Abstract
We describe a postgenomic in silico approach for identifying genes that are likely to be essential and estimate their proportion in haploid genomes. With the knowledge of all sites eligible for mutagenesis and an experimentally determined partial list of nonessential genes from genome mutagenesis, a Bayesian statistical method provides reasonable predictions of essential genes with a subsaturation level of random mutagenesis. For mutagenesis, a transposon such as Himar1 is suitable as it inserts randomly into TA sites. All of the possible insertion sites may be determined a priori from the genome sequence and with this information, data on experimentally hit TA sites may be used to predict the proportion of genes that cannot be mutated. As a model, we used the Mycobacterium tuberculosis genome. Using the Himar1 transposon, we created a genetically defined collection of 1,425 insertion mutants. Based on our Bayesian statistical analysis using Markov chain Monte Carlo and the observed frequencies of transposon insertions in all of the genes, we estimated that the M. tuberculosis genome contains 35% (95% confidence interval, 28%–41%) essential genes. This analysis further revealed seven functional groups with high probabilities of being enriched in essential genes. The PE-PGRS (Pro-Glu polymorphic GC-rich repetitive sequence) family of genes, which are unique to mycobacteria, the polyketide/nonribosomal peptide synthase family, and mycolic and fatty acid biosynthesis gene families were disproportionately enriched in essential genes. At subsaturation levels of mutagenesis with a random transposon such as Himar1, this approach permits a statistical prediction of both the proportion and identities of essential genes of sequenced genomes.
Numerous genomes have been completely sequenced, and the location, length, and sequence identities of the genes have been defined. However, parallel high-throughput identification of essential genes has been difficult. Traditional methods for identifying putative essential genes, such as creating conditional knockouts, are not feasible for large-scale evaluation. In vivo allelic exchange of fragments of genomic DNA fragments that are mutagenized in vitro with transposon insertion has been used for identification of essential genes by a process called GAMBIT (genomic analysis and mapping by in vitro transposition) (1), but this approach requires that the microbe be naturally competent for transformation. Systematic antisense expression of gene fragments has also been used to identify essential genes on a global scale (2). With genome sequence information and knowledge of all target sites for disruption by well-characterized transposons, a biostatistical approach is now feasible for prediction of essential genes and their proportion at subsaturation levels of mutagenesis. Here we present a method to predict the proportion and identity of essential genes in a genome that is based on the sequence information of a genome, experimentally determined identities of a subset of the organism's nonessential target sites, and a Bayesian statistical analysis.
We used the Mycobacterium tuberculosis CDC1551 strain containing 4,250 annotated ORFs as a model genome (3). For mutagenesis we sought a transposon that inserts randomly into genomes of a broad array of species and has a small but defined target site. The Himar1 transposon of the mariner family requires only the dinucleotide TA for insertion (4) and thus is suited to transpose into nearly all ORFs. It has been used successfully to create insertion mutations in numerous species (5). We have initiated a random insertion mutagenesis of M. tuberculosis using the Himar1 transposon. Data from our initial genetic characterization of mutants have directly identified 770 nonessential genes. We demonstrate a Bayesian statistical analysis technique to estimate the overall percentage of essential genes and identify specific genes with high probabilities of being essential. Using this method we estimated 35% of the genes in M. tuberculosis to be essential. This analysis further revealed functional groups that are enriched in essential genes. The PE-PGRS (Pro-Glu polymorphic GC-rich repetitive sequence) gene family, the polyketide/nonribosomal peptide synthase family, and the mycolic and fatty acid biosynthesis gene families are disproportionately enriched in essential genes. This information is valuable for rational drug design because genes essential for bacterial growth are promising targets for the development of new antimicrobial agents against tuberculosis.
Methods
Statistical Analysis. To estimate the proportion of essential genes, we performed a Bayesian statistical analysis using Markov chain Monte Carlo. Complete details and computer simulations to verify the proper performance of the method are available at www.biostat.jhsph.edu/~kbroman/publications/ms0220.pdf. In brief, we assumed a priori that essential genes were uniformly distributed and that all genes were equally likely to be essential. We used a Gibbs sampler (6) to estimate the posterior distribution. We used the results of every 50th of 500,000 Markov chain Monte Carlo steps, after a burn-in period of 500 steps. Computer software implementing this method has been constructed as an add-on package for the R statistical software (7) and is available at www.biostat.jhsph.edu/~kbroman/software.
