Abstract
Nucleotide excision repair (NER) prevents skin cancer by eliminating highly genotoxic cyclobutane pyrimidine dimers (CPDs) induced in DNA by the UVB component of sunlight. NER consists of two distinct but overlapping subpathways, i.e., global NER, which removes CPD from the genome overall, and transcription-coupled NER (TCNER), which removes CPD uniquely from the transcribed strand of active genes. Previous investigations have clearly established that the p53 tumor suppressor plays a crucial role in the NER process. Here we used the ligation-mediated PCR technique to demonstrate, at nucleotide resolution along two chromosomal genes in human cells, that the requirement for functional p53 in TCNER, but not in global NER, depends on incident UV wavelength. Indeed, relative to an isogenic p53 wild-type counterpart, p53-deficient human lymphoblastoid strains were shown to remove CPD significantly less efficiently along both the transcribed and nontranscribed strands of the c-jun and hprt loci after exposure to polychromatic UVB (290–320 nm). However, in contrast, after irradiation with 254-nm UV, p53 deficiency engendered less efficient CPD repair only along the nontranscribed strands of these target genes. The revelation of this intriguing wavelength-dependent phenomenon reconciles an apparent conflict between previous studies which used either UVB or 254-nm UV to claim, respectively, that p53 is required for, or plays no role whatsoever in, TCNER of CPD. Furthermore, our finding highlights a major caveat in experimental photobiology by providing a prominent example where the extensively used “nonsolar” model mutagen 254-nm UV does not accurately replicate the effects of environmentally relevant UVB.
Nucleotide excision repair (NER) forestalls the accumulation of genetic mutations, thus guarding against neoplastic transformation, by eliminating helix-distorting “bulky” DNA adducts induced by diverse environmental carcinogens. Such adducts include highly genotoxic (replication- and transcription-blocking) cyclobutane pyrimidine dimers (CPDs), which form through covalent linkage of adjacent pyrimidine bases subsequent to direct absorption of UV photons by DNA. Misreplication of CPDs induced by the UVB component of natural sunlight is the primary cause of mutations prerequisite to the development of skin cancer, the most frequent neoplasia in Caucasian populations (1, 2). Individuals afflicted with the rare autosomal recessive disorder xeroderma pigmentosum (XP) carry mutations in any of seven different NER genes (designated XP-A through -G) and are therefore defective in CPD repair (3). As a consequence, XP patients exhibit extreme photosensitivity and UV hypermutability, coupled with a striking predisposition to both melanoma and nonmelanoma tumors (4).
NER is comprised of two distinct subpathways, which manifest strong mechanistic overlap, i.e., differing only in the lesion-recognition step (see ref. 5 for a comprehensive review of the NER pathway). Global NER (GNER) removes CPD from virtually anywhere in the genome and is initiated when the XP complementation group-E gene product, in conjunction with the XPC-hHR23B protein complex, recognizes and binds the helical distortion introduced into DNA by CPDs or other bulky lesions. This initial event is followed by recruitment to the damaged site of the core NER pathway, which faithfully restores the integrity of the DNA through sequential steps of (i) strand separation mediated by the XP-B and -D helicases; (ii) incision on either side of the lesion via the structure-specific endonuclease activities of XP-G and XP-F/ERCC1; (iii) excision of the damaged base as part of a single-stranded oligonucleotide ≈30 bp in length; and (iv) DNA resynthesis (gap filling) and ligation, using normal DNA replication factors and the intact complementary strand as template.
In contrast to the situation for GNER, the transcription-coupled NER (TCNER) subpathway removes only those lesions occurring along the transcribed strand (TS) of active genes and is triggered when RNA polymerase II becomes stalled at positions where transcription-blocking CPDs occur (6). The CS-B and -A proteins then converge at the damaged sites, followed by removal or retraction of the polymerase. This facilitates completion of the repair process by the core NER pathway, as described immediately above for GNER. Deficiency in either CS-B or -A causes Cockayne's syndrome (CS), which, unlike XP, is characterized by defective TCNER but normal GNER, developmental abnormalities, and no predisposition to cutaneous tumors (7, 8).
