Abstract
Follicular lymphoma (FL) is the most common form of low-grade non-Hodgkin's lymphoma. Transformation to diffuse large B cell lymphoma (DLBCL) is an important cause of mortality. Using cDNA microarray analysis we identified 113 transformation-associated genes whose expression differed consistently between serial clonally related samples of FL and DLBCL occurring within the same individual. Quantitative RT-PCR validated the microarray results and assigned blinded independent group of 20 FLs, 20 DLBCLs, and five transformed lymphoma-derived cell lines with 100%, 70%, and 100% accuracy, respectively. Notably, growth factor cytokine receptors and p38β-mitogen-activated protein kinase (MAPK) were differentially expressed in the DLBCLs. Immunohistochemistry of another blinded set of samples demonstrated expression of phosphorylated p38MAPK in 6/6 DLBCLs and 1/5 FLs, but not in benign germinal centers. SB203580 an inhibitor of p38MAPK specifically induced caspase-3-mediated apoptosis in t(14;18)+/p38MAPK+-transformed FL-derived cell lines. Lymphoma growth was also inhibited in SB203580-treated NOD-SCID mice. Our results implicate p38MAPK dysregulation in FL transformation and suggest that molecular targeting of specific elements within this pathway should be explored for transformed FL therapy.
Follicular lymphoma (FL) is the most common form of low-grade non-Hodgkin's lymphoma in the Western hemisphere (1). The disease course is typically indolent, but 25–60% of cases may transform to an aggressive diffuse large B cell lymphoma (DLBCL) (2). FL is characterized by t(14;18)(q32;q21) (3, 4), resulting in dysregulated expression of the antiapoptotic BCL-2 (5) and accumulation of follicle center cells with prolonged survival (6). Secondary genetic alterations involving p53, (7, 8), p16INK4A (9–11), c-myc (12), and BCL-6 (13) have been associated with transformation to DLBCL.
The advent of microarray analysis provides an opportunity to assess genomewide perturbations involved in FL transformation. Lossos et al. (14) recently showed that transformed FLs may exhibit germinal center B cell (GCB)-like or activated B cell (ABC)-like profiles analogous to those originally described for de novo DLBCL (15). They also determined that transformed FLs are biologically distinct from de novo DLBCL. However, functional studies were not performed. Nevertheless, that and other studies have established the important role of expression profiling for class prediction (16) and discovery of prognostically relevant classes (15). The next important challenge is the utilization of expression profiles in the discovery of therapeutic targets.
Here, we have used expression profiling for the identification of important genes/pathways involved in FL transformation. We studied an initial cohort of matched pairs of FLs and their transformed counterparts (DLBCLs) occurring within the same individual. We show that dysregulation of several growth factor and cytokine receptors connected to p38-mitogen-activated protein kinase (MAPK) may be relevant to FL transformation. We validated the results by quantitative RT-PCR (qRT-PCR) and immunohistochemistry (IHC) and further demonstrated that selected differentially expressed genes could serve as robust classifiers of a blinded and independent validation set of FL and DLBCL samples. Finally, we showed that pharmacologic targeting of phosphorylated-p38MAPK in t(14;18)+/p38MAPK-overexpressing lymphoma-derived cell lines inhibits their growth in vitro and in vivo, confirming its importance to FL progression.
Materials and Methods
Samples. We studied a group of 12 matched pairs of FL and their DLBCL counterparts and an additional validation group comprising 20 FLs and 20 DLBCLs. The patient samples were obtained from the Sunnybrook and Women's College Health Sciences Centre and the Cross Cancer Institute. We also studied five t(14;18)-positive cell lines (SUDHL-4, SUDHL-6, SUDHL-16, Karpas 422, and OCI-LY1) and two t(14;18)-negative DLBCL cell lines (SUDHL-5 and OCI-LY10). Purified lymphocyte subpopulations including naïve B cells, memory B cells, GCBs, ABCs, CD2+ T cells, and CD4+ T cells were also studied (17). We also studied paraffin-embedded tissue sections of reactive hyperplasia (n = 6), FL (n = 5), and DLBCL (n = 6). Clinical samples were obtained with informed consent and institutional review board approval.
Cell Culture and Treatment. Cell lines were cultured under standard conditions. SUDHL-4, SUDHL-5, and OCI-LY1 cell lines (1.5 × 105 cells per ml) were treated with SB203580, a selective p38MAPK inhibitor (Upstate Biotechnology, Lake Placid, NY), or U0126, a selective p42/44MAPK inhibitor (Cell Signaling, Beverly, MA) dissolved in DMSO. Cells were collected at specified time points for viability, proliferation, cell cycle, and apoptosis analysis. Control cells were exposed to equal volumes of DMSO and maintained under the same conditions as the treated cells.
Assessment of Clonal Relationships. PCR analysis for Ig heavy chain gene (IgH) rearrangements and for detection of t(14;18)(bcl-2/ JH) were performed as described (18). Fluorescent in situ hybridization for t(14;18) was performed on paraffin-embedded tissues with a probe against t(14;18) (Vysis, Downers Grove, IL).