Because a transposon insertion in the distal portion of an essential gene may not be sufficiently disruptive to eliminate activity in the gene product, we excluded TA sites that fell within the distal 20% or 100 bp of a gene (smaller of the two: this was designated as the “5′80%–3′100-bp” rule) from the list of TA sites, which, if hit, could define a gene as nonessential. This rule was used to determine the number of nonessential genes and subsequently predict the essential genes. We defined essential genes to be those that cannot tolerate disruptive transposon insertion. Thus, in our analyses essential genes include those that encode essential gene products and also those which, if mutated, might be polar upon the expression a distal of gene whose product is essential.
Bioinformatics. Sequences of the transposon junction sites were analyzed for similarity by using BLASTN search. The site of transposition, as a numerical coordinate, was compiled for each mutant. Genome sequence information of CDC1551 was used to count TA dinucleotide insertion sites for all 4,250 ORFs. This information along with functional classification of H37Rv genome (Sanger database: www.sanger.ac.uk/Projects/M_tuberculosis/Gene_list) was used to construct a transposon mutation functional classification table.
Bacterial Strains, Phage, and Construction of Transposon Mutants. CDC1551, an M. tuberculosis clinical isolate (8), was used as the host for mutagenesis. It was grown in Middlebrook 7H9 medium (Difco) supplemented with 0.2% glycerol, 0.05% Tween 80, 10% ADC (0.5% BSA, 0.2% dextrose, and 0.085% NaCl) at 37°C. Mycobacterium smegmatis mc2155, used as a host to grow the phage carrying the Himar1 transposon, was also cultured in complete 7H9 liquid medium. Phage lysates were prepared as described (9). Mycobacterial cell cultures grown to an A600 of ≈1.0 were washed twice with an equal volume of wash medium (7H9 + 0.2% glycerol + 10% ADC); the pellet was resuspended in wash medium and infected with the phage at a multiplicity of infection of 1:10. The infection mix was incubated at 37°C for 2–3 h, and an aliquot was plated on selection media [Middlebrook 7H10 solid medium, 0.5% glycerol, 10% vol/vol supplemented with OADC (Becton Dickinson), 0.05% Tween 80 + 50 μg/ml cycloheximide, and 20 μg/ml kanamycin]. The remaining infection mixture was frozen, and its complexity was estimated from colony-forming unit counts on plates incubated at 37°C in sealed bags. Individual colonies were picked and grown separately in 7H9 liquid media with 20 μg/ml kanamycin, and genomic DNA was harvested as described (10). Each mutant has been preserved in liquid culture at -80°C.
Identification of Transposon Insertion Sites. Genomic DNA was digested with AluI and ligated to blunt-ended adaptors, a duplex made of oligo AdBTM (5′ P-ACCAGCCCGGGC-NH2) and AdTOP (5′-GTAATACGACTCACTATAGGGCACGCGTGGTCGACGGCCCGGGCTGGT). The 3′ amino modification blocks extension, permitting only the de novo-synthesized strands to serve as templates for primer AP1 to anneal and extend (11). The transposon junction site DNA was PCR-amplified. Transposon-specific primer HimarSP1 (5′-ACCA ATAGGCCGAAATCGGCAAAATCC) and adaptor-specific primer AP1 (5′-GTAATACGACTCACTATAGGGCAC) were used for the first PCR with the following cycles: 94°C for 3 min, 28 cycles of 94°C for 30 s + 55°C for 30 s + 72°C for 50 s, and 72°C for 5 min. Next, nested PCR was performed with primers HimarSP2 (5′-CCGAGATAGGGTGAGTGTTGTTCCAG) and AP1a (5′-ACTATAGGGCACGCGTGGTCGACG) using the following cycles: 94°C for 3 min, 30 cycles of 94°C for 30 sc + 58°C for 30 s + 72°C for 50 s, and 72°C for 5 min. PCR products were precipitated and sequenced by using the transposon-specific primer SeqPr (5′-CCGAGATAGGGTGAGTGTTG).
Results
Construction and Description of the Transposon Insertion Mutant Library. The M. tuberculosis CDC1551 genome contains 65,649 intragenic TA sites. Of the 4,250 ORFs, only 16 ranging in length from 99 to 300 bp (mean length = 160 bp) lack TA sites altogether. We conducted phage-mediated transposon mutagenesis multiple times and picked 200–300 individual mutants from platings of each mutant pool. To avoid selection bias all colonies on a particular plate were picked, including large (3.1%) and small (4.1%) colony phenotypes that were different from the WT colony morphology. Individual mutants were grown in pure culture and archived. Mutants were characterized by PCR amplification of the transposon junction site by a genome walking technique followed by DNA sequencing of the resulting PCR product.
