Abstract
In this study, we show that LIM kinase 1 (LIMK1), a critical regulator of actin dynamics, plays a regulatory role in tumor cell invasion. We found that the level and activity of endogenous LIMK1 is increased in invasive breast and prostate cancer cell lines in comparison with less invasive cells. Overexpression of LIMK1 in MCF-7 and in MDA-MB-231 human breast cancer cell lines increased their motility, whereas the specific ROCK and Rho inhibitors Y-27632 and C3, respectively, attenuated this effect. In addition, inhibition of LIMK1 activity in the MDA-MB-231 cells by expression of dominant-negative LIMK1 resulted in decreased motility and formation of osteolytic bone lesions in an animal model of tumor invasion. This study shows an important role for LIMK1 signaling in invasion of cancer, demonstrating its potential as a therapeutic molecular target to decrease metastasis.
Tumor invasion and metastasis is a critical event for cancer patients as it often results in death. Current therapies are of limited value in most patients with disseminated disease, leaving us with the goal of identifying genes that regulate the metastatic process and designing drugs that target their function. During progression of tumor cells to a metastatic phenotype, they undergo a series of changes that begin with loss of contact inhibition and increased motility, allowing them to migrate from the primary tumor site, invade distant organs, and induce neo-vascularization resulting in metastasis (1). Many of these changes are associated with dynamic actin reorganization and activation of signaling pathways through transmembrane receptors, including receptor tyrosine kinases and phosphatidylinositol 3-kinases (2, 3), G-protein-coupled receptors (4), chemokine receptors (5), and transforming growth factor-β receptor (6). In association with cell adhesion molecules at the plasma membrane, the cytoskeleton affects the nature of cell-to-cell and cell-to-substrate interactions via clustered transmembrane integrins that are associated with extracellular matrix proteins (7). These complexes provide the driving force for cell movement and surface remodeling, including neurite extension and axon formation.
Members of the small guanosine triphosphatase (GTPase) family control cell adhesion and motility through reorganization of the actin cytoskeleton and regulation of actomyosin contractility (8). We have previously demonstrated the role of the Rho-actomyosin system in tumor cell invasion (9). Both RhoA (10) and the related RhoC (11) are expressed at a relatively higher level in metastatic tumors, and their expression levels positively correlate with the stage of the tumors (12). However, mutations in the Rho gene have not yet been found in human tumors; rather, the overexpression of RhoA in the cell facilitates its translocation from the cytosol to the plasma membrane, where its activation results in stimulation of the actomyosin system, followed by cellular invasion both in vitro and in vivo (13).
One of the target molecules of Rho is the family of Rho-associated serine-threonine protein kinases (ROCK) (14), which also participates in cell-to-substrate adhesions, stress fiber formation, and stimulation of actomyosin-based cellular contractility (15). We have demonstrated that ROCK, like Rho, is involved in tumor invasion, and that a specific ROCK inhibitor, Y-27632 (16), markedly attenuates the invasion and dissemination of active RhoA-expressing rat hepatoma cells after implantation into the peritoneal cavity of syngeneic rats (17). Recent studies showed that ROCK can phosphorylate and activate LIM kinase 1 and 2 (LIMK1 and LIMK2) (18, 19) as well as myosin light chain 20 (MLC-20; ref. 20). We and others have shown that LIMK1 regulates actin dynamics by inhibiting the activity of the actin depolymerizing protein cofilin (21, 22). When cofilin is phosphorylated by LIMK, it no longer binds to and depolymerizes actin, resulting in net actin polymerization. Because LIMK1 activity is regulated by Rho and ROCK, which have a role in regulating tumor invasion, we postulate that LIMK1 is also involved in mediating this phenotype.
Here, we demonstrate that the expression level of LIMK1 and its activity are increased in highly invasive breast and prostate cancer cell lines in comparison with less invasive cells. Furthermore, the overexpression of LIMK1 in MCF-7 and in MDA-MB-231 cells results in their conversion to more motile cells, whereas the overexpression of dominant-negative LIMK1 in MDA-MB-231 cells decreases their ability to form osteolytic lesions and thus invade bone.
Materials and Methods
Animals. Female BALB/c-nu/nu mice (SLC, Shizuoka, Japan), 5 wk old, were housed under specific pathogen-free conditions. All animal experiments were approved by the Animal Ethics Committee of the Osaka Medical Center for Cancer and Cardiovascular Diseases.
Cell Culture. MDA-MB-231 human breast cancer cells were cultured as described (23). MCF-7 human breast cancer cells (JCRB0134; ref. 24) were obtained from Health Science Research Resources Bank (Osaka) and cultured in MEM (Invitrogen) with nonessential amino acids (Invitrogen), 0.1 mM sodium pyruvate (Invitrogen), 10 μg/ml insulin (Invitrogen), and 10% FCS (Equitech-Bio, Kerrville, TX). Mouse olfactory epithelial cells (25), NIH 3T3 cells, and Ras-NIH 3T3 cells were maintained in DMEM and 10% FCS. Human prostate PC-3 and LNCaP cancer cell lines were a gift of E. Williams (St. Vincent Medical Research Institute, Melbourne) and were maintained in RPMI medium 1640 with 10% FCS and 10 μg/ml insulin. Human fibrosarcoma cell line HT1080 (JCRB9113) was obtained from Health Science Research Resources Bank and was used for the production of chemoattractant (26).