Gene Expression Profiling. Total RNA was extracted by using TRIzol Reagent (Life Technologies, Gaithersburg, MD). cDNA synthesis and linear amplification were performed as described (19). cDNA microarray analysis was performed at the Huntsman Cancer Institute Microarray Core Facility (Salt Lake City) by using a two-color hybridization (Cy5/Cy3) scheme as described (20) and a 6,912-clone set (Research Genetics, Huntsville, AL). Test sample RNAs were hybridized against a reference composed of RNA pooled from five cell lines (Jurkat, SKW3, L428, Raji, and NCEB). Reproducibility of test to reference ratios across four replicates was accomplished by using GENESPRING software (Silicon Genetics, Redwood City, CA). Hierarchical clustering analysis was performed by using CLUSTER and visualized with TREEVIEW (21) (http://rana.lbl.gov). The expression profiles of the FLs and their DLBCL counterparts were compared with those of the purified lymphocyte subpopulations, ABCs, the t(14;18)+ SUDHL-4 cell line, and the ABC-like cell line OCI-LY10 (15). We selected 76 genes in our clone set, including some genes previously shown to distinguish GCB and ABC programs for hierarchical clustering of our FLs and DLBCLs (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). For the identification of genes involved in FL transformation, we performed (indirect) pairwise analysis of the data obtained from the FLs and DLBCLs, both compared with the reference RNA. The raw data for the indirect FL and the DLBCL experiments are provided at www.path.utah.edu/labs/kojo. We also performed experiments involving (direct) hybridization of RNA obtained from a FL against RNA obtained from the DLBCL from same patient (raw data available at www.path.utah.edu/labs/kojo. The results of the indirect and direct analyses were consistent (Fig. 7, which is published as supporting information on the PNAS web site). Details of our array data processing and a description of our gene discovery algorithm are provided in Supporting Text, which is published as supporting information on the PNAS web site.
Real-Time qRT-PCR. qRT-PCR for C-MET, N-RAS, antioxidant protein 2/acidic phospholipase A2 (PLA2), α-1 and erythrocyte membrane protein 4.1-like 1 (EMP 4.1-L1) was performed by using SYBR Green I and the LightCycler (Roche Applied Science) (PCR protocols are provided in Tables 2 and 3, which are published as supporting information on the PNAS web site). The qRT-PCR to microarray data equivalence comparison and Bayesian analysis for class assignment using qRT-PCR data are provided in Supporting Text.
IHC. IHC was performed by using standard protocols with the following antibodies at the dilutions indicated: anti-human phospho-p38MAPK (1:60) and phospho-p42/p44 MAPK (1:200) (Cell Signaling).
In Vitro Cell Viability Assay. Viability of drug-treated and control cells were determined by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide assay (22). Three replicates were tested for each experimental condition. Statistical comparisons were made by using the two-tailed Student's t test.
Apoptosis Analysis. Annexin V and propidium iodide staining for detection of early and late apoptosis was analyzed by flow cytometry as described (23). Caspase-3 activity was measured by using the CaspACE Assay System (Promega). Cells (1 × 105 cells per ml) were cultured with and without SB203580 at specified concentrations. The pan-caspase inhibitor Z-VAD-FMK was added at a final concentration of 50 μM. Negative controls were prepared by using untreated cells. We performed terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) for the detection of apoptotic nuclear fragments in the xenografted lymphomas, using the In Situ Cell Death Detection (Peroxidase) kit (Roche Molecular Biochemicals).
Immunoblot Analysis. Western blotting was performed following standard protocols using the following antibodies: p42/ p44MAPK, p38MAPK, phospho-p42/p44MAPK (Thr-202/ Tyr-204) E10 mAb, and phospho-p38MAPK 28B10 mAb (Cell Signaling). Immunoreactive bands were visualized by enzyme chemiluminescence (Amersham Pharmacia).
In Vivo Antitumor Activity. Six- to 8-week-old NOD/SCID mice were implanted s.c. with 2.5 × 106 SUDHL-4 cells in the flanks bilaterally. When the tumor implants were palpable, treatment was started with SB2083580 (three animals) at a dose of 4 μmol/kg or an equal volume of vehicle (three animals) injected i.p. five times a week. Perpendicular tumor diameters were measured, and tumor volumes were calculated by using the formula: 0.5(width2 × length). Four weeks after the initiation of drug treatment, the animals were killed, and the tumor implants were studied by histology and in situ TUNEL assay. The Institutional Animal Care and Use Committee of the University of Utah approved the experiments.