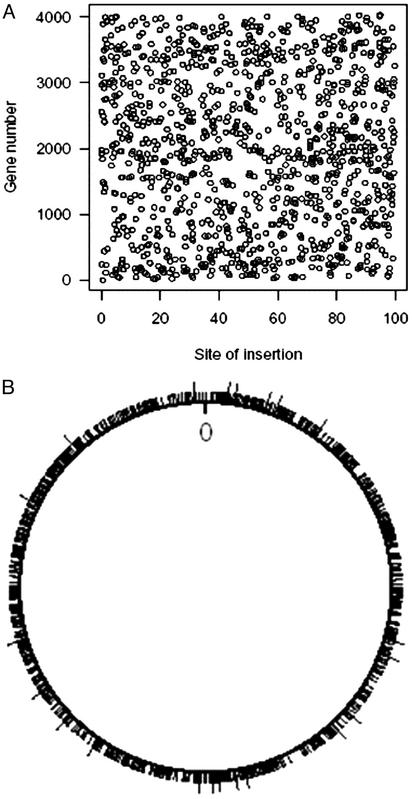
As listed in Table 1, of 1,425 insertions, 1,183 occurred within 878 genes, and 242 insertions occurred within intergenic sequences. A total of 661 genes were interrupted only once by transposon insertion, whereas 217 genes were mutated more than one time with the maximum number of transposon insertions per gene being eight. Thus far, 3,356 of the 4,234 genes that contain a TA site have not been hit by transposon insertion. As may be seen in Fig. 1A, when plotted as a percentage of length of an ORF, the insertions are uniformly distributed within intragenic regions. Fig. 1B shows the exact locations of transposon insertions within the circular chromosome and indicates a uniform distribution of insertion within coding sequences.
Table 1. Summary of Himar1 transposon insertion mutagenesis of M. tuberculosis CDC1551.
| Transposon insertion library statistics | |
|---|---|
| No. of insertions sequenced | 1,425 |
| No. of unique insertions | 1,403 |
| No. of hits in genes | 1,183 |
| No. of unique genes hit | 878 |
| No. of intergenic spaces hit | 242 |
| No. of genes hit one time | 661 |
| No. of genes hit two times | 158 |
| No. of genes hit three times | 43 |
| No. of genes hit four times | 9 |
| No. of genes hit five times | 2 |
| No. of genes hit six times | 4 |
| No. of genes hit eight times | 1 |
| No. of genes not hit | 3,356 |
Fig. 1.
(A) Distribution of transposon insertions within the ORFs of M. tuberculosis by percentage of each ORF's total length. The MT numbers for each of the 1,183 genes containing a transposon insertion is plotted versus the percent distance from the 5′ end of the ORF. Insertions in the TA sites comprising stop codons have been excluded. The insertions are uniformly distributed, suggesting intragenic Himar1 insertion is random. (B) Distribution of transposon insertions in the 4.4-Mb circular chromosome of M. tuberculosis. The origin is marked as 0. The line segments around the circle indicate the locations of the 1,183 intragenic insertion mutants: 1,161 distinct locations and 22 double hits (indicated by longer line segments).
Our collection of mutants permits the identification of nonessential genes for in vitro growth on rich media. For the purposes of defining essential and nonessential genes, we excluded insertions near the 3′ end of genes (using the 5′80%–3′100-bp rule, see Methods) because these would be expected to lead to short truncations that might not inactivate the gene product. A total of 4,204 genes had at least one sufficiently proximal and hence, potentially inactivating, TA insertion site, totaling 57,934 TA sites, including 77 sites in regions of overlap between adjacent genes. Of the 1,183 intragenic insertion mutants, 1,025 were proximal enough by our rule to be inactivating, and of the 878 unique M. tuberculosis genes mutated, 770 contained at least one insertion in the proximal, inactivating portion of the gene using the rule. Hence, our collection of 878 intragenic insertions defines 770 M. tuberculosis genes that are highly likely to be nonessential for in vitro growth on complete Middlebrook 7H10 medium (see Table 5, which is published as supporting information on the PNAS web site, www.pnas.org).