Transfections. Myc-tagged DNA constructs encoding mouse LIMK1 and LIMK1(-) [LIMK1-short, which lacks 20 aa in the catalytic domain; dominant-negative LIMK1 (DN-LIMK1)] are described elsewhere (21, 25). cDNAs encoding activated human RhoA (V14RhoA; ref. 9) and the catalytic domain of ROCK (Δ4ROCK; ref. 17) were subcloned into FLAG-tagged pcDNA3 vector (Invitrogen). Stable cell lines were generated by transfection of constructs or empty vector by using Lipofectamine Plus reagent (Invitrogen), and selected in the presence of 800 μg/ml G418 for 4 wk.
In Vitro Invasion Assay. The chemoinvasion assay was carried out as described (27), except that MDA-MB-231 and MCF-7 cells were incubated for 6 and 20 h, respectively.
Purification of C3 Exoenzyme. Bacterially expressed GST-C3 exoenzyme was purified as described (28).
Immunoblotting. For the analysis of endogenous LIMK1 expression in different cell lines, the immunoblot analysis was performed essentially as described (21). One hundred micrograms of protein lysates from each cell line were separated by 7% SDS/PAGE, electroblotted to Hybond C extra membrane (Amersham Pharmacia), and probed with a rat monoclonal anti-LIMK1 antibody that specifically recognizes LIMK1, but not the related LIMK2 protein (V.F. and O.B., unpublished results), at a 1:3,000 dilution. For alkaline phosphatase treatment, 50 μg of protein lysates were incubated with 10 units of calf intestinal alkaline phosphatase (Roche Diagnostics) for 1 h at 37°C before electrophoresis. To detect HSP70 protein, used as loading control, a mouse monoclonal anti-HSP70 antibody was used at 1:1,000. The secondary antibodies were horseradish peroxidase-conjugated anti-rat or anti-mouse IgG used at a 1:20,000 dilution (Chemicon). The final signal was revealed by ECL chemiluminescence (Pierce).
For detection of LIMK1 in the cell transfectants, cells (5 × 104) were lysed in Laemmli sample buffer (29), separated by 7% SDS/PAGE, and transferred to a nitrocellulose membrane (Bio-Rad) with semi-dry method. The blot was probed with rat monoclonal anti-LIMK1 antibody (1:3,000), followed by incubation with anti-rat IgG alkaline phosphatase conjugate (Promega, 1:2,500). The signal was developed by using nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl-phosphate (Promega) as substrate.
Estimation of Phospho-Cofilin and Phospho-MLC-20 Levels. Cells (3 × 104 per sample) were starved in medium containing 0.1% BSA for 24 h before harvesting and separated on 12.5% SDS/PAGE. For estimation of phospho-cofilin levels, the blots were probed with rabbit anti-phospho-cofilin (1:1,000), and rabbit anti-cofilin (1:200) polyclonal antibodies, followed by goat anti-rabbit IgG alkaline phosphatase conjugate (Promega, 1:7,500). For estimation of phospho-MLC-20 (PMLC20) levels, the blot was probed with anti-PMLC20 polyclonal antibodies (1:100), followed by goat anti-rabbit IgG alkaline phosphatase conjugate (Promega, 1:7,500). The relative phosphorylation level of MLC-20 in each cell lysate was estimated from the signal of PMLC20 normalized with the signal of MLC-20, which was obtained by immunoblotting using MY21 monoclonal Abs on the duplicate blot. Protein levels were measured by using a flat scanner (GT-9500, Epson) and analyzed with NIH IMAGE software (Version 1.62). Data are representative of three separate experiments. Statistical analysis was performed with STATVIEW software package (Hulinks, Tokyo).
Immunofluorescence. Cells were grown on fibronectin-treated glass coverslips, fixed in 1% paraformaldehyde/PBS for 20 min, and blocked in 2% normal goat serum, 5% FCS, and 0.1% Tween 20 in PBS for 1 h at room temperature. Primary antibodies, including rat monoclonal anti-LIMK1 (1:500) and mouse monoclonal anti-vinculin (Sigma, 1:100), were diluted in fresh blocking solution and added to the cells for 1 h at room temperature or overnight at 4°C. After three washes in PBS, secondary antibodies including Alexa 488 anti-mouse or anti-rat IgG and Alexa 594 anti-mouse IgG (Molecular Probes) were diluted in the blocking solution at 1:700 and incubated with the cells for 1 h at room temperature. To stain for F-actin, Alexa 594 phalloidin (Molecular Probes) at 1:100 dilution was included in the secondary antibody dilution mix. Cells were washed three times in PBS and mounted in an anti-fade mounting medium (DAKO), and images were taken by using a confocal microscope.