Results
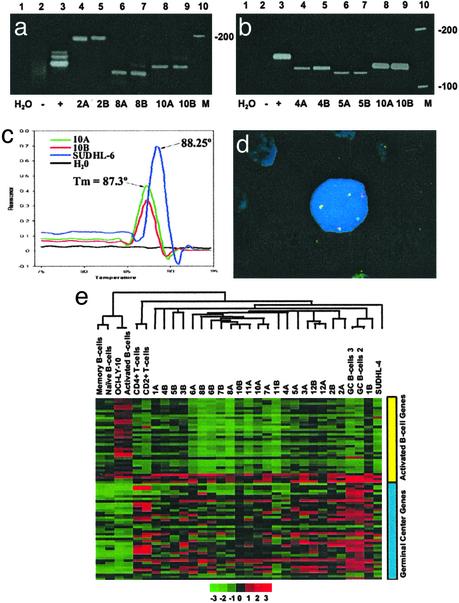
Low-Grade FLs and Their Transformed DLBCL Counterparts Are Clonally Related and Exhibit a GCB-Like Profile. We demonstrated evidence of clonal identity between 11 of the matched pairs of FL and their corresponding DLBCL samples either by demonstrating clonal IgH and bcl-2/JH PCR bands of identical size or sequences, bcl-2/JH product melting temperature, or fluorescent in situ hybridization demonstration of t(14;18) (Fig. 1 a–d and Table 4, which is published as supporting information on the PNAS web site). Hierarchical clustering of array data revealed that the FLs and their DLBCL counterparts exhibited a GCB-like profile (Fig. 1e) distinct from the naïve B cells, memory B cells, and ABCs.
Fig. 1.
(a– d) Clonal relationship between matched pairs of FL and DLBCL. (a) IgH PCR. Lane 1, H2O control; lane 2, polyclonal control (reactive tonsil); lane 3, monoclonal control from the Raji cell line; lanes 4 and 5, monoclonal bands of identical size in the FL (2A) and the subsequent DLBCL (2B); lanes 6 and 7, monoclonal bands of identical size in the FL (8A) and DLBCL (8B) from patient 8; lanes 8 and 9, monoclonal bands of identical size in the FL (10A) and DLBCL (10B) from patient 10; and lane 10, DNA ladder. (b) bcl-2/JH (major breakpoint region, MBR) translocation. Lane 1, H2O control; lane 2, negative control (reactive tonsil); lane 3, positive control (SUDHL-6 cell line); lanes 4 and 5, bcl-2/JH products of identical size in a FL (4A) and the subsequent DLBCL (4B); lanes 6 and 7, identical bcl-2/JH product sizes in the FL and DLBCL from patient 5; lanes 8 and 9, identical bcl-2/JH product sizes in the FL (10A) and DLBCL (10B) obtained from patient 10; and lane 10, DNA ladder. (c) bcl-2(MBR)/JH product detection by fluorescence melting peak analysis. The peak at melting temperature 88.25°C represents the bcl-2/JH product from the positive control (SUDHL-6). bcl-2/JH products with a distinct melting peak with melting temperature 87.3°C is observed for both the FL (10A) and DLBCL (10B) samples from patient 10. No peak is observable in the negative control (reactive tonsil) or the H2O control. (d) t(14;18) Fluorescent in situ hybridization. One bcl-2 (green), one IgH (red), and three bcl-2/IgH (yellow) fusion signals are present. Two of the bcl-2/IgH fusion signals (yellow) represent the derivative chromosomes resulting from the reciprocal translocation. The third yellow signal represents duplication of the derivative chromosome. This fluorescent in situ hybridization pattern was detected in both the FL and DLBCL samples from patient 6. (e) Hierarchical clustering of FL and transformed FL. A total of 76 genes distinguishing GCB from the ABC-like profiles were selected from our 6,912-gene array. Clustering was performed with cluster and visualized by using tree view. Each row represents a gene and each column represents a sample. Red represents higher relative expression of a particular gene and green represents lower relative expression. The color scale at the bottom varies from -3 to +3 in log base 2 units. Consistent with their origin from GCB, the FLs and their corresponding DLBCLs exhibit a GCB-like profile.
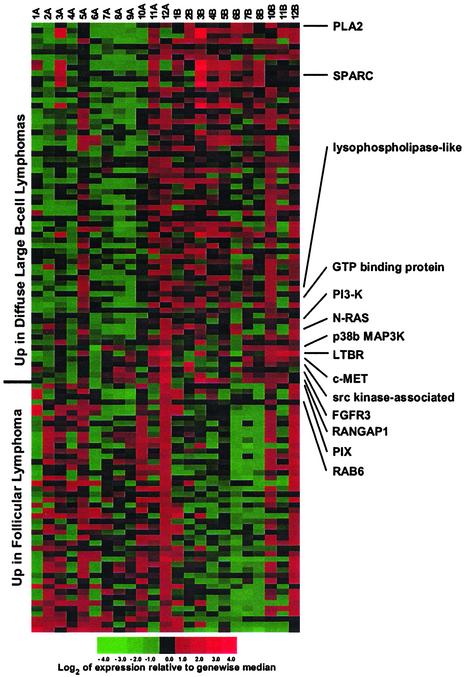
Growth Factor and Cytokine Receptors, Members of the RAS Family, and p38MAPK Are Differentially Expressed in Transformed FL. Our algorithm identified genes that showed consistent increasing or decreasing trends in the transition from FL to DLBCL within each matched pair. It also ranked genes that distinguished the FLs from the DLBCLs. Thus, we identified 113 genes that served as effective classifiers. Of these, 67 showed increased expression, and 46 showed decreased expression in the DLBCLs (Fig. 2 and Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 2.