A Statistical Method Permits Prediction of the Proportion of Essential Genes in M. tuberculosis. Based on a Bayesian statistical analysis using Markov chain Monte Carlo, in which the number of essential genes was assumed a priori to be uniformly distributed, we estimated the overall number of essential genes in the M. tuberculosis genome from the observed data with 1,025 transposon insertions within the proximal portions of genes (see Methods). The aim of the algorithm is to estimate the joint distribution of the essential status of all genes, given the observed data. The algorithm proceeds by first assigning all genes for which a mutant was observed to be nonessential and all others to be essential. At each iteration of the algorithm, each gene is considered one at a time, and the probability that it is essential, given the observed data and the assumed essential status of all other genes, is calculated. The gene is then randomly assigned to be essential or nonessential according to that probability. After many iterations, the probability that a gene is essential may be estimated by the proportion of iterations at which it was assigned to be essential, and the distribution of the proportion of essential genes may be estimated by the distribution of that proportion across the iterations (Table 2).
Table 2.
Proportion of essential genes in M. tuberculosis
| Rule | Estimate, % | 95% Confidence interval |
|---|---|---|
| 100% | 34 | 27-39 |
| 90% | 36 | 29-42 |
| 5′80%-3′100 bp | 35 | 28-41 |
| 80% | 40 | 33-46 |
| 70% | 42 | 35-49 |
| 60% | 42 | 33-50 |
With a conservative assumption that a proportion of distal region of genes may be dispensable and transposon insertion may produce a functional product, we estimated percentage of essential genes for various rules. The 100% rule assumes that all TA sites (excluding the two stop codons) within a gene are able to inactivate the gene if disrupted. The 90% rule assumes that only TAs in the proximal (5′) 90% of the gene to be able to inactivate gene and so on
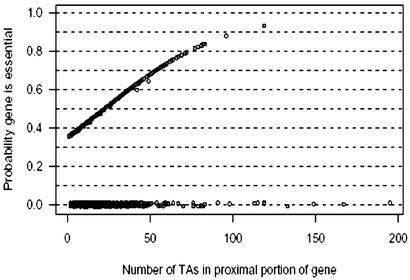
Our method permitted us to assign a nonzero probability of being essential to each of the 3,434 protein-encoding genes containing a proximal insertion site by our rule but not yet hit by the transposon. As may be seen in Fig. 2, there are 43 genes with 62 or more insertion sites in the proximal portion of their length. No mutant was observed for about half of these. Some of these long genes with numerous TA sites that have not been mutated thus far have the highest probabilities of being essential as may be seen in Table 3, and a complete list is shown in Table 6, which is published as supporting information on the PNAS web site. As shown, the majority of genes on this list have known functional roles; these include genes presumably involved in lipid biosynthesis (fatty acid synthase 1 and an acyl CoA synthase) and a number of genes expected to be essential, including three aminoacyl-tRNA synthase genes and pyruvate carboxylase. Additionally, several genes with unknown function are on the list of likely essential genes including a PPE (Pro-Pro-Glu polymorphic repetitive gene) family member, two probable membrane proteins, and two conserved hypothetical proteins.
Fig. 2.
Posterior probability that an M. tuberculosis gene is essential as a function of the number of TA sites determined by the 5′80%–3′100-bp rule. The 770 disrupted genes have a probability of zero to be essential and are jittered vertically so that the points may be distinguished. The vertical scatter in the remaining points is largely caused by Markov chain Monte Carlo sampling error. The identities of 15 genes whose probabilities of being essential were ≥75% are shown in Table 3.
Table 3. M. tuberculosis genes with high probabilities of being essential.
| MT no. | Rv no. | Gene description | Probability, % |
|---|---|---|---|
| 0417 | 0404 | Acyl-CoA synthase (fadD30) | 83 |
| 2062 | 2006 | Trehalose-6-phosphatase | 83 |
| 3285 | 3193c | Probable integral membrane protein | 83 |
| 2082 | 2024c | Conserved hypothetical protein | 82 |
| 3974 | 3859c | Glutamate synthase (gltB) | 82 |
| 1587 | 1536 | Isoleucyl-tRNA synthase | 81 |
| 1198 | 1161 | Nitrate reductase[α] subunit (narG) | 79 |
| 0047 | 0041 | Leucyl-tRNA synthase (leuS) | 79 |
| 2600 | 2524c | Fatty acid synthase (Fasl) | 78 |
| 0070 | 0064 | Probable membrane protein | 77 |
| 1678 | 1640c | C-term Lysyl-tRNA synthase (lysX) | 76 |
| 2551 | 2476c | Conserved hypothetical protein | 75 |
| 1796 | 1753c | PPE-family protein | 75 |
| 0116 | 0107c | Probable Mg transport ATPase | 75 |
| 3045 | 2967c | Pyruvate carboxylase | 75 |
Gene Families Enriched in Essential Genes. M. tuberculosis genes have been grouped into 109 categories, each group having genes involved in one or more related biochemical functions. We evaluated the functional categories by the number of transposon insertions per eligible site. As seen in Table 4, seven gene families, together accounting for 201 genes, sustained relatively fewer insertion mutations per eligible site than would be expected. These families could be assigned probabilities of being enriched in essential genes of >75%. As might be expected, aminoacyl tRNA synthases and modifying enzymes and key metabolic functions (such as purine nucleotide biosynthesis) appeared on this list. However, we were surprised that the PE-PGRS, polyketide and nonribosomal peptide biosynthesis, and fatty and mycolic acid biosynthesis gene families showed high likelihoods of being enriched of essential genes.