Bone Invasion of MDA-MB-231 Transfectants in Nude Mice. Intracardiac injections of LIMK1-transfected MDA-MB-231 cells were performed as described (23, 30). Cells from clone 3 of the wild-type LIMK1 (wtLIMK1) or DN-LIMK1 transfectants or from untransfected cells (2 × 105 in 0.1 ml of PBS) were injected into the left ventricle of 5-wk-old anesthetized (with pentobarbital 0.25 mg/kg) female BALB/c-nu/nu mice. Three weeks later, anesthetized mice were exposed to an x-ray at 45 kV, 3 mA for 1.5 min by using a soft x-ray apparatus (Softex Type, Osaka). The radiographs were assessed independently by two orthopedic surgeons. The number and area of osteolytic lesions, recognized as demarcated radiolucent lesions in the bone, were quantitatively assessed by using a MAC SCOPE Version 2.51 image analysis system (Mitani, Fukui, Japan).
Statistical Analysis. Data are expressed as mean ± SD unless otherwise noted. Significance (P ≤ 0.05) was determined by using a two-sided Student t analysis.
Results
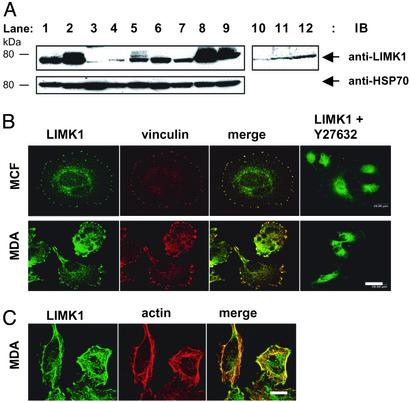
LIMK1 Expression and Activity Are Increased in Cancer Cell Lines. LIMK1 is activated by phosphorylation of threonine 508 in the activation loop of the kinase domain by ROCK (18), suggesting that the activation of ROCK results in increased LIMK1 activity. In addition, like many other protein kinases, LIMK1 is further activated by transphosphorylation on serine residues (21). Accordingly, we examined the levels and the phosphorylation state of endogenous LIMK1 protein in cell lines with varying potential to form tumors and metastasize in animal models. Endogenous LIMK1 protein was observed primarily as an ≈70-kDa protein in all of the cell lines tested (Fig. 1A). Low to moderate levels of LIMK1 protein were observed in mouse olfactory epithelial cells, human 293T cells, and mouse NIH 3T3 cells (lanes 1, 3, and 4), and in the low invasive human breast cancer MCF-7 (lane 6) and prostate cancer LNCaP (lane 7) cell lines. However, both elevated levels and apparent slower moving forms of LIMK1 were detected in the SV40-transformed monkey COS-7 cells (lane 2) and in the invasive prostate and breast cancer cell lines PC-3 (lane 8) and MDA-MB-231 (lane 9), respectively. In addition, Ras-transformed NIH 3T3 cells (lane 5), known to be highly invasive, express higher level of LIMK1 than normal 3T3 cells (lane 4). The apparent higher molecular weight forms of LIMK1 in these cell lines correspond to phosphorylated forms of LIMK1 as they essentially disappeared after treatment with alkaline phosphatase (lanes 10–12).
Fig. 1.
Immunoblot and immunofluorescence analyses of endogenous LIMK1 expression in cell lines. (A Upper) Endogenous LIMK1 expression in various cell lines (lanes 1–9), and after alkaline phosphatase treatment of cell lysates from transformed and invasive cancer cell lines (lanes 10–12). (Lower) HSP70 expression to indicate loading of proteins. Lane 1, mouse olfactory epithelial cells; lane 2, monkey COS-7 cells; lane 3, 293T cells; lane 4, mouse NIH 3T3 fibroblasts; lanes 5 and 10, Ras-transformed NIH 3T3 fibroblasts; lane 6, breast cancer MCF-7 cells; lane 7, prostate cancer LNCaP cells; lanes 8 and 12, invasive prostate cancer PC-3 cells; lanes 9 and 11, invasive breast cancer MDA-MB-231 cells. (B) LIMK1 and vinculin expression in MCF-7 (Upper) and MDA-MB-231 cells (Lower), and LIMK1 localization in both cell-types after a 30-min treatment with 10 μM Y-27632 (Right). (C) LIMK1 and F-actin expression in nontreated MCF-7 and MDA-MB-231 cells. (Bars = 20 μm.)
Immunofluorescence analysis of endogenous LIMK1 expression in breast cancer cell lines revealed predominant colocalization, with focal adhesion complexes as shown by colabeling with an anti-vinculin antibody in MCF-7 cells (Fig. 1B Upper) and MDA-MB-231 cells (Fig. 1B Lower). After the treatment with 10 μM of the ROCK inhibitor Y-27632 for 30 min, LIMK1 localization at the focal adhesion complexes changed and was found dispersed throughout the cytoplasm in both cell types (Fig. 1B), identifying a role for the Rho-ROCK pathway in the formation of focal adhesion complexes, as suggested (31). Colocalization of LIMK1 with F-actin is also evident in MDA-MB-231 (Fig. 1C). Overall, the level of LIMK1 staining seemed higher in MDA-MB-231 cells compared with MCF-7 cells where the majority of the protein is evident at the cells' periphery in addition to prominent cytoplasmic localization.