Gene expression differences between FL and transformed FLs (DLBCLs). The FLs are designated A, and their corresponding DLBCLs are designated B. Several genes associated with MAPK signaling are differentially expressed in the transformation from FL to DLBCL.
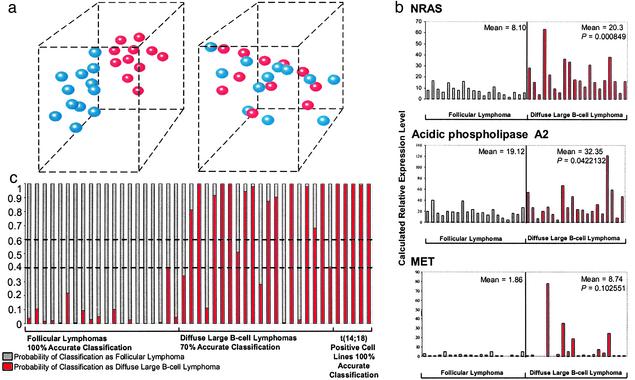
The differentially expressed genes included growth factor and cytokine receptors such as hepatocyte growth factor receptor (C-MET), fibroblast growth factor receptor 3 (FGFR3), lymphotoxin β receptor (LTβR), and platelet-derived growth factor receptor β (PDGFRβ), all of which have been described to effect known growth-promoting activity in various cellular systems (24–27). N-RAS and various RAS-related genes were also differentially expressed in the transformed FL (Fig. 2). In addition, p38βMAPK was up-regulated in the DLBCLs (Fig. 2). This finding is interesting because p38MAPK is an important mediator of cytokine-induced, stress-induced, and cellular responses. Interestingly, similarity analysis of expression patterns revealed that the profiles most similar to p38βMAPK were generated by several of the known p38MAPK target genes such as ATF-2 and several cytokines/receptors (Fig. 9, which is published as supporting information on the PNAS web site). Up-regulation of phosphatidylinositol 3-kinase was also observed in some of the transformed FL (Fig. 2). We further trimmed the differentially expressed genes to three genes that effectively separated the FLs from the DLBCLs in three dimensions, based on the microarray data obtained from the training set of samples (Fig. 3a). The separator genes that emerged included additional genes associated with RAS and MAPK signaling, i.e., PLA2 (x axis), PDGFRβ (y axis), and Rab6 (z axis) (Fig. 3a). Interactive 3D animation is accessible at www.path.utah.edu/labs/kojo. The instructions for the interactive 3D animation are in Supporting Text.
Fig. 3.
(a) 3D separation of FL from DLBCL using three genes. The blue spheres represent the FLs and the red spheres represent their transformed counterparts (DLBCLs). (Left) Clear separation is shown in 3D using the microarray-derived expression values from PLA2 (x axis), PDGFRβ (y axis), and Rab6 (z axis). Interactive 3D animation is at www.path.utah.edu/labs/kojo. (Right) Three randomly selected genes do not separate the FL from the DLBCL. ESTs similar to KIAA0147 mRNA (x axis), human clone 23773 sequence (y axis), and diastrophic dysplasia gene (z axis). (b) Real-time PCR validation of microarray data using an independent group of FLs (n = 20) and DLBCLs (n = 20). The qRT-PCR results corroborate the results of cDNA microarray analysis and show significantly higher expression levels of N-RAS, PLA2, and C-MET in the DLBCLs. (c) Bayesian classification using a minimal discriminative gene set and qRT-PCR. Three highly ranked genes were used for classification of a blinded set of FLs and DLBCLs. The estimated probability of classification as FL (θ) ranges from 0 to 1 (gray bars). Conversely, the estimated probability of classification of the same sample as DLBCL is 1–θ (red bars). Estimated probabilities from 0.4 to 0.6 are considered indeterminate (broken horizontal lines). The accuracy rates for classification of FLs, DLBCLs, and t(14;18)-transformed lymphoma-derived cell lines were 100%, 70%, and 100%, respectively.
Quantitative Fluorescent RT-PCR Validation of Microarray Data. We performed qRT-PCR for quantitation of PLA2, N-RAS, and C-MET to validate our microarray data. The microarray results obtained with the initial test set were corroborated by qRT-PCR (data not shown). We further validated the expression trends observed in the training set of 12 matched pairs of transformed FL by analyzing a validation set consisting of a group of unrelated FLs (n = 20) and DLBCLs (n = 20). The trends observed by microarray were corroborated in the unrelated samples. The calculated transcript levels for our selected genes were significantly higher in the DLBCL, N-RAS (P = 0.000849), antioxidant protein (PLA2) (P = 0.0422), and C-MET (P = 0.102) (Fig. 3b).
qRT-PCR Analysis Using a Minimized But Highly Discriminative Gene Set Provides Accurate Class Assignment into FL or DLBCL Categories. We selected PLA2, N-RAS, and α-1 transcripts for qRT-PCR to test the accuracy of class assignment on an independent “test” set of FL (n = 20), DLBCL (n = 20), and t(14;18)+ cell lines (n = 5). These genes were selected based on their high ranking by our algorithm as strong discriminators of FL from DLBCL. qRT-PCR amplification of these transcripts yielded a classification accuracy of 100% for the FLs, 70% for the DLBCLs, and 100% for the t(14;18)+ cell lines. One case each within the FL and DLBCL categories was scored as indeterminate (Fig. 3c).