Table 4. M. tuberculosis gene families enriched and deficient in essential genes.
| Probability enriched, % | Est. % essential | Functional group |
|---|---|---|
| 97 | 54 (32-76) | Aminoacyl tRNA synthases and their modification |
| 94 | 45 (30-60) | PE family: PGRS subfamily |
| 82 | 46 (21-68) | Purine ribonucleotide biosynthesis |
| 80 | 40 (28-52) | Polyketide and nonribosomal peptide synthesis |
| 78 | 42 (23-62) | Synthesis of fatty and mycolic acids |
| 75 | 43 (21-64) | Ser/Thr protein kinases and phosphoprotein phosphotases |
| 75 | 42 (20-65) | Biosynthesis of molybdopterin |
| 4 | 32 (25-39) | Unknown proteins |
| 4 | 20 (7-40) | Metabolism of sulphur |
| 4 | 27 (17-36) | PPE family |
| 0 | 10 (0-24) | Conserved membrane proteins |
The probability with which the gene families are enriched in essential genes and the estimated percentages of essential genes in each family (along with 95% confidence intervals) are shown
The PE-PGRS family, one of several protein families unique to mycobacteria, contains polymorphic repeat elements that were clearly defined and quantified by the M. tuberculosis genome sequence (12). The PE-PGRS genes contain a common N-terminal Pro-Glu motif and a central PGRS. The function of the PE-PGRS family members is unknown, although there is some evidence that these family members are cell wall-associated (13, 14), may play a role in the modifying host immune responses (15, 16), and are preferentially expressed by bacilli in macrophages and granulomas (17). Our observations suggest a critical role for certain PE-PGRS proteins as the analysis predicts this group to be enriched in essential genes.
It was also surprising that our analysis identified the polyketide and nonribosomal peptide synthase family and the gene family involved in biosynthesis of fatty and mycolic acids to have high probabilities of being enriched in essential genes (Table 4). M. tuberculosis has a large number of genes devoted to synthesis of polyketides. Recently, an M. tuberculosis gene involved in mycobactin synthesis, which is a nonribosomal peptide pathway, was shown to be nonessential for in vitro growth, but required for survival in macrophages (18). Moreover, a related species, Mycobacterium ulcerans, was recently shown to produce a 12-memberedring polyketide toxin known as mycolactone, which is responsible for a progressive and painless dermal ulceration syndrome that may involve host cell apoptosis (19, 20). The M. tuberculosis polyketide synthase 2 gene has recently been shown to play a key role in the synthesis of sulfolipids that have been associated with the pathogenesis of tuberculosis by potentially blocking phagosome-lysosome fusion in infected macrophages (21, 22). Thus there is evidence that polyketide and nonribosomal synthesis pathways are involved in mycobacterial virulence, and our observations suggest that genes within these pathways are also indispensable. Fatty and mycolic acids are prominent constituents of the cell wall of mycobacteria, and two bactericidal antituberculous drugs, isoniazid and ethionamide, target and inhibit the FAS-II system (23). Our prediction that the fatty and mycolic acid synthesis functional group is enriched in essential genes is supported by the bactericidal nature of drugs that inhibit mycolic acid biosynthesis.