The p53 tumor suppressor protein plays a critical role in the inhibition of multistage photocarcinogenesis in part by transactivating proapoptotic genes that stimulate the elimination of UV-damaged precancerous “sunburn” cells (9, 10). Moreover, p53 would be expected to inhibit skin cancer development in view of its demonstrated essential participation in NER. Indeed, a well characterized Southern blot-based assay that measures DNA strand-specific repair at the level of the gene was used to show that genetically p53-deficient skin fibroblasts derived from Li–Fraumeni syndrome patients, or skin fibroblasts expressing the human papillomavirus E6 (HPV-E6) oncoprotein that functionally inactivates p53 by accelerating its proteosomal degradation, are each defective in GNER (11–13). This defect was manifested by the inability of these p53-deficient strains to efficiently remove CPD from the nontranscribed strand (NTS) of the transcriptionally active dihydrofolate reductase and p53 genes after irradiation with the model mutagen 254-nm UV. At the same time, these latter investigations demonstrated that functional p53 was not required for TCNER, because p53-deficient skin fibroblasts were fully proficient in the removal of 254-nm UV-induced CPD from the TS of either dihydrofolate reductase or p53. Equivalent results on NER of UV-induced CPD were subsequently obtained for mammalian strains carrying homozygous null p53 mutations, including human colorectal carcinoma cells (14) and murine embryonic fibroblasts (15). In addition, it was demonstrated that human skin fibroblasts lacking functional p53 are defective only in GNER, and not TCNER, of bulky DNA adducts induced by the environmental carcinogen benzo(a)pyrene diolepoxide (16). In providing some potential mechanistic explanation for the above findings, it was reported by using 254-nm UV-exposed human cells that up-regulation of the XP-C and -E proteins (required for lesion recognition during GNER only) depends on the presence of functional p53 (17, 18).
Notwithstanding the above investigations on 254-nm UV-exposed cells, which convincingly demonstrated an essential role for p53 in GNER but not in TCNER, we originally postulated that p53 might actually regulate both NER subpathways. Indeed, p53 had been shown to interact with the XP-B and -D helicases, which are required for strand separation during GNER and TCNER, as well as with the CS-B protein that participates in lesion recognition during TCNER only (19). In addition, after exposure to genotoxic agents, p53 transactivates a gene encoding the ribonucleotide reductase subunit p53R2 (20), which is essential for the DNA resynthesis step common to both NER subpathways. Finally, cells lacking functional p53 are deficient in the recovery of mRNA synthesis after UV treatment, indicating the relative inability of such cells to efficiently clear transcription-blocking CPD from the TS of active genes (21). With the above rationale in mind, we initially used the ligation-mediated PCR (LMPCR) technique to demonstrate at nucleotide resolution that p53-deficient human fibroblasts, either derived from Li–Fraumeni patients or expressing HPV-E6, exhibit a substantial deficiency in both GNER and TCNER as measured at the endogenous c-jun and p53 loci after treatment with polychromatic UVB (290–320 nm) (22). We reasoned that the apparent discrepancy between these latter data showing p53 dependence for TCNER in human skin fibroblasts and those cited in the preceding paragraph, which seemingly demonstrated the opposite, might be explained by the circumstance that in each case cells were irradiated with different UV sources, i.e., emitting, respectively, either environmentally relevant polychromatic UVB or monochromatic 254-nm (“germicidal”) UV, which is virtually absent from terrestrial sunlight. Although important similarities have been documented regarding the types of premutagenic DNA photoproducts and concomitant genotoxic stress responses elicited by 254-nm UV vs. UVB (23), these wavelengths manifest differential capacities to alter the cellular redox state and, as a consequence, to influence patterns of gene activation (see Discussion). To evaluate the intriguing possibility that p53 might regulate NER in a wavelength-dependent manner, we used well characterized isogenic human lymphoblastoid strains differing only in p53 status to study the kinetics of strand-specific CPD removal along two chromosomal genes, i.e., c-jun and hypoxanthine phosphoribosyltransferase (hprt), after treatment with either 254-nm UV or UVB.
Materials and Methods
Cell Strains. The p53+/+ human lymphoblastoid strain TK6 and its functionally p53-deficient counterpart TK6-5E (constitutively expressing the HPV-E6 oncoprotein derived from the high-risk HPV16 subtype) were kindly provided by J. B. Little (Harvard School of Public Health, Harvard University, Boston). NH32, a homozygous p53-knockout derivative of TK6, was a generous gift of H. L. Liber (Massachusetts General Hospital, Boston). All three strains were routinely maintained in suspension culture in a 5% CO2 atmosphere at 37°C in RPMI medium 1640 (GIBCO/BRL) supplemented with 10% inactivated horse serum and 100 units/ml penicillin/streptomycin.
UV Irradiation. Approximately 5 × 106 exponentially growing lymphoblastoid cells were washed once with PBS (containing Ca2+ and Mg2+), resuspended in 10 ml PBS, and added directly to 100-mm Petri dishes. Replicate cultures were then irradiated with acute doses of either polychromatic UVB (290–320 nm) or monochromatic 254-nm UV at room temperature. The UVB source consisted of two fluorescent tubes (F15T8 UVB lamp; Ultraviolet Products) generating a dose rate of 6.5 J/m2/s. The incident UVB was filtered by using a sheet of cellulose acetate (Kodacel TA-407 0.015 in; Eastman Kodak) to virtually eliminate contaminating wavelengths below 290 nm. In the case of 254-nm UV, cells were irradiated with a G25T8 germicidal lamp (Sankyo Denki) at a dose rate of 0.2 J/m2/s. UVB and 254-nm UV fluences were measured with a Spectroline DRC 100× digital radiometer (Spectronics, Westbury, NY) equipped with DIX 300 and 254 sensors, respectively. It should be noted that the UVB lamp emitted some measurable incident energy within the UVA range (320–400 nm), which could not be eliminated. Although we cannot categorically rule out that this UVA component exerted some effect, the total UVA output under our exposure conditions, i.e., ≈50 J/m2, was unlikely to elicit any significant biological response given that UVA is ≈50,000-fold less genotoxic than UVB on a per-joule basis.