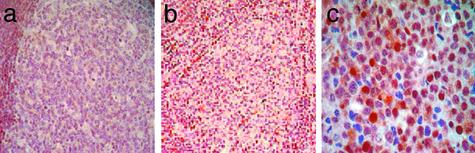
Immunohistochemical Validation: p38MAPK Protein Is Activated and Constitutively Expressed in Transformed FLs. We analyzed the expression of phosphorylated p38MAPK and p42/p44MAPK by using IHC of cell-line microarrays containing the five t(14;18)+ cell lines and biopsies from a cohort of FLs (n = 5) and DLBCLs (n = 6) independent from the samples used for microarray analysis. We detected nuclear phosphorylated (active) p38MAPK expression in 6/6 (100%) DLBCLs, 3/5 (60%) t(14;18)+ cell lines, and 1/5 (20%) FLs. We observed no reactivity for phosphorylated p38MAPK in the reactive germinal centers of 6/6 benign lymph nodes (Fig. 4). IHC for phosphorylated p42/44MAPK protein revealed no reactivity in benign GC B cells and 5/5 FLs. Low levels were detectable in one case of DLBCL and 1/5 t(14;18)+ cell lines (data not shown).
Fig. 4.
Expression of phosphorylated p38MAPK in transformed FL. (a) IHC demonstrates lack of phosphorylated p38MAPK expression in benign germinal centers, whereas the mantle cells demonstrate basal phosphorylated p38MAPK expression. (b) The FL also shows lack of phosphorylated p38MAPK. (c) High levels of nuclear and cytoplasmic phosphorylated p38MAPK expression are seen in the DLBCL. (Magnifications: ×40, a and b; ×400, c.)
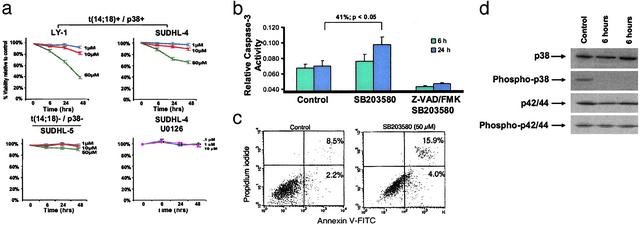
p38MAPK Blockade Inhibits the Growth of t(14;18)+ Cell Lines. We investigated the functional consequence of pharmacological inhibition of the MAPK pathway by using selective inhibitors of p38MAPK (SB203580) and p42/p44MAPK (U0126). Treatment of two t(14;18)+/phospho-p38MAPK+ cells (SUDHL-4, OCI-LY1) with SB203580 (1–50 μM) led to a marked dose- and time-dependent decrease in cell viability as determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide assay (Fig. 5a). Significant decrease in cell viability was observed with 50 μM SB203580 as early as 6 h (89.8 ± 1.3% of control; P < 0.05) with increasing viability loss at 24 h (76.3 ± 3.1%; P < 0.05) and 48 h (66.1 ± 3.1%; P < 0.05) for SUDHL-4 cells. Similar results were obtained for OCI-LY1 cells (Fig. 5a). In contrast, the SUDHL-4 cells were resistant to p42/44MAPK blockade using the p42/44-specific inhibitor U0126. Similarly, SB203580 treatment of SUDHL-5 cells that are t(14;18) negative and phospho-p38MAPK negative yielded negligible reduction of cell viability.
Fig. 5.
