fitch, D. H., Bailey, W., Czelusniak, J., Koop, B. F., Benson, P. & Slightom, J. L. (1990) J. Mol. Evol. 30, 260-266. [DOI] [PubMed] [Google Scholar]
- 31.Sibley, C. G., Comstock, J. A. & Ahlquist, J. E. (1990) J. Mol. Evol. 30, 202-236. [DOI] [PubMed] [Google Scholar]
- 32.Chen, F. C., Vallender, E. J., Wang, H., Tzeng, C. S. & Li, W.-H. (2001) J. Hered. 92, 481-489. [DOI] [PubMed] [Google Scholar]
- 33.Chen, F. C. & Li, W.-H. (2001) Am. J. Hum. Genet. 68, 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'hUigin, C., Satta, Y., Takahata, N. & Klein, J. (2002) Mol. Biol. Evol. 19, 1501-1513. [DOI] [PubMed] [Google Scholar]
- 35.Locke, D. P., Segraves, R., Carbone, L., Archidiacono, N., Albertson, D. G., Pinkel, D. & Eichler, E. E. (2003) Genome Res. 13, 347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Britten, R. J. (2002) Proc. Natl. Acad. Sci. USA 99, 13633-13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujiyama, A., Watanabe, H., Toyoda, A., Taylor, T. D., Itoh, T., Tsai, S. F., Park, H. S., Yaspo, M. L., Lehrach, H., Chen, Z., et al. (2002) Science 295, 131-134. [DOI] [PubMed] [Google Scholar]
- 38.Li, W.-H. (1997) Mol. Evol. (Sinauer, Sunderland, MA).
- 39.Felsenstein, J. (1985) Evol. Int. J. Org. Evol. 39, 783-791. [DOI] [PubMed] [Google Scholar]
- 40.Huchon, D., Madsen, O., Sibbald, M. J., Ament, K., Stanhope, M. J., Catzeflis, F., de Jong, W. W. & Douzery, E. J. (2002) Mol. Biol. Evol. 19, 1053-1065. [DOI] [PubMed] [Google Scholar]
- 41.Springer, M. S., Murphy, W. J., Eizirik, E. & O'Brien, S. J. (2003) Proc. Natl. Acad. Sci. USA 100, 1056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu, X. & Li, W.-H. (1992) Mol. Phylogenet. Evol. 1, 211-214. [DOI] [PubMed] [Google Scholar]
- 43.Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- 44.Yi, S., Ellsworth, D. L. & Li, W.-H. (2002) Mol. Biol. Evol. 19, 2191-2198. [DOI] [PubMed] [Google Scholar]
- 45.Endo, T., Ikeo, K. & Gojobori, T. (1996) Mol. Biol. Evol. 13, 685-690. [DOI] [PubMed] [Google Scholar]
- 46.Fay, J. C., Wyckoff, G. J. & Wu, C. I. (2001) Genetics 158, 1227-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fay, J. C., Wyckoff, G. J. & Wu, C. I. (2002) Nature 415, 1024-1026. [DOI] [PubMed] [Google Scholar]
- 48.Mishmar, D., Ruiz-Pesini, E., Golik, P., Macaulay, V., Clark, A. G., Hosseini, S., Brandon, M., Easley, K., Chen, E., Brown, M. D., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fitch, W. M. & Markowitz, E. (1970) Biochem. Genet. 4, 579-593. [DOI] [PubMed] [Google Scholar]
- 50.Miyamoto, M. M. & Fitch, W. M. (1995) Mol. Biol. Evol. 12, 503-513. [DOI] [PubMed] [Google Scholar]
- 51.Huelsenbeck, J. P. (2002) Mol. Biol. Evol. 19, 698-707. [DOI] [PubMed] [Google Scholar]
- 52.Pupko, T. & Galtier, N. (2002) Proc. R. Soc. London Ser. B 269, 1313-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bininda-Emonds, O. R., Gittleman, J. L. & Purvis, A. (1999) Biol. Rev. Cambridge Philos. Soc. 74, 143-175. [DOI] [PubMed] [Google Scholar]
- 54.Norman, J. E. & Ashley, M. V. (2000) J. Mol. Evol. 50, 11-21. [DOI] [PubMed] [Google Scholar]
- 55.Querouil, S., Hutterer, R., Barriere, P., Colyn, M., Kerbis Peterhans, J. C. & Verheyen, E. (2001) Mol. Phylogenet. Evol. 20, 185-195. [DOI] [PubMed] [Google Scholar]
- 56.Mercer, J. M. & Roth, V. L. (2003) Science 299, 1568-1572. [DOI] [PubMed] [Google Scholar]
- 57.Simpson, G. G. (1961) Principles of Animal Taxonomy (Columbia Univ. Press, New York). [DOI] [PubMed]
- 58.McGrew, W. C. (1992) Chimpanzee Material Culture: Implications for Human Evolution (Cambridge Univ. Press, Cambridge, U.K.).
- 59.Wrangham, R. W., McGrew, W. C., de Waal, F. B. & Heltne, P., eds. (1994) Chimpanzee Cultures (Harvard Univ. Press, Cambridge, MA).
- 60.de Waal, F. B. (1995) Sci. Am. 272, 82-88. [DOI] [PubMed] [Google Scholar]
- 61.de Waal, F. B. (1998) Chimpanzee Politics: Power and Sex Among Apes (Johns Hopkins Univ. Press, Baltimore, MD).
- 62.Goldberg, T. L. (1998) Int. J. Primatol. 19, 237-254. [Google Scholar]
- 63.Whiten, A., Goodall, J., McGrew, W. C., Nishida, T., Reynolds, V., Sugiyama, Y., Tutin, C. E., Wrangham, R. W. & Boesch, C. (1999) Nature 399, 682-685. [DOI] [PubMed] [Google Scholar]
- 64.Fouts, R. & Mills, S. T. (1997) Next of Kin: What Chimpanzees Have Taught Me About Who We Are (Morrow, New York).
- 65.Savage-Rumbaugh, E. S., Shanker, S. & Taylor, T. J. (1998) Apes, Language, and the Human Mind (Oxford Univ. Press, New York).
- 66.de Waal, F. B. (2001) Tree of Origin: What Primate Behavior Can Tell Us About Human Social Evolution (Harvard Univ. Press, Cambridge, MA).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.