Enzymatic CPD Cleavage and LMPCR. LMPCR is a genomic sequencing method that allows quantification, at nucleotide resolution along chromosomal genes, of any DNA adduct that can be revealed either chemically or enzymatically as a ligatable strand break. The basic LMPCR protocol used here to investigate CPD repair rates along the TS and NTS of either c-jun or hprt has been described in detail (24, 25). Briefly, replicate cultures were irradiated with either 20 J/m2 254-nm UV or 450 J/m2 UVB and incubated for varying times (0–24 h) to allow repair. Genomic DNA was then purified and digested first with T4 endonuclease V, which efficiently induces single-strand breaks immediately adjacent to CPD sites with high specificity, and secondly with Escherichia coli photolyase to convert these breaks to ligatable 5′ termini. In a manner described previously (26), aliquots of this enzyme-digested DNA were run on denaturing agarose gels to show that 20 J/m2 of 254-nm UV and 450 J/m2 of UVB induced initial global CPD frequencies of 1.6 and 2.4 CPD/10 kb, respectively (data not shown). After denaturation of the DNA, a gene-specific oligonucleotide was annealed downstream of the region to be analyzed, and a set of genomic cleavage products (i.e., terminating precisely at sites where T4 endonuclease V incised the DNA adjacent to CPDs) was generated via primer extension with cloned Pfu polymerase. An asymmetric double-stranded oligonucleotide linker was ligated to the phosphate groups at the fragment termini, thus providing a common sequence on the 5′ end of all fragments. An oligonucleotide primer complementary to this linker, in conjunction with another gene-specific primer, was then used in a PCR reaction to amplify the gene-specific cleavage products of interest. These products were subjected to electrophoresis on 8% polyacrylamide gels alongside a Maxam and Gilbert sequencing ladder, transferred to nylon membranes, hybridized to a 32P-labeled gene-specific probe, and visualized by autoradiography. All bands corresponding to dipyrimidine sites and yielding a measurable signal above background were quantified using a Bio-Rad GS-525 phosphorimager (Bio-Rad Canada). Variations in DNA sample loading were carefully controlled for by quantitatively assessing the relative intensities of nonspecific signals between lanes. The LMPCR primer sets used for quantification of NER rates at the c-jun locus have been described (27, 28), whereas those used at the hprt locus have not been previously published and are listed in Table 1.
Table 1. Oligonucleotide primers sets for LMPCR analysis of the human hprt gene.
| Primer* | Sequence | Strand/exon | Tm, °C |
|---|---|---|---|
| HA-1 | 5′-ATCCAATCAAATGTTTGTATC | TS, exon 2 | 50.2 |
| HA-2 | 5′-GTTTGTATCCTGTAATGCTCTCATTGAAAC | 60.6 | |
| HB-1 | 5′-GAAGATTTTAGAAAGCATCAG | NTS, exon 2 | 52.1 |
| HB-2 | 5′-CCTAGTTTATGTTCAAATAGCAAGTACTCAG | 60.7 | |
| HC-1 | 5′-TTGGTGTGGAAGTTTAATG | TS, exon 3 | 51.0 |
| HC-2 | 5′-GTGGAAGTTTAATGACTAAGAGGTGTTTG | 60.6 | |
| HD-1 | 5′-GAAAATATAAGAAAACCTACTG | NTS, exon 3 | 50.7 |
| HD-2 | 5′-GAAAACCTACTGTTGCCACTAAAAAGAATC | 60.6 |
Tm, melting temperatures used for primer annealing reactions
The first oligonucleotide of each pair is used for primer extension, whereas the second one is used for the amplification step (see Materials and Methods). Primers are located within introns closely adjacent to the indicated exon
Results and Discussion
Despite the general accord that functional p53 is essential for efficient GNER of UV-induced CPD, apparently divergent claims initially emerged that this tumor suppressor either is also required for TCNER (22) or plays no role whatsoever in this process (11). Although essentially the same p53-proficient vs. -deficient human skin fibroblast strains were compared in these two sets of studies, there were some potentially significant differences in experimental design. For example, in each case, a distinct strand-specific repair assay was used, i.e., LMPCR, a PCR-based genomic sequencing method that has been adapted to measure repair of DNA adducts at nucleotide resolution (29), vs. an extremely well characterized Southern blot-based assay that measures repair at the level of the gene (30). We reasoned that this methodological difference is unlikely to account for the observed contrariety in experimental outcome, because each of the aforementioned assays has been rigorously validated as a means to evaluate NER rates along the TS and NTS of active genes in living cells. For example, by using LMPCR, we previously reproduced the expected result, initially obtained via the Southern blot-based method (8), that CS-B cells are deficient in TCNER but not GNER of UV-induced CPD (31). Finally, we note that each of the above-mentioned assays quantifies precisely the same event, i.e., the lesion recognition/incision step of NER as manifested by the disappearance of T4 endonuclease V-sensitive sites.