(a) Growth inhibition of lymphoma cell lines in response to blockade of p38MAPK activity. Growth inhibition was measured by a decrease in 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide conversion at 6, 24, and 48 h in comparison to DMSO-treated control cells. The results are expressed as inhibition of growth compared with control DMSO-treated cells and represent the mean ± SD of the results obtained in triplicate. Blockade of p38MAPK activity resulted in significant dose-dependent and time-dependent growth inhibition in OCI LY-1 and SUDHL-4 cells, which are t(14;18)+/p38MAPK+ (Upper). By contrast, the t(14;18)-negative/p38MAPK-negative cell line SUDHL-5 was relatively unaffected by similar doses of SB203580 (Lower Left). The specificity of p38MAPK as a target was confirmed by the lack growth inhibition by U0126 the p42/44MAPK inhibitor in the t(14;18)+/p38+ SUDHL-4 cell line (Lower Right). One representative experiment of six independent repetitions is illustrated. (b) SB203580 treatment induces caspase-3 activity. Treatment of SUDHL-4 cells with 30 μM SB203580 resulted in significant time-dependent induction of caspase-3 activity (12.6% at 6 h and 41% at 24 h; P = 0.02). Caspase-3 induction was inhibited by Z-VAD-FMK, a selective caspase-3 inhibitor. One representative experiment of three independent repetitions is illustrated. (c) Inhibition of p38MAPK induces apoptosis in t(14;18)+ cell lines. Cells were cultured in the presence of DMSO or SB203580 (50 μM) and stained for annexin V binding at the times indicated. The control represents DMSO-treated cells at 72 h. (Right) We illustrate a representative histogram for t(14;18) cell lines cultured in the presence of DMSO or SB203580 at the indicated concentrations for 72 h. Apoptosis was determined by annexin V binding and expressed as net apoptosis induction [percentage of apoptosis in treated cells minus percentage of apoptosis in control (DMSO-treated) cells]. One representative experiment of two independent repetitions is illustrated. (d) SB203580 results in selective inhibition of p38MAPK activation. Western blot analysis revealed high basal levels of phosphorylated p38MAPK in SUDHL-4 cells. Treatment with 30 μM SB203580 resulted in a significant decrease in the level of phospho-p38MAPK at 6 h. The levels of total p38MAPK and total and phospho-p42/p44 MAPK were unaffected by SB203580 treatment.
Exposure of SUDHL-4 cells to SB203580 (30 μM) resulted in significant time-dependent induction of caspase-3 activity (12.6% at 6 h and 41% at 24 h; P = 0.02), which was inhibited by the pan-caspase inhibitor Z-VAD-FMK (Fig. 5b). Furthermore, SB203580 treatment also led to increased numbers of annexin V-FITC-positive cells compared with control DMSO-treated cells (15.9% vs. 8.5%), indicative of early apoptosis (Fig. 5c).
Western blot analysis of SUDHL-4 cells revealed high basal levels of phospho-p38MAPK. Treatment with 30 μm of SB203580 resulted in specific decrease in the level of phospho-p38MAPK by 6 h. The levels of total p38MAPK remained unchanged and served as protein loading controls. The levels of total and phospho-p42/44 were also unaffected by SB203580 treatment (Fig. 5d).
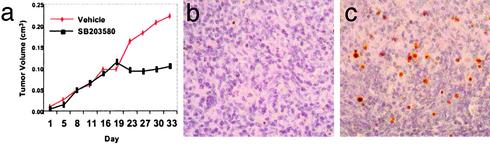
In Vivo Inactivation of p38MAPK Inhibits Tumor Growth by Inducing Apoptosis. We tested the efficacy of SB203580 in inhibiting the growth of SUDHL-4 cells xenografted into NOD-SCID mice. Tumors typically developed 2 weeks after s.c. inoculation of 2.5 × 106 cells into each flank. SB203580 was administered i.p. at a dose of 4 μmol/kg five times a week. This dose was chosen to achieve peak levels that were above the 50% growth inhibitory concentration deduced from the in vitro experiments. Our studies showed inhibition of tumor growth in the SB203580-treated animals from days 18 to 30. By contrast, mice receiving vehicle treatment exhibited unabated tumor growth until day 30 when the mice were killed (Fig. 6a). Microscopic examination of xenografts revealed significantly increased levels of apoptosis and karyorrhexis in the SB203580-treated animals as compared with the vehicle-treated animals. Correspondingly, in situ TUNEL assays revealed significantly increased apoptotic signals in the tumors exposed to SB203580 treatment when compared with the vehicle-treated tumors (Fig. 6 b and c).
Fig. 6.
SB203580 treatment inhibits the growth of SUDHL-4 cells by apoptosis in vivo. (a) There is a significant difference in the growth of tumors in the treated versus the untreated animals. (b) In situ TUNEL assay shows only rare apoptotic (red-brown) signals in SUDHL-4 xenografts of untreated animals. (c) In situ TUNEL assay shows a significant number of apoptotic signals in comparison to untreated animals.
Discussion
This study reveals a role for dysregulation of several growth factor/cytokine receptors and the p38MAPK in FL transformation. Importantly our functional studies strongly suggest that pharmacologic targeting of p38MAPK can be an effective strategy for transformed FL therapy.
We used a longitudinal approach comparing the global transcriptional profiles of clonally related FL and DLBCLs occurring within the same individual. This approach offers significant advantages by focusing on changes distinctively associated with the transformation process, rather than random differences associated with comparison of tumors from different hosts. We observed higher levels of C-MET, FGFR3, LTβR, and PDGFRβ transcripts in the DLBCLs in comparison to the low-grade FLs. Dysregulation of these genes have been implicated in the pathogenesis of a variety of human tumors including primary leukemia and lymphoma cell lines and multiple myeloma (27, 28). However, our study suggests a role in FL transformation. Additionally, PDGFRβ was differentially up-regulated in our DLBCLs and emerged as a useful discriminator of FL from DLBCL in the 3D analysis. This finding is interesting because PDGF is a known mitogen that induces the transcription of IL-6, a potent promoter of B cell proliferation (29). Of additional interest was our observation of up-regulation of phosphatidylinositol 3-kinase, which is also known to exert antiapoptotic and proliferative effects induced by specific growth factors and cytokines (25, 30).