Biochemical Inhibitors of Genes Products Likely To Be Essential Have Antibiotic Properties. Beyond the fact that our statistical method identified many genes and gene families expected to be essential, we investigated whether small molecule inhibitors of some likely essential genes from Table 3 would, in fact, be bactericidal. MT1587 (encoding isoleucyl-tRNA synthase) showed an 81% probability of being essential. The product of this gene is inhibited by the antibiotic mupirocin, which is currently used clinically as a broad spectrum, topical antibiotic (24). In addition, our study identified MT3974 (gltB, encoding ferredoxin-dependent glutamate synthase) as having an 82% probability of being an essential gene. Azaserine is a known potent inhibitor of this enzyme in other species (25). We tested the inhibitory activity of azaserine for M. tuberculosis CDC1551 by using the proportion method and found that it has an MIC90 (minimal inhibitory concentration) of <1 μg/ml. Thus, there is evidence that pharmacologic blockade of proteins predicted to be essential by our statistical method is inhibitory to bacterial growth.
Other Considerations. We considered a number of potential biases in our sampling that might skew our analysis. One possibility is that essential genes may have a lower density of TA target sites than nonessential genes. Although M. tuberculosis genes vary in the density of TA targets per 100 bp, our analysis should not be affected by this variable because it assumes only that all nonessential TA targets are equally eligible and thus stratifies the likelihood of insertion into short genes with few TAs or longer genes with low TA density as may be seen in Fig. 2. A further concern is that of polar effects because transposon insertion into a polycistronic message might prevent translation of distal genes. We did not attempt to model functional polarity in our algorithm (which would have allowed us to make stronger predictions under the assumption that insertion into a proximal gene defined all distal genes to be nonessential) for several reasons. First, there are promoters within the Himar1 transposon that might rescue distal genes from a polar mutation, and indeed in our own data set we observe instances of Himar1 insertion upstream of a predicted essential genes (insertions in MT2999 and MT3000: see Table 5) within a defined operon (26). Second, distal promoters within coding sequences of operons have been reported in bacteria that may negate some polar effects when they occur [e.g., DNA gyrase genes in M. tuberculosis (27)]. Third, without experimental transcriptional analysis, defining operons from DNA sequence data alone is speculative and depends on several arbitrary assumptions regarding the maximal permitted intergenic length.
The possibility that Himar1 insertion in M. tuberculosis is nonrandom is another factor that might bias our results. We assessed the distribution of transposon insertion in both intragenic and intergenic regions. As seen in Fig. 1B, the global distribution of insertions was uniform and showed no evidence of insertion hot spots, and the handful of small apparent cold spots in Fig. 1B could reasonably be regions rich in putative essential genes. The location of intragenic mutations failed to reveal preferences for insertion (Fig. 1 A). Intergenic spaces, which are more permissive than coding sequences for insertion mutation and hence are a good sentinel for hot or cold spots for mutation, also did not show any detectable high- or low-density regions of insertion, and the percentage of available intergenic TA sites (13%) closely matched the observed transposon insertion frequency (17%). We assessed our collection of 1,425 mutants for multiple transposon insertions at the same site that might indicate the presence of insertion hot spots. As strong evidence against the presence of hot spots, there were no instances of hitting the same TA three times or more. Among the 1,183 intragenic insertions, 22 pairs of identical-site insertion mutants were found. These mutants were nonclonal as they were derived from different transposition experiments performed on different days. With a 35% abundance of essential genes ≈42,672 TA sites are eligible for insertions, and we calculated that the anticipated mean number of duplicate insertions by a fully random transposon after 1,183 intragenic insertions would be 16.1 (with a 95% chance of having between 9 and 24 duplicate insertions). Thus the observed number of duplicate insertions, 22, was within the range to be expected for a fully random transposon. These data indicate that at our current level of sensitivity (1,425 insertions in ≈42,672 eligible TA sites), Himar1 appears to transpose randomly into eligible TA sites globally within the M. tuberculosis CDC1551 genome.
Discussion
Our findings suggest that 35% of ORFs in M. tuberculosis are essential. Comprehensive targeted mutagenesis of the Saccharomyces cerevisiae genome (12 million bp) revealed 19% of genes to be essential (28). Site-directed mutagenesis of Bacillus subtilis (4.2 million bp) predicted 9% of the genome to be essential (29), and conditional antisense expression in Staphylococcus aureus (2.8 million bp) identified ≈150 of its 2,594–2,714 genes (6%) as being critical for growth (2). In contrast, two small-genome microbes have demonstrated higher proportions of essential genes: in Mycoplasma genitalium, the free-living microorganism with the smallest genome, transposon-mediated mutagenesis estimated the minimal set of essential genes as being 265–350 of the 480 protein coding genes (55–73%) (30). And recently, with a systematic in vitro transposition mutagenesis technique known as GAMBIT, Haemophilus influenzae (1.8 million bp) was found to contain 478 essential genes among 1,272 ORFs that were scored (38%) (31). As an obligate pathogen with a midsized genome of 4.4 million bp, M. tuberculosis was generally considered to possess a large complement of nonessential virulence genes. One would anticipate that such genes would be conditionally expressed during infection to mediate bacterial survival in different phases of its disease cycle, but be dispensable for replication on rich laboratory medium. In fact, however, our data suggest that one-third of M. tuberculosis genes are necessary for simple viability.