A more likely and indeed intriguing explanation to account for the perceived discrepancy regarding the role of p53 in strand-specific NER concerned the use of different CPD-inducing wavelengths, i.e., either 254-nm UV or polychromatic UVB in the case of studies showing no role or an important role, respectively, for p53 in TCNER. To investigate this possibility, we used the p53+/+ human lymphoblastoid strain TK6 and two isogenic p53-deficient derivatives, i.e., NH32, carrying a homozygous knockout of p53; and TK6-5E, which constitutively expresses the HPV-E6 oncoprotein that functionally inactivates p53. The p53 status and phenotype of these lymphoblastoid strains have been verified in our laboratory (32) and by others (33, 34). Specifically, in response to DNA damaging agents, it was shown that NH32 and TK6-5E manifest clear defects in hallmark p53-regulated functions including cell cycle arrest, apoptosis, and induction of the p53 downstream effector p21waf1.
After treatment of TK6, TK6-5E, and NH32 with polychromatic UVB, or TK6 and NH32 with 254-nm UV, we used LMPCR to compare the rate of CPD removal in each strain at nucleotide resolution along the TS and NTS of the autosomal c-jun protooncogene, a locus routinely used for LMPCR studies. However, c-jun is known to be transcriptionally up-regulated in a variety of cell types at early times after UV exposure (35), thereby introducing a potential bias when this particular gene target is used for investigations on transcription-coupled repair of UV-induced DNA damage. In addition, it must be taken into account that NER rates may predictably be modulated in a locus-specific manner, possibly due to variations in local chromatin structure. We therefore also developed and applied the LMPCR technique to investigate the kinetics of strand-specific CPD repair along the X-linked hprt locus, i.e., a nonessential housekeeping gene, which to our knowledge is not regulated by DNA damage.
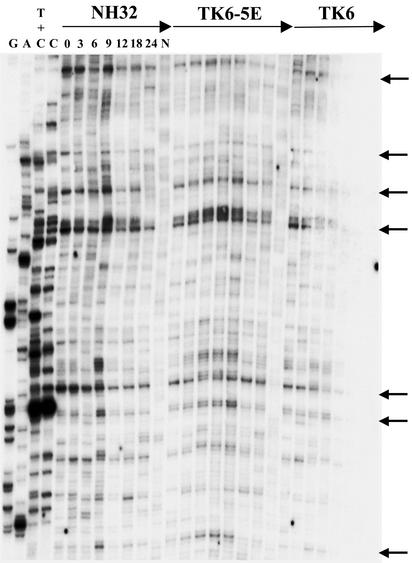
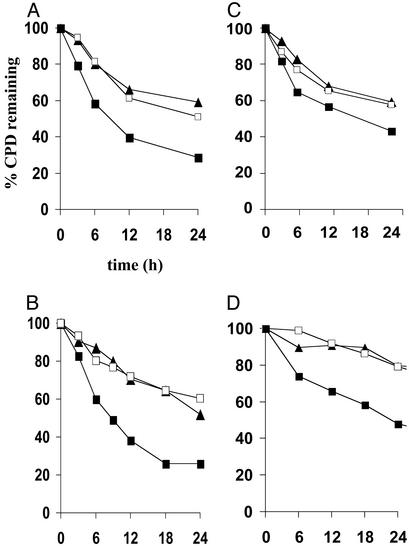
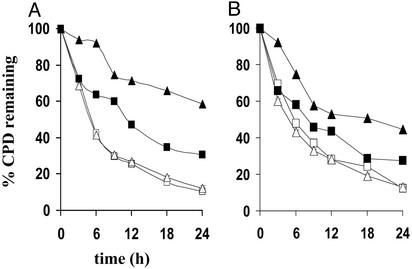
Fig. 1 shows a sample LMPCR autoradiogram reflecting CPD repair rates at individual nucleotide positions along a portion of the TS of the hprt gene in NH32, TK6-5E, and TK6 after irradiation with 450 J/m2 of UVB. (Autoradiograms depicting repair along other regions of the TS or NTS of either c-jun or hprt in UVB-exposed cells are not shown.) The arrows on the right side of this representative figure indicate dipyrimidine sites manifesting a clear signal above background, i.e., which could be precisely quantified by phosphorimager analysis. For any given site, the percentage of CPD remaining at various times post-irradiation was calculated by comparing the intensities of bands produced at these times to that of the corresponding band at time 0 (i.e., no opportunity for repair, 100% of CPD remaining). Graphical compilations of all repair rate determinations along the TS and NTS of both hprt and c-jun for each UVB-exposed lymphoblastoid strain are presented in Fig. 2. At least two different regions along each strand of each gene were independently evaluated on separate autoradiograms, and 10–20 dipyrimidine sites were quantified per region to arrive at a total of 25–40 sites analyzed per strand per gene. Each data point on the graphs represents a mean value calculated for these 25–40 sites. The results clearly show that after irradiation with polychromatic UVB, human lymphoblastoid cells wherein p53 is genetically or functionally inactivated are significantly less proficient relative to an isogenic wild-type counterpart in the removal of CPD from the TS of either c-jun or hprt (Fig. 2 A and B, respectively), as well as from the NTS of these target genes (Fig. 2 C and D). This provides critical confirmation for our previous finding that p53 regulates TCNER (in addition to GNER) at the c-jun and p53 loci in human skin fibroblasts (22).