The most consistently up-regulated gene identified by the pairwise comparisons of the matched pairs of FL and DLBCLs in our study was PLA2. Growth factor and cytokine-mediated activation of cytosolic PLA2 (cPLA2) plays a critical role in α-2 macroglobulin-induced cell proliferation in macrophages (31). Our study implicates cPLA2 dysregulation in the pathogenesis of lymphoma.
A unifying feature of the up-regulated growth factor and cytokine receptors identified in our study is their connection to the RAS/MAPK pathway (Fig. 10, which is published as supporting information on the PNAS web site). Interestingly, our microarray and qRT-PCR studies also revealed up-regulation of N-RAS in the DLBCLs (Figs. 2 and 3b). The RAS family proteins are key mediators of cell growth, apoptosis, and responses to extracellular signals. In addition to being significantly overexpressed in our validation set of DLBCLs (P = 0.000849), N-RAS, together with PLA2 and the α-1 transcript, served as a robust classifier of a blinded group of FLs and DLBCLs with 86.7% accuracy. Our studies also revealed that a RAS-related gene, Rab6, was down-regulated in the DLBCLs, and in combination with two other RAS pathway genes provided clear segregation of the FL and DLBCLs in a 3D model (Fig. 3a). This finding is in keeping with previous observations that down-regulation of Rab6 in simian virus 40-tranformed keratinocytes correlates with the transformed phenotype (32). Taken together, our findings indicate a role for transcriptional dysregulation of RAS pathway-related genes in FL transformation.
We observed increased levels of p38βMAPK in the transformed FLs. Moreover, we demonstrated that several p38 target genes including growth factors and cytokines showed coordinately expression profiles across our paired samples. Importantly, we demonstrated increased expression and nuclear localization of phosphorylated p38MAPK protein in DLBCLs as compared with the reactive germinal centers and FLs. In addition to its role in stress and cytokine-induced cellular responses, p38MAPK activation is central to the antiapoptotic and growth-promoting effects of many of the up-regulated growth factor and cytokine receptors identified in this study (25, 27, 31).
We thus hypothesized that p38MAPK may be a rational pharmacologic target for transformed FL therapy. We demonstrated growth inhibition in two p38MAPK-overexpressing t(14;18)+ cell lines by using SB203580, a selective inhibitor of p38MAPK activity. The growth inhibitory effect of SB203580 was specific and was observed only in p38MAPK-overexpressing cell lines. Furthermore, growth inhibition was preceded by decreased levels of phospho-p38MAPK and was affected via caspase-3-mediated apoptosis. The potential role of inhibition of p38MAPK activity for transformed FL therapy is further strengthened by our demonstration of inhibition of tumor growth in an in vivo model by using NOD-SCID mice xenografted with the t(14;18)+ and p38MAPK-overexpressing cell line.
In summary, our studies implicate dysregulation of several growth factor receptors connected to p38MAPK in FL transformation. Dysregulation of p38MAPK target genes may induce growth factors and cytokine genes and thus exert an autocrine-paracrine effect that further promotes lymphoma progression. Our studies also highlight the powerful applications of microarray analysis in the identification of potential targets for transformed FL therapy.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant CA 83984-01 (to K.S.J.E.-J.) and the Associated Regional and University Pathologists Institute for Clinical and Experimental Pathology.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FL, follicular lymphoma; DLBCL, diffuse large B cell lymphoma; GCB, germinal center B cell; ABC, activated B cell; MAPK, mitogen-activated protein kinase; IHC, immunohistochemistry; qRT-PCR, quantitative RT-PCR; IgH, Ig heavy chain gene; PLA, phospholipase A; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling.