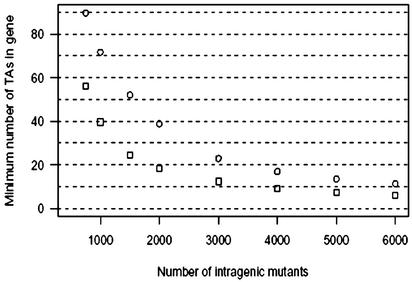
Although comprehensive random mutagenesis experiments require isolation and characterization of tens of thousands of mutants, our statistical method, which capitalizes on full a priori knowledge of available mutagenesis sites, offers surprisingly powerful functional predictions with a relatively small number of mutants characterized. In this study, after sequencing only 1,425 mutants, we identified 15 M. tuberculosis genes with a high probability of being essential, and we found seven gene families (containing 201 genes) with a high probability of being enriched in essential genes. Although our data have permitted the identification of ≈770 nonessential genes, it is important to recognize that our method assigns only probabilities for being essential to the remaining genes. As seen in Fig. 3, with data on another 1,500 mutants, resulting in a total of ≈2,400 mutants with intragenic insertions, genes with as few as 15 TA sites and for which no mutant was observed could be identified as essential with high probability. Hence, with knowledge of the genome sequence plus the nature and location of eligible targets, a large amount of functional information may be gained at the outset of such random transposon mutagenesis experiments without achieving saturation. This postgenomic approach marries genome sequence information and experimental data for high-throughput prediction of the proportion and identity of essential genes in a haploid genome.
Fig. 3.
The minimum number of TA sites an M. tuberculosis gene must contain to have >75% (□) or 90% (○) probability to be essential, if no mutant is observed with insertion in the gene, as a function of the total number of intragenic mutants observed.
Identification of both essential and nonessential genes has significant implications as it paves a way to discover linkages between genes that are functionally related and beyond. In addition, the availability of large collections of mutants in nonessential genes and information on the identity of essential M. tuberculosis genes will serve as valuable tools for comparative functional genomic studies of this and other closely related human pathogens.
Supplementary Material
Acknowledgments
The assistance of Naomi Gauchet is gratefully acknowledged. We thank Drs. Eric J. Rubin and Christopher M. Sassetti of the Harvard School of Public Health (Cambridge, MA) for providing the Himar1-containing mycobacteriophage and technical advice and for sharing results before publication. We also thank Sandeep Tyagi and Ian Rosenthal of The Johns Hopkins Bloomberg School of Public Health and Bozho Todorich of Elizabethtown College (Elizabethtown, PA) for technical assistance. This work was supported by National Institutes of Health Grants AI36973, AI37856, and AI43846.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: PE-PGRS, Pro-Glu polymorphic GC-rich repetitive sequence.
References
- 1.Akerley, B. J., Rubin, E. J., Camilli, A., Lampe, D. J., Robertson, H. M. & Mekalanos, J. J. (1998) Proc. Natl. Acad. Sci. USA 95, 8927-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ji, Y., Zhang, B., Van Horn, S. F., Warren, P., Woodnutt, G., Burnham, M. K. & Rosenberg, M. (2001) Science 293, 2266-2269. [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann, R. D., Alland, D., Eisen, J. A., Carpenter, L., White, O., Peterson, J., DeBoy, R., Dodson, R., Gwinn, M., Haft, D., et. al. (2002) J. Bacteriol. 184, 5479-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lampe, D. J., Churchill, M. E. & Robertson, H. M. (1996) EMBO J. 15, 5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin, E. J., Akerley, B. J., Novik, V. N., Lampe, D. J., Husson, R. N. & Mekalanos, J. J. (1999) Proc. Natl. Acad. Sci. USA 96, 1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geman, S. & Geman, D. (1984) IEEE Trans. Pattern Anal. Machine Intelligence 6, 721-741. [DOI] [PubMed] [Google Scholar]
- 7.Ihaka, R. & Gentleman, R. R. (1995) J. Comp. Graph. Stat. 5, 299-314. [Google Scholar]
- 8.Valway, S. E., Sanchez, M. P., Shinnick, T. F., Orme, I., Agerton, T., Hoy, D., Jones, J. S., Westmoreland, H. & Onorato, I. M. (1998) N. Engl. J. Med. 338, 633-639. [DOI] [PubMed] [Google Scholar]
- 9.Hatfull, G. F. (2000) in Molecular Genetics of Mycobacteria, eds. Hatfull, G. F. & Jacobs, W. R. J. (Am. Soc. Microbiol. Press, Washington, DC), p. 318.