Fig. 1.
Repair of UVB-induced CPD at nucleotide resolution along a portion of the TS of the human hprt gene in TK6, TK6-5E, and NH32. Repair rates at individual sites (exon 3, primer HC, nucleotides 16740–16880; GenBank accession no. M26939) are depicted for NH32 (Left), TK6-5E (Center), and TK6 (Right). The first four lanes on the left show LMPCR of DNA treated with standard Maxam–Gilbert cleavage reactions. For each strain, the following seven lanes show LMPCR of DNA isolated from UVB-irradiated cells that have undergone repair for the indicated times. The last lane for each strain shows LMPCR of unirradiated DNA followed by T4 endonuclease V/photolyase digestion (background). The arrows indicate dipyrimidine sites that were quantified using a GS-25 phosphorimager, equipped with multianalyst, Ver. 1.1 (Bio-Rad Canada).
Fig. 2.
Influence of p53 status on CPD removal along the TS and NTS of the c-jun and hprt loci in TK6, TK6-5E, and NH32 treated with polychromatic UVB. (A) TS of c-jun; each data point represents the mean of 39 sites using primers JE (nucleotides +100 to +190; GenBank accession no. J09111), JX (nucleotides +1748 to +1910), and JS (nucleotides +233 to +389). (B) TS of hprt; mean of 25 sites using primers HA (exon 2, nucleotides +14890 to +14980) and HC (exon 3, nucleotides +16740 to +16880). (C) NTS of c-jun; mean of 39 sites using primers JY (nucleotides +1644 to +1820) and JB (nucleotides -360 to -215). (D) NTS of hprt; mean of 20 sites using primer HB (exon 2, nucleotides +14850 to +14967). The standard error of the mean, calculated for each data point, was less than ±3.0% in all cases. ▪, TK6; □, TK6-5E; ▴, NH32.
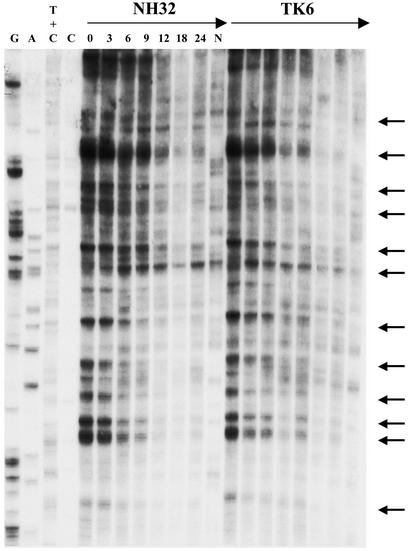
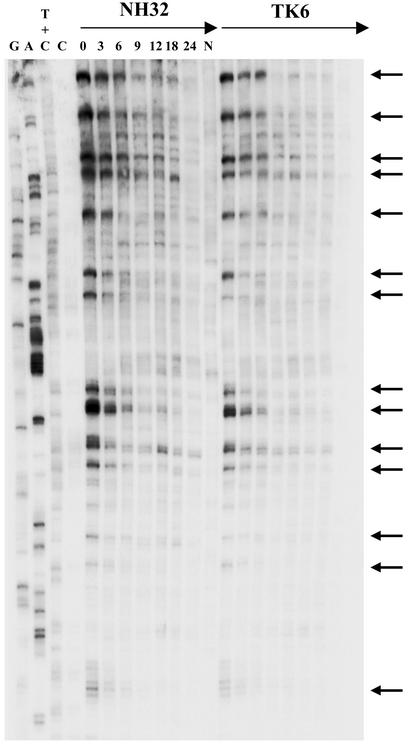
Sample LMPCR autoradiograms are shown in Figs. 3 and 4, which depict CPD repair rates along portions of the NTS and TS, respectively, of the hprt gene in TK6 vs. NH32 after treatment with 20 J/m2 of 254-nm UV. In essentially the identical manner as for the UVB studies described above, the kinetics of strand-specific repair at the c-jun and hprt loci were determined and displayed graphically (Fig. 5). Relative to wild-type TK6, p53-null NH32 manifested defective CPD removal from the NTS of either c-jun or hprt after 254-nm UV exposure, whereas these strains displayed no difference whatsoever in repair along the TS of either target gene (Fig. 5 A and B). This result is in complete accord with the previous investigations (cited in the Introduction) on various human and murine strains exposed to 254-nm UV but is in contrast to the situation described in the preceding paragraph for UVB-irradiated human lymphoblastoid or fibroblast strains where efficient repair of both the TS and NTS display strict p53 dependence.