References
- 1.Armitage, J. O. & Weisenburger, D. D. (1998) J. Clin. Oncol. 16, 2780-2795. [DOI] [PubMed] [Google Scholar]
- 2.Horning, S. J. & Rosenberg, S. A. (1984) N. Engl. J. Med. 311, 1471-1475. [DOI] [PubMed] [Google Scholar]
- 3.Rowley, J. D. (1988) J. Clin. Oncol. 6, 919-925. [DOI] [PubMed] [Google Scholar]
- 4.Tsujimoto, Y., Finger, L. R., Yunis, J., Nowell, P. C. & Croce, C. M. (1984) Science 226, 1097-1099. [DOI] [PubMed] [Google Scholar]
- 5.Hockenbery, D., Nunez, G., Milliman, C., Schreiber, R. D. & Korsmeyer, S. J. (1990) Nature 348, 334-336. [DOI] [PubMed] [Google Scholar]
- 6.McDonnell, T. J., Deane, N., Platt, F. M., Nunez, G., Jaeger, U., McKearn, J. P. & Korsmeyer, S. J. (1989) Cell 57, 79-88. [DOI] [PubMed] [Google Scholar]
- 7.Sander, C. A., Yano, T., Clark, H. M., Harris, C., Longo, D. L., Jaffe, E. S. & Raffeld, M. (1993) Blood 82, 1994-2004. [PubMed] [Google Scholar]
- 8.Lo Coco, F., Gaidano, G., Louie, D. C., Offit, K., Chaganti, R. S. & Dalla-Favera, R. (1993) Blood 82, 2289-2295. [PubMed] [Google Scholar]
- 9.Elenitoba-Johnson, K. S., Gascoyne, R. D., Lim, M. S., Chhanabai, M., Jaffe, E. S. & Raffeld, M. (1998) Blood 91, 4677-4685. [PubMed] [Google Scholar]
- 10.Pinyol, M., Cobo, F., Bea, S., Jares, P., Nayach, I., Fernandez, P. L., Montserrat, E., Cardesa, A. & Campo, E. (1998) Blood 91, 2977-2984. [PubMed] [Google Scholar]
- 11.Dreyling, M. H., Roulston, D., Bohlander, S. K., Vardiman, J. & Olopade, O. I. (1998) Genes Chromosomes Cancer 22, 72-78. [PubMed] [Google Scholar]
- 12.Yano, T., Jaffe, E. S., Longo, D. L. & Raffeld, M. (1992) Blood 80, 758-767. [PubMed] [Google Scholar]
- 13.Lossos, I. S. & Levy, R. (2000) Blood 96, 635-639. [PubMed] [Google Scholar]
- 14.Lossos, I. S., Alizadeh, A. A., Diehn, M., Warnke, R., Thorstenson, Y., Oefner, P. J., Brown, P. O., Botstein, D. & Levy, R. (2002) Proc. Natl. Acad. Sci. USA 99, 8886-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alizadeh, A. A., Eisen, M. B., Davis, R. E., Ma, C., Lossos, I. S., Rosenwald, A., Boldrick, J. C., Sabet, H., Tran, T., Yu, X., et al. (2000) Nature 403, 503-511. [DOI] [PubMed] [Google Scholar]
- 16.Golub, T. R., Slonim, D. K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J. P., Coller, H., Loh, M. L., Downing, J. R., Caligiuri, M. A., et al. (1999) Science 286, 531-537. [DOI] [PubMed] [Google Scholar]
- 17.Morgan, J. W., Kouttab, N., Ford, D. & Maizel, A. L. (2000) Endocrinology 141, 3225-3234. [DOI] [PubMed] [Google Scholar]
- 18.Robetorye, R. S., Bohling, S. D., Medeiros, L. J. & Elenitoba-Johnson, K. S. (2000) Lab. Invest. 80, 1593-1599. [DOI] [PubMed] [Google Scholar]
- 19.Wang, E., Miller, L. D., Ohnmacht, G. A., Liu, E. T. & Marincola, F. M. (2000) Nat. Biotechnol. 18, 457-459. [DOI] [PubMed] [Google Scholar]
- 20.Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. (1995) Science 270, 467-470. [DOI] [PubMed] [Google Scholar]
- 21.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, Z., Lim, S., Viani, M. A., Sapp, M. & Lim, M. S. (2001) Am. J. Pathol. 159, 711-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vermes, I., Haanen, C., Steffens-Nakken, H. & Reutelingsperger, C. (1995) J. Immunol. Methods 184, 39-51. [DOI] [PubMed] [Google Scholar]
- 24.Weidner, K. M., Sachs, M. & Birchmeier, W. (1993) J. Cell Biol. 121, 145-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao, G. H., Jeffers, M., Bellacosa, A., Mitsuuchi, Y., Vande Woude, G. F. & Testa, J. R. (2001) Proc. Natl. Acad. Sci. USA 98, 247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plowright, E. E., Li, Z., Bergsagel, P. L., Chesi, M., Barber, D. L., Branch, D. R., Hawley, R. G. & Stewart, A. K. (2000) Blood 95, 992-998. [PubMed] [Google Scholar]
- 27.Liu, R. Y., Fan, C., Liu, G., Olashaw, N. E. & Zuckerman, K. S. (2000) J. Biol. Chem. 275, 21086-21093. [DOI] [PubMed] [Google Scholar]
- 28.Chesi, M., Nardini, E., Brents, L. A., Schrock, E., Ried, T., Kuehl, W. M. & Bergsagel, P. L. (1997) Nat. Genet. 16, 260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franchimont, N., Durant, D., Rydziel, S. & Canalis, E. (1999) J. Biol. Chem. 274, 6783-6789. [DOI] [PubMed] [Google Scholar]
- 30.Recio, J. A. & Merlino, G. (2002) Oncogene 21, 1000-1008. [DOI] [PubMed] [Google Scholar]
- 31.Misra, U. K. & Pizzo, S. V. (2002) J. Biol. Chem. 277, 4069-4078. [DOI] [PubMed] [Google Scholar]
- 32.Gromov, P. S. & Celis, J. E. (1994) Electrophoresis 15, 474-481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.