- 10.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1998) Current Protocols in Molecular Biology (Wiley, New York), unit 2.4.
- 11.Siebert, P. D., Chenchik, A., Kellogg, D. E., Lukyanov, K. A. & Lukyanov, S. A. (1995) Nucleic Acids Res. 23, 1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, et. al. (1998) Nature 393, 537-544. [DOI] [PubMed] [Google Scholar]
- 13.Banu, S., Honore, N., Saint-Joanis, B., Philpott, D., Prevost, M. C. & Cole, S. T. (2002) Mol. Microbiol. 44, 9-19. [DOI] [PubMed] [Google Scholar]
- 14.Brennan, M. J., Delogu, G., Chen, Y., Bardarov, S., Kriakov, J., Alavi, M. & Jacobs, W. R., Jr. (2001) Infect. Immun. 69, 7326-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delogu, G. & Brennan, M. J. (2001) Infect. Immun. 69, 5606-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh, K. K., Zhang, X., Patibandla, A. S., Chien, P., Jr., & Laal, S. (2001) Infect. Immun. 69, 4185-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramakrishnan, L., Federspiel, N. A. & Falkow, S. (2000) Science 288, 1436-1439. [DOI] [PubMed] [Google Scholar]
- 18.De Voss, J. J., Rutter, K., Schroeder, B. G., Su, H., Zhu, Y. & Barry, C. E., 3rd (2000) Proc. Natl. Acad. Sci. USA 97, 1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.George, K. M., Chatterjee, D., Gunawardana, G., Welty, D., Hayman, J., Lee, R. & Small, P. L. (1999) Science 283, 854-857. [DOI] [PubMed] [Google Scholar]
- 20.George, K. M., Pascopella, L., Welty, D. M. & Small, P. L. (2000) Infect. Immun. 68, 877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirakova, T. D., Thirumala, A. K., Dubey, V. S., Sprecher, H. & Kolattukudy, P. E. (2001) J. Biol. Chem. 276, 16833-16839. [DOI] [PubMed] [Google Scholar]
- 22.Goren, M. B., D'Arcy Hart, P., Young, M. R. & Armstrong, J. A. (1976) Proc. Natl. Acad. Sci. USA 73, 2510-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marrakchi, H., Laneelle, G. & Quemard, A. (2000) Microbiology 146, 289-296. [DOI] [PubMed] [Google Scholar]
- 24.Sutherland, R., Boon, R. J., Griffin, K. E., Masters, P. J., Slocombe, B. & White, A. R. (1985) Antimicrob. Agents Chemother. 27, 495-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marques, S., Florencio, F. J. & Candau, P. (1992) Eur. J. Biochem. 206, 69-77. [DOI] [PubMed] [Google Scholar]
- 26.Camacho, L. R., Constant, P., Raynaud, C., Laneelle, M. A., Triccas, J. A., Gicquel, B., Daffe, M. & Guilhot, C. (2001) J. Biol. Chem. 276, 19845-19854. [DOI] [PubMed] [Google Scholar]
- 27.Unniraman, S., Chatterji, M. & Nagaraja, V. (2002) J. Bacteriol. 184, 5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et. al. (2002) Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
- 29.Itaya, M. (1995) FEBS Lett. 362, 257-260. [DOI] [PubMed] [Google Scholar]
- 30.Hutchison, C. A., Peterson, S. N., Gill, S. R., Cline, R. T., White, O., Fraser, C. M., Smith, H. O. & Venter, J. C. (1999) Science 286, 2165-2169. [DOI] [PubMed] [Google Scholar]
- 31.Akerley, B. J., Rubin, E. J., Novick, V. L., Amaya, K., Judson, N. & Mekalanos, J. J. (2002) Proc. Natl. Acad. Sci. USA 99, 966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.