Fig. 3.
Repair of 254-nm UV-induced CPD at nucleotide resolution along a portion of the NTS of the human hprt gene in TK6 and NH32. Repair rates at individual sites using primer HD (exon 3, nucleotides, i.e., +16510 to +16630) are depicted for NH32 (Left) and TK6 (Right). The lane designations and arrow indications are the same as for Fig. 1.
Fig. 4.
Repair of 254-nm UV-induced CPD at nucleotide resolution along a portion of the TS of the human hprt gene in TK6 and NH32. Repair rates at individual sites using primer HA (exon 2, nucleotides +14890–+14980) are depicted for NH32 (Left) and TK6 (Right). The lane designations and arrow indications are the same as for Fig. 1.
Fig. 5.
Influence of p53 status on CPD removal along the TS and NTS of c-jun and hprt in TK6 and NH32 treated with monochromatic 254 nm-UV. (A) TS and NTS of c-jun using primers JE (nucleotides +100 to +190) and JS (nucleotides +233 to +389) for the TS and primers JT (nucleotides +230 to +310) and JY (nucleotides +1644 to +1820) for the NTS. Each data point represents the mean of 25 sites for the TS and 27 sites for the NTS. (B) TS and NTS of hprt using primers HA (exon 2, nucleotides +14890–+14980) and HC (exon 3, nucleotides +16740 to +16880) for the TS, and primers HB (exon 2, nucleotides +14850 to +14967) and HD (exon 3, nucleotides +16510 to +16630) for NTS. Each data point represents the mean of 38 sites for the TS and 45 sites for the NTS. The standard error of the mean, calculated for each data point, was less than ±3.0% in all cases. Open and filled symbols depict repair of the TS and NTS, respectively. □, ▪ TK6; ▵, ▴ NH32.
Our overall data clearly demonstrate, in a well defined isogenic system at two chromosomal loci, that loss of functional p53 significantly reduces the efficiency of GNER and TCNER in human cells exposed to polychromatic UVB; however, after treatment with monochromatic 254-nm UV, p53 is essential for efficient GNER but absolutely dispensable for TCNER. The revelation of this striking wavelength-dependent effect apparently explains the perceived discrepancy discussed in detail earlier regarding the role of p53 in transcription-coupled repair. Although the underlying mechanism remains unknown, we suggest a plausible general model based on the capacity of photonic energy in the form of 254-nm UV, but not of UVB, to compensate for loss of functional p53 by stimulating the induction/activation of one or more proteins (normally also regulated by p53) that are required for TCNER. We further postulate that this process would constitute part of the so-called “mammalian UV response,” which comprises a plethora of protective UV-inducible signaling cascades triggered via autophosphorylation of plasma membrane-associated growth-factor receptors (36). Evidence to support such a model has been provided by the demonstration that activation of jun-NH2 terminal kinase, i.e., a major early event in the mammalian UV response, may be required for efficient removal of cisplatin-induced DNA damage, which is an NER-dependent process (37, 38). In addition, primary CS-B fibroblasts but not XP fibroblasts were shown to be impaired in jun-NH2 terminal kinase activation (39), suggesting a possible role for this event specifically in TCNER.
In further support of the above model, previous investigations have demonstrated that UV-induced signal transduction originating at the plasma membrane can be subject to wavelength-dependent regulation. Of particular significance given the example rendered in the preceding paragraph is that 254-nm UV was shown to be considerably more effective than UVB in the activation of mitogen-activated protein kinases, including jun-NH2-terminal kinase, in human and murine cells (40–43). The occurrence of such phenomena may be explained based on the fact that, to a far greater extent than 254-nm UV, polychromatic UVB is able to significantly alter the cellular redox state through the production of reactive oxygen species (44). Furthermore, it is well established that shifts in cellular redox potential per se can exert highly significant effects at the level of activation or inhibition of major stress-responsive signaling pathways (45). In any event, eventual characterization of the precise basis for the UV wavelength-dependent regulation of CPD removal observed here may be expected to reveal novel mechanistic information regarding the control of transcription-coupled repair in humans.
Finally, our results have important implications regarding experimental photobiology. UV at 254 nm is virtually completely absorbed by the atmosphere and therefore not a biologically significant component of terrestrial sunlight. On the other hand, UVB is present in the natural environment and constitutes the primary mediator for the most deleterious effects of solar radiation, including erythema, immunosuppression, mutagenesis, and skin cancer (46, 47). Despite the above, since the inception of modern-day photobiological research, a vast in vitro and in vivo database has accumulated on the genotoxic and carcinogenic effects of 254-nm UV. Considerably fewer investigations have focused on UVB, and fewer still have rigorously compared the effects of UVB vs. 254-nm UV. The convenience and ready availability of germicidal lamps emitting 254-nm UV, coupled with the popular assumption that 254-nm UV and polychromatic UVB elicit similar biological responses, has perpetuated this situation. We note that the majority of previous studies using the “nonsolar” model mutagen 254-nm UV may indeed stand the test of time with respect to physiological relevance. Nonetheless, in presenting one prominent example where 254-nm UV does not accurately replicate the effects of UVB, our data strongly emphasize the need to exercise much greater caution when choosing appropriate experimental models for skin cancer development in humans.