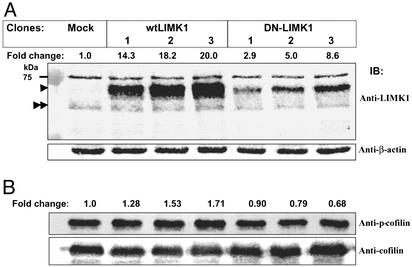
LIMK1 Expression and Activity Are Altered in MDA-MB-231 Transfectants. After the expression data, we next analyzed whether changes in LIMK1 expression or activity in MDA-MB-231 cells affected their motility. We generated three MDA-MB-231 cell lines stably expressing myc-tagged wtLIMK1 or DN-LIMK1 proteins. The level of endogenous and overexpressed LIMK1 proteins was determined by immunoblotting. Whereas all of the clones expressed a similar level of endogenous LIMK1 protein, the expression level of overexpressed wtLIMK1 and DN-LIMK1 proteins differed among the different clones. The three cell lines overexpressing wtLIMK1 showed ≈14- to 20-fold more wtLIMK1 whereas the three DN-LIMK1 transfectants revealed ≈3- to 9-fold more DN-LIMK1 when compared with endogenous LIMK1 levels in the mock transfectants (Fig. 2A). Clone 3 from both wtLIMK1 and DN-LIMK1 transfectants showed the highest levels of transgene overexpression.
Fig. 2.
LIMK1 expression and activity in MDA-MB-231 transfectants. (A) Immunoblots of cell lysates prepared from mock, wtLIMK1, and DN-LIMK1 MDA-MB-231 transfectants probed with rat monoclonal anti-LIMK1 (Upper) or β-actin antibodies, as loading control (Lower). The fold-change in the level of LIMK1 protein compared with the mock transfectants is indicated above Upper. The positions of endogenous and overexpressed tagged-LIMK1 proteins are indicated by double and single arrowheads, respectively. (B) Immunoblots of cell lysates prepared from mock, wtLIMK1, and DN-LIMK1 MDA-MB-231 transfectants probed with anti-phospho-cofilin (Upper) and pancofilin (Lower) antibodies. The fold-change in phosphorylated cofilin level is indicated above Upper. The data are representative of three independent experiments.
To investigate whether changes in the level of LIMK1 protein also resulted in changes in its activity, we estimated the level of phosphorylation of the endogenous substrate, cofilin, by immunoblotting and probing with anti-phospho-cofilin antibodies (Fig. 2B). The stable transfectants expressing wtLIMK1 showed 1.3- to 1.7-fold increase in the level of phospho-cofilin compared with that of mock transfectants (clone 1, 1.28 ± 0.13-fold; clone 2, 1.53 ± 0.13-fold, P ≤ 0.05; and clone 3, 1.71 ± 0.03-fold, P ≤ 0.05). Conversely, DN-LIMK1 transfectants showed a small but significant decrease (0.9- to 0.7-fold change) in the level of phospho-cofilin compared with that of mock transfectants (clone 1, 0.89 ± 0.08-fold; clone 2, 0.79 ± 0.09-fold; and clone 3, 0.68 ± 0.08-fold, P ≤ 0.05; Fig. 2B). The activity of LIMK1 as judged by the level of phospho-cofilin positively correlated with the expression level of overexpressed wtLIMK1, and negatively correlated with the expression level of DN-LIMK1.
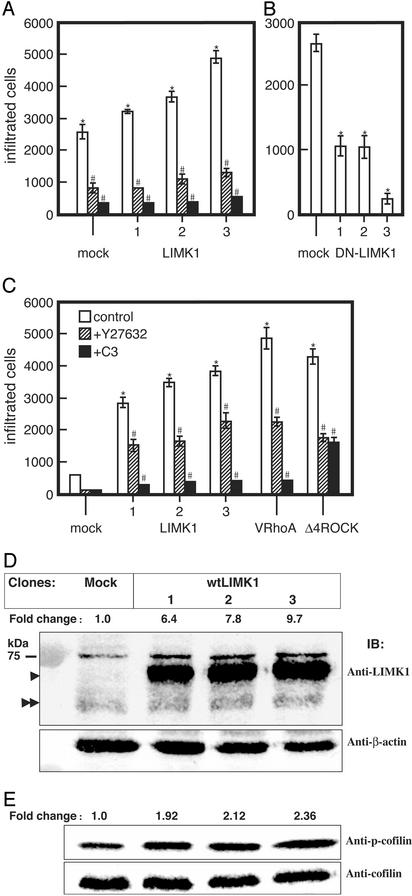
LIMK1 Is Important for Invasiveness. The invasiveness of each cell line was determined in the in vitro Matrigel invasion assay. MDA-MB-231 cells overexpressing wtLIMK1 showed increased motility (1.2-, 1.3-, and 1.8-fold, respectively) than mock transfectants (Fig. 3A). Pretreatment of the cells with 10 μg/ml C3 for 24 h or with 10 μM Y-27632 for 6 h resulted in marked inhibition of their migration (86–90% and 70–75%, respectively; Fig. 3A). In contrast, the migration of MDA-MB-231 cells expressing DN-LIMK1 was reduced compared with the mock transfected cells (Fig. 3B). Clones expressing higher levels of DN-LIMK1 (Fig. 3B) exhibited lower invasiveness, indicating that endogenous LIMK1 activity is required for the invasiveness of MDA-MB-231 cells.
Fig. 3.