Acknowledgments
This work was supported by grants (to E.A.D.) from the National Cancer Institute of Canada (with funds from the Canadian Cancer Society) and from the Canadian Institutes of Health Research. E.A.D. and R.D. are scholars of the Fonds de la Recherche en Santé du Québec. J.-P.T. is the recipient of a postdoctoral fellowship from the National Cancer Institute of Canada.
Abbreviations: CPD, cyclobutane pyrimidine dimer; CS, Cockayne's syndrome; NER, nucleotide excision repair; GNER, global NER; HPV, human papillomavirus; hprt, hypoxanthine phosphoribosyltransferase; LMPCR, ligation-mediated PCR; NTS, nontranscribed strand; TCNER, transcription-coupled NER; TS, transcribed strand; XP, xeroderma pigmentosum.
References
- 1.Brash, D. E., Rudolph, J. A., Simon, J. A., Lin, A., McKenna, G. J., Baden, H. P., Halperin, A. J. & Ponten, J. (1991) Proc. Natl. Acad. Sci. USA 88, 10124-10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.You, Y. H., Lee, D. H., Yoon, J. H., Nakajima, S., Yasui, A. & Pfeifer, G. P. (2001) J. Biol. Chem. 276, 44688-44694. [DOI] [PubMed] [Google Scholar]
- 3.Ford, J. M. & Hanawalt, P. C. (1997) Curr. Top. Microbiol. Immunol. 221, 47-70. [DOI] [PubMed] [Google Scholar]
- 4.Kraemer, K. H., Lee, M. M. & Scotto, J. (1984) Carcinogenesis 5, 511-514. [DOI] [PubMed] [Google Scholar]
- 5.de Laat, W. L., Jaspers, N. G. & Hoeijmakers, J. H. (1999) Genes Dev. 13, 768-785. [DOI] [PubMed] [Google Scholar]
- 6.Svejstrup, J. Q. (2002) Nat. Rev. Mol. Cell Biol. 3, 21-29. [DOI] [PubMed] [Google Scholar]
- 7.Nance, M. A. & Berry, S. A. (1992) Am. J. Med. Genet. 42, 68-84. [DOI] [PubMed] [Google Scholar]
- 8.Venema, J., Mullenders, L. H., Natarajan, A. T., van Zeeland, A. A. & Mayne, L. V. (1990) Proc. Natl. Acad. Sci. USA 87, 4707-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziegler, A., Jonason, A. S., Leffell, D. J., Simon, J. A., Sharma, H. W., Kimmelman, J., Remington, L., Jacks, T. & Brash, D. E. (1994) Nature 372, 773-776. [DOI] [PubMed] [Google Scholar]
- 10.Jonason, A. S., Kunala, S., Price, G. J., Restifo, R. J., Spinelli, H. M., Persing, J. A., Leffell, D. J., Tarone, R. E. & Brash, D. E. (1996) Proc. Natl. Acad. Sci. USA 93, 14025-14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford, J. M. & Hanawalt, P. C. (1995) Proc. Natl. Acad. Sci. USA 92, 8876-8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ford, J. M. & Hanawalt, P. C. (1997) J. Biol. Chem. 272, 28073-28080. [DOI] [PubMed] [Google Scholar]
- 13.Ford, J. M., Baron, E. L. & Hanawalt, P. C. (1998) Cancer Res. 58, 599-603. [PubMed] [Google Scholar]
- 14.Adimoolam, S., Lin, C. X. & Ford, J. M. (2001) J. Biol. Chem. 30, 25813-25822. [DOI] [PubMed] [Google Scholar]
- 15.Smith, M. L., Ford, J. M., Hollander, M. C., Bortnick, R. A., Amundson, S. A., Seo, Y. R., Deng, C. X., Hanawalt, P. C. & Fornace, A. J., Jr. (2000) Mol. Cell. Biol. 20, 3705-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wani, M. A., Zhu, Q., El-Mahdy, M., Venkatachalam, S. & Wani, A. A. (2000) Cancer Res. 60, 2273-2280. [PubMed] [Google Scholar]
- 17.Hwang, B. J., Ford, J. M., Hanawalt, P. C. & Chu, G. (1999) Proc. Natl. Acad. Sci. USA 96, 424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adimoolam, S. & Ford, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 12985-12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, X. W., Yeh, H., Schaeffer, L., Roy, R., Moncollin, V., Egly, J. M., Wang, Z., Freidberg, E. C., Evans, M. K., Taffe, B. G., et al. (1995) Nat. Genet. 10, 188-195. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka, H., Arakawa, H., Yamaguchi, T., Shiraishi, K., Fukuda, S., Matsui, K., Takei, Y. & Nakamura, Y. (2000) Nature 404, 42-49. [DOI] [PubMed] [Google Scholar]
- 21.McKay, B. C., Francis, M. A. & Rainbow, A. J. (1997) Carcinogenesis 18, 245-249. [DOI] [PubMed] [Google Scholar]
- 22.Therrien, J. P., Drouin, R., Baril, C. & Drobetsky, E. A. (1999) Proc. Natl. Acad. Sci. USA 96, 15038-15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyrrell, R. M. (1996) EXS 77, 255-271. [PubMed] [Google Scholar]
- 24.Drouin, R., Therrien, J. P., Angers, M. & Ouellette, S. (2001) in Methods in Molecular Biology: DNA–Protein Interactions, Principles, and Protocols, ed. Moss, T. (Humana, Totawa, NJ), pp. 175-219.
- 25.Angers, M., Cloutier, J. F., Castonguay, A. & Drouin, R. (2001) Nucleic Acids Res. 29, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drouin, R., Gao, S. & Holmquist, G. P. (1996) in Technologies for Detection of DNA Damage and Mutations, ed. Pfeifer, G. P. (Plenum, New York), pp. 37-43.
- 27.Tu, Y., Tornaletti, S. & Pfeifer, G. P. (1996) EMBO J. 15, 675-683. [PMC free article] [PubMed] [Google Scholar]
- 28.Rozek, D. & Pfeifer, G. P. (1993) Mol. Cell. Biol. 13, 5490-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeifer, G. P., Drouin, R., Riggs, A. D. & Holmquist, G. P. (1991) Proc. Natl. Acad. Sci. USA 88, 1374-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellon, I., Spivak, G. & Hanawalt, P. C. (1987) Cell 51, 241-249. [DOI] [PubMed] [Google Scholar]
- 31.Rochette, P. J., Bastien, N., McKay, B. C., Therrien, J. P., Drobetsky, E. A. & Drouin, R. (2002) Oncogene 21, 5743-5752. [DOI] [PubMed] [Google Scholar]
- 32.Leger, C. & Drobetsky, E. A. (2002) Carcinogenesis 23, 1631-1640. [DOI] [PubMed] [Google Scholar]
- 33.Chuang, Y. Y., Chen, Q. & Liber, H. L. (1999) Cancer Res. 59, 3073-3076. [PubMed] [Google Scholar]
- 34.Yu, Y., Li, C. Y. & Little, J. B. (1997) Oncogene 14, 1661-1667. [DOI] [PubMed] [Google Scholar]
- 35.Devary, Y., Gottlieb, R. A., Lau, L. F. & Karin, M. (1991) Mol. Cell. Biol. 11, 2804-28011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bender, K., Blattner, C., Knebel, A., Iordanov, M., Herrlich, P. & Rahmsdorf, H. J. (1997) J. Photochem. Photobiol. 37, 1-17. [DOI] [PubMed] [Google Scholar]
- 37.Gjerset, R. A., Lebedeva, S., Haghighi, A., Turla, S. T. & Mercola, D. (1999) Cell Growth Differ. 10, 545-554. [PubMed] [Google Scholar]
- 38.Potapova, O., Haghighi, A., Bost, F., Liu, C., Birrer, M. J., Gjerset, R. & Mercola, D. (1997) J. Biol. Chem. 272, 14041-14044. [DOI] [PubMed] [Google Scholar]
- 39.Dhar, V., Adler, V., Lehmann, A. & Ronai, Z. (1996) Cell Growth Differ. 7, 841-846. [PubMed] [Google Scholar]
- 40.Adler, V., Polotskaya, A., Kim, J., Dolan, L., Davis, R., Pincus, M. & Ronai, Z. (1996) Carcinogenesis 17, 2073-2076. [DOI] [PubMed] [Google Scholar]
- 41.Adler, V., Fuchs, S. Y., Kim, J., Kraft, A., King, M. P., Pelling, J. & Ronai, Z. (1995) Cell Growth Differ. 6, 1437-1446. [PubMed] [Google Scholar]
- 42.Medrano, E. E., Im, S., Yang, F. & Abdel-Malek, Z. A. (1995) Cancer Res. 55, 4047-4052. [PubMed] [Google Scholar]
- 43.Dhanwada, K. R., Dickens, M., Neades, R., Davis, R. & Pelling, J. C. (1995) Oncogene 11, 1947-1953. [PubMed] [Google Scholar]
- 44.Tyrrell, R. M. (1996) BioEssays 18, 139-148. [DOI] [PubMed] [Google Scholar]
- 45.Adler, V., Yin, Z., Tew, K. D. & Ronai, Z. (1999) Oncogene 18, 6104-6111. [DOI] [PubMed] [Google Scholar]
- 46.Matsumura, Y. & Ananthaswamy, H. N. (2002) Front. Biosci. 7, d765-783. [DOI] [PubMed] [Google Scholar]
- 47.Ullrich, S. E. (2002) Front. Biosci. 7, d684-703. [DOI] [PubMed] [Google Scholar]