The invasiveness of LIMK1 MDA-MB-231 and MCF-7 transfectants. Cells from each cell line (5 × 105 cells) were added onto the upper chamber of Matrigel-coated PET membrane. (A) For MDA-MB-231 mock transfectants or overexpressing wtLIMK1 (LIMK1), cells were left untreated or were treated with Y-27632 (10 μM) and added directly to the assay system, or pretreated for 24 h with 10 μg/ml C3. The number of migrating cells was determined after 6h.(B) Migration of MDA-MB-231 DN-LIMK1 or mock transfectants. (C) MCF-7 transfectants expressing wtLIMK1 (LIMK1) or V14RhoA and Δ4ROCK were analyzed after 20 h. The numbers at the bottom of each panel indicate the clone's number. (D) Immunoblots of cell lysates prepared from mock and wtLIMK1 MCF-7 transfectants probed with rat monoclonal anti-LIMK1 (Upper) and β-actin (Lower) antibodies. The fold-change in the level of LIMK1 protein compared with the mock transfectants is indicated above Upper. The positions of endogenous and overexpressed tagged-LIMK1 proteins are indicated by double and single arrowheads, respectively. (E) Immunoblots of cell lysates prepared from mock and wtLIMK1 MCF-7 transfectants probed with anti-phospho-cofilin (Upper) and pan-cofilin (Lower) antibodies. The fold-change in phosphorylated cofilin compared with the mock transfectants is indicated above Lower. All data are expressed as the number of migrated cells and are the average of three independent experiments. Error bars indicate SD. *, P < 0.01 to mock transfectants; #, P < 0.01 to Y (-) each transfectant.
To further ascertain that the higher levels of LIMK1 are responsible for the increased invasiveness of MDA-MB-231 cells, we generated clones of MCF-7 cells permanently expressing different levels of wtLIMK1. In general, the invasiveness of the MDA-MB-231 cells is ≈16-fold higher than that of MCF-7 cells (compare mock in Fig. 3A with mock in Fig. 3C). However, the MCF-7 clones expressing higher levels of LIMK1 (clone 3 > 2 > 1) had a greater number of cells traversing the Matrigel than those expressing lower levels of LIMK1, and their invasiveness was comparable to those expressing activated Rho (V14RhoA) or ROCK (Δ4ROCK) (Fig. 3C). Pretreatment of the wtLIMK1 transfectants with 10 μg/ml C3 for 24 h or with 10 μM Y-27632 for 20 h resulted in inhibition of their migration. The MCF-7 clones were found to express 6- to 10-fold more wtLIMK1 compared with endogenous LIMK1 (Fig. 3D), and correspondingly displayed increased levels of phospho-cofilin (Fig. 3E; clone 1, 1.92 ± 0.19-fold; clone 2, 2.11 ± 0.17-fold; and clone 3, 2.36 ± 0.07-fold; P ≤ 0.05).
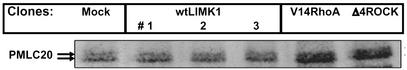
To verify that the overexpression of LIMK1 had no effect on other known signaling pathways of ROCK, the level of phosphorylated MLC-20 was measured in MCF-7 transfectants by using anti-phospho-MLC-20 polyclonal antibodies. An immunoblot analysis revealed that the level of phosphorylated MLC-20 in wtLIMK1 MCF-7 transfectants was similar to that in mock transfectants, whereas V14RhoA and Δ4ROCK transfectants showed marked increase in phosphorylated MLC-20 (Fig. 4). The overexpression of wtLIMK1 or DN-LIMK1 in MDA-MB-231 also had no effect on the level of phosphorylated MLC-20 (data not shown).
Fig. 4.
Phosphorylated MLC-20 levels in MCF-7 transfectants. Immunoblots of cell lysates prepared from mock, wtLIMK1, V14RhoA, and Δ4ROCK MCF-7 transfectants probed with anti-phospho-MLC-20 antibodies. The position of phosphorylated MLC-20 (PMLC20) is indicated by arrows.
Immunofluorescence analysis of LIMK1 MDA-MB-231 transfectants stained for F-actin and with anti-vinculin antibody demonstrated increased stress fibers and enlarged focal adhesion complexes compared with mock transfectants (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org.) where treatment with the ROCK and Rho inhibitors Y-27632 and C3, respectively, alleviated the appearance of the wtLIMK1 MDA-MB-231 and wtLIMK1 MCF-7 transfectants. In contrast, expression of DN-LIMK1 in MDA-MB-231 cells resulted in flattening and rounding of the cells and loss of F-actin. Analysis of the wtLIMK1 MCF-7 transfectants demonstrated similar morphological changes to those induced by overexpression of activated Rho and ROCK, such as increased size and number of focal adhesions and increased F-actin staining.
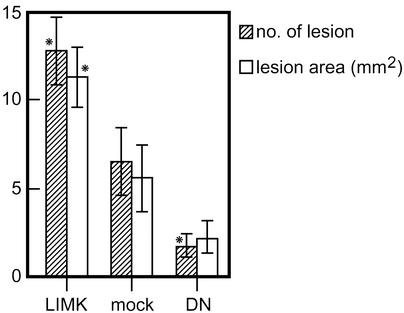
Formation of Osteolytic Lesions by LIMK1 Mutants of MDA-MB-231 Cells. To demonstrate that the Matrigel chamber assay data reflect the ability of the cells to invade in vivo, we tested their capacity to form osteolytic lesions by injecting them into the left heart ventricle of female nude mice (23). Nude mice injected with wtLIMK1-overexpressing MDA-MB-231 cells (clone 3) showed cachexia (body weight, 18.7 ± 3.4 g) compared with mice injected with mock transfectants (body weight, 21.5 ± 2.0 g). Osteolytic lesions were assessed by x-rays, and, 3 wk after injection, they appeared primarily in the lower extremities close to the knee joint. Mice injected with wtLIMK1-expressing MDA-MB-231 cells showed a significantly increased number (12.8 ± 1.9) and area (11.3 ± 1.7 mm2) of the osteolytic lesions compared with those injected with mock transfectants (6.5 ± 1.9 and 5.5 ± 1.9 mm2, respectively; Fig. 5; P < 0.01). In contrast, mice injected with clone 3 of DN-LIMK1 MDA-MB-231 cells resulted in a significantly reduced number (1.6 ± 0.7) and area (2.1 ± 1.0 mm2; P < 0.01) of the lesions (Fig. 5), and showed no change in body weight.
Fig. 5.
Osteolytic lesions caused by LIMK1-MDA-MB-231 cells in nude mice. The number and area of osteolytic lesions in nude mice bearing wtLIMK1-MDA-MB-231 (clone 3; 11 mice), DN-LIMK1 (clone 3; 11 mice), and mock (14 mice) transfected cells. The number and area of osteolytic lesions were scored 3 wk after cell inoculation on the radiographs by using quantitative image analysis.
Discussion
Our previous studies of ROCK in tumor cell dissemination (17) did not delineate whether both ROCK's effectors, MLC-20 and LIMK1, were involved. Cell body contraction depends on actomyosin, which is regulated by ROCK as it affects MLC phosphorylation (20, 32). However, overexpression of wtLIMK1 or DN-LIMK1 in MDA-MB-231 and of wtLIMK1 in MCF-7 cells had no effect on MLC phosphorylation but affected the invasiveness of these cells, indicating that the ROCK-LIMK pathway is independent of the ROCK-MLC-20 pathway in these cells' motility.
In agreement with our study, Nishita et al. (33) have shown that SDF-1α-induced actin-dependent T cell migration is mediated by increased LIMK1 activity, whereas p21-activated kinase 4 (PAK4), a known LIMK1 activator (34), interacts with αvβ5 and selectively promotes integrin αvβ5-mediated cell migration (35). However, our data also contrast with past studies where cell motility was inhibited by LIMK1 overexpression in Ras-transformed Swiss 3T3 fibroblasts (36), and transient expression of the kinase domain of LIMK1 completely inhibited epidermal growth factor-induced lamellipodial extension in metastatic rat mammary adenocarcinoma (37). The reason for these differences is not clear but could be due to different cell types used or to the level and activity of overexpressed LIMK1. We generated stable cell lines overexpressing varying levels of wtLIMK1 and DN-LIMK1 proteins; however, we have consistently observed that transient transfections of high levels of wtLIMK1 or of the kinase domain alone results in massive actin-clumping followed by cell death 24–48 h posttransfection in multiple cell types (ref. 21 and unpublished observations). Thus, we propose that the level of LIMK1 is critical for cell survival. Too much or too little LIMK1 may differentially impact the actin-depolymerizing activity of cofilin in different cell types.
One of the important findings of this study is that the motility of tumor cells correlates with the level of LIMK1 expression and activity. We correlated the capacity of these cells to invade by counting the number of osteolytic lesions after intracardiac injection. During the formation of osteolytic lesions, a number of cytokines are secreted and stimulate osteoclast activity, which leads to local osteolysis (38). This process requires the invasion of tumor cells into the bone marrow cavity; therefore, the number and the area of the lesions correlate with the invasive ability of the cancer cells. The development of cachexia only in mice injected with wtLIMK1 MDA-MB-231 cells is likely due to the increased number of osteolytic lesions. The overall well-being of the mice may be affected due to decreased food intake, which is often associated with cancer patients and/or increased levels of tumor cell-derived growth factors (reviewed by Tisdale in ref. 39). For example, parathyroid hormone (PTH)-related peptide (PTHrP) has been associated with the development of osteolytic lesions from breast cancer metastatic cells and cachexia in the presence of hypercalcemia (40, 41). The increased infiltration of MDA-MB-231 cells containing increased levels of wtLIMK1 may have led to increased PTHrP production in the bone (41).
We have also observed that LIMK1 is highly expressed in multiple human tumors, including melanoma, ovarian carcinoma, lung, breast, and prostate, all of which are highly invasive (data not shown). Potentially, the level and activity of endogenous LIMK1 could correlate with the invasive and therefore metastatic potential of the tumor. Thus, measurement of LIMK1 levels could be important in the management of prostate cancer where routine treatment is prostatectomy regardless of the cancer's condition. The availability of a reliable marker for cancer's ability to invade would be helpful in avoiding surgical intervention. Another important observation is that the inhibition of LIMK1 activity results in a dramatic reduction in cell motility. This finding indicates the potential to treat invasive cancers with compounds that inhibit LIMK1 activity specifically, as opposed to those such as the ROCK inhibitor, Y-27632, as ROCK is involved in several signal transduction pathways including those affecting blood pressure control (16), penile erection (42), apoptosis (43), and malignant transformation (44). Furthermore, the structure of the LIMK1 kinase domain is unique and its activation loop is longer than that of many kinases, increasing the possibility of designing a drug that will inhibit specifically the activity of LIMK1 in cancer invasion and metastasis. To date, the only known target of LIMK1 is cofilin, whose only known function is the regulation of actin dynamics. In addition, it was recently shown that LIMK1 is not essential for cell survival because mice lacking LIMK1 gene exhibit only minor abnormalities in spine morphology and in synaptic function (45), making this molecule an attractive target for drug design.
Supplementary Material
Acknowledgments
We thank T. Ueda and A. Myoi for quantitation of the osteolytic lesions, J. Toshima for anti-phospho-cofilin antibody, H. Aizawa for anti-cofilin antibody, F. Matsumura for anti-phospho MLC-20 antibody, D. Huang for anti-HSP70 antibody, T. Yoneda for MDA-MB-231 human breast cancer cells, and L. Feig for pGEX2T-C3 expression vector. Y-27632 was a generous gift from Mitsubishi Pharma. We also thank R. Anderson and A. Kraft for critical reading of the manuscript. O.B. was supported by the Australian National Health and Medical Research Council and the Anti-Cancer Council of Victoria. K.I. was supported by Grants-in-Aid for Cancer Research from the Ministry of Health and Welfare of Japan, by the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by the Princess Takamatsu Cancer Research Fund.
Abbreviations: LIMK, LIM kinase; MLC, myosin light chain; DN-LIMK1, dominant-negative LIMK1; wtLIMK1, wild-type LIMK1.
References
- 1.Chambers, A. F., Groom, A. C. & MacDonald, I. C. (2002) Nat. Rev. Cancer 2, 563-572. [DOI] [PubMed] [Google Scholar]
- 2.Vivanco, I. & Sawyers, C. L. (2002) Nat. Rev. Cancer 2, 489-501. [DOI] [PubMed] [Google Scholar]
- 3.Roymans, D. & Slegers, H. (2001) Eur. J. Biochem. 268, 487-498. [DOI] [PubMed] [Google Scholar]
- 4.Seasholtz, T. M., Majumdar, M. & Brown, J. H. (1999) Mol. Pharmacol. 55, 949-956. [DOI] [PubMed] [Google Scholar]
- 5.Muller, A., Homey, B., Soto, H., Ge, N., Catron, D., Buchanan, M. E., McClanahan, T., Murphy, E., Yuan, W., Wagner, S. N., et al. (2001) Nature 410, 50-56. [DOI] [PubMed] [Google Scholar]
- 6.Oft, M., Heider, K. H. & Beug, H. (1998) Curr. Biol. 8, 1243-1252. [DOI] [PubMed] [Google Scholar]
- 7.Schoenwaelder, S. M. & Burridge, K. (1999) Curr. Opin. Cell Biol. 11, 274-286. [DOI] [PubMed] [Google Scholar]
- 8.Etienne-Manneville, S. & Hall, A. (2002) Nature 420, 629-635. [DOI] [PubMed] [Google Scholar]
- 9.Yoshioka, K., Matsumura, F., Akedo, H. & Itoh, K. (1998) J. Biol. Chem. 273, 5146-5154. [DOI] [PubMed] [Google Scholar]
- 10.Fritz, G., Just, I. & Kaina, B. (1999) Int. J. Cancer 81, 682-687. [DOI] [PubMed] [Google Scholar]
- 11.Clark, E. A., Golub, T. R., Lander, E. S. & Hynes, R. O. (2000) Nature 406, 532-535. [DOI] [PubMed] [Google Scholar]
- 12.Nakamori, S., Tamura, S., Arai, I., Kameyama, M., Furukawa, F., Ishikawa, O., Imaoka, S., Yoshioka, K., Mukai, M., Shinkai, K. & Akedo, H. (1996) Recent Adv. Gastroenterol. Carcinogen. 1, 901-904. [Google Scholar]
- 13.Yoshioka, K., Nakamori, S. & Itoh, K. (1999) Cancer Res. 59, 2004-2010. [PubMed] [Google Scholar]
- 14.Leung, T., Manser, E., Tan, L. & Lim, L. (1995) J. Biol. Chem. 270, 29051-29054. [DOI] [PubMed] [Google Scholar]
- 15.Narumiya, S., Ishizaki, T. & Watanabe, N. (1997) FEBS Lett. 410, 68-72. [DOI] [PubMed] [Google Scholar]
- 16.Uehata, M., Ishizaki, T., Satoh, H., Ono, T., Kawahara, T., Morishita, T., Tamakawa, H., Yamagami, K., Inui, J., Maekawa, M. & Narumiya, S. (1997) Nature 389, 990-994. [DOI] [PubMed] [Google Scholar]
- 17.Itoh, K., Yoshioka, K., Akedo, H., Uehata, M., Ishizaki, T. & Narumiya, S. (1999) Nat. Med. 5, 221-225. [DOI] [PubMed] [Google Scholar]
- 18.Ohashi, K., Nagata, K., Maekawa, M., Ishizaki, T., Narumiya, S. & Mizuno, K. (2000) J. Biol. Chem. 275, 3577-3582. [DOI] [PubMed] [Google Scholar]
- 19.Amano, T., Tanabe, K., Eto, T., Narumiya, S. & Mizuno, K. (2001) Biochem. J. 354, 149-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amano, M., Ito, M., Kimura, K., Fukata, Y., Chihara, K., Nakano, T., Matsuura, Y. & Kaibuchi, K. (1996) J. Biol. Chem. 271, 20246-20249. [DOI] [PubMed] [Google Scholar]
- 21.Arber, S., Barbayannis, F. A., Hanser, H., Schneider, C., Stanyon, C. A., Bernard, O. & Caroni, P. (1998) Nature 393, 805-809. [DOI] [PubMed] [Google Scholar]
- 22.Yang, N., Higuchi, O., Ohashi, K., Nagata, K., Wada, A., Kangawa, K., Nishida, E. & Mizuno, K. (1998) Nature 393, 809-812. [DOI] [PubMed] [Google Scholar]
- 23.Hiraga, T., Williams, P. J., Mundy, G. R. & Yoneda, T. (2001) Cancer Res. 61, 4418-4424. [PubMed] [Google Scholar]
- 24.Soule, H. D., Vazquez, J., Long, A., Albert, S. & Brennan, M. (1973) J. Natl. Cancer Inst. 51, 1409-1416. [DOI] [PubMed] [Google Scholar]
- 25.Bernard, O. (1993) Neuroprotocols 3, 200-213. [Google Scholar]
- 26.Watanabe, H., Carmi, P., Hogan, V., Raz, T., Silletti, S., Nabi, I. R. & Raz, A. (1991) J. Biol. Chem. 266, 13442-13448. [PubMed] [Google Scholar]
- 27.Albini, A., Iwamoto, Y., Kleinman, H. K., Martin, G. R., Aaronson, S. A., Kozlowski, J. M. & McEwan, R. N. (1987) Cancer Res. 47, 3239-3245. [PubMed] [Google Scholar]
- 28.Dillon, S. T. & Feig, L. A. (1995) Methods Enzymol. 256, 174-184. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. (1970) Nature 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 30.Yoneda, T., Sasaki, A., Dunstan, C., Williams, P. J., Bauss, F., De Clerck, Y. A. & Mundy, G. R. (1997) J. Clin. Invest. 99, 2509-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amano, M., Chihara, K., Kimura, K., Fukata, Y., Nakamura, N., Matsuura, Y. & Kaibuchi, K. (1997) Science 275, 1308-1311. [DOI] [PubMed] [Google Scholar]
- 32.Kimura, K., Ito, M., Amano, M., Chihara, K., Fukata, Y., Nakafuku, M., Yamamori, B., Feng, J., Nakano, T., Okawa, K., et al. (1996) Science 273, 245-248. [DOI] [PubMed] [Google Scholar]
- 33.Nishita, M., Aizawa, H. & Mizuno, K. (2002) Mol. Cell. Biol. 22, 774-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dan, C., Kelly, A., Bernard, O. & Minden, A. (2001) J. Biol. Chem. 276, 32115-32121. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, H., Li, Z., Viklund, E. K. & Stromblad, S. (2002) J. Cell Biol. 158, 1287-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahai, E., Olson, M. F. & Marshall, C. J. (2001) EMBO J. 20, 755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zebda, N., Bernard, O., Bailly, M., Welti, S., Lawrence, D. S. & Condeelis, J. S. (2000) J. Cell Biol. 151, 1119-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bendre, M. S., Gaddy-Kurten, D., Mon-Foote, T., Akel, N. S., Skinner, R. A., Nicholas, R. W. & Suva, L. J. (2002) Cancer Res. 62, 5571-5579. [PubMed] [Google Scholar]
- 39.Tisdale, M. J. (2002) Nat. Rev. Cancer 2, 862-871. [DOI] [PubMed] [Google Scholar]
- 40.Iguchi, H., Onuma, E., Sato, K. & Ogata, E. (2001) Int. J. Cancer 94, 24-27. [DOI] [PubMed] [Google Scholar]
- 41.Guise, T. A., Yin, J. J., Thomas, R. J., Dallas, M., Cui, Y. & Gillespie, M. T. (2002) Bone 30, 670-676. [DOI] [PubMed] [Google Scholar]
- 42.Chitaley, K., Wingard, C. J., Webb, R. C., Branam, H., Stopper, V. S., Lewis, R. W. & Mills, T. M. (2001) Nat. Med. 7, 119-122. [DOI] [PubMed] [Google Scholar]
- 43.Coleman, M. L., Sahai, E. A., Yeo, M., Bosch, M., Dewar, A. & Olson, M. F. (2001) Nat. Cell Biol. 3, 339-345. [DOI] [PubMed] [Google Scholar]
- 44.Sahai, E., Ishizaki, T., Narumiya, S. & Treisman, R. (1999) Curr. Biol. 9, 136-145. [DOI] [PubMed] [Google Scholar]
- 45.Meng, Y., Zhang, Y., Tregoubov, V., Janus, C., Cruz, L., Jackson, M., Lu, W. Y., MacDonald, J. F., Wang, J. Y., Falls, D. L. & Jia, Z. (2002) Neuron 35, 121-133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.