Abstract
HIV pseudotypes bearing native hepatitis C virus (HCV) glycoproteins (strain H and Con1) are infectious for the human hepatoma cell lines Huh-7 and PLC/PR5. Infectivity depends on coexpression of both E1 and E2 glycoproteins, is pH-dependent, and can be neutralized by mAbs mapping to amino acids 412–447 within E2. Cell-surface expression of one or all of the candidate receptor molecules (CD81, low-density lipoprotein receptor, scavenger receptor class B type 1, and dendritic cell-specific intercellular adhesion molecule 3 grabbing nonintegrin) failed to confer permissivity to HIV–HCV pseudotype infection. However, HIV–HCV pseudotype infectivity was inhibited by a recombinant soluble form of CD81 and a mAb specific for CD81, suggesting that CD81 may be a component of a receptor complex.
Hepatitis C virus (HCV) is an enveloped, positive-stranded RNA virus classified in the family Flaviviridae. Infection is often associated with chronic disease, sometimes resulting in cirrhosis and hepatocellular carcinoma. The principal site of replication is thought to be the liver, although several laboratories have suggested that HCV may infect a wider range of cell types including monocytes/macrophages and B cells (1, 2).
HCV encodes two putative envelope glycoproteins (gps), E1 and E2, which are believed to be type I integral transmembrane proteins. In vitro expression studies have shown that both gps associate to form heterodimers, which accumulate in the endoplasmic reticulum (ER), the proposed site for HCV assembly and budding (reviewed in ref. 3). The lack of in vitro systems for HCV propagation has hampered biological and physiochemical studies on the virion and its mechanism(s) of cell entry, and the cellular receptors remain unknown. HCV purified from plasma has been reported to exist in association with plasma lipoproteins, suggesting that the virus may use the low-density lipoprotein receptor (LDLR) to gain entry into cells (4–6).
The selective association of a virus with a target cell is usually determined by an interaction between the viral gps and specific cell-surface receptor(s) and is an essential step in the initiation of infection. Such interaction(s) often define the host range and cellular or tissue tropism of a virus and have a role in determining virus pathogenicity. In the absence of native HCV particles, truncated version(s) of the E2 gp (7, 8), E1E2 gp-liposomes (9), and virus-like particles expressed in insect cell systems (10, 11) have been used as mimics to study virus–cell interactions. Truncated soluble versions of E2 have been reported to bind specifically to human cells and were used to identify interactions with CD81 (7, 8), scavenger receptor class B type 1 (SR-B1) (12), and dendritic cell-specific intercellular adhesion molecule 3 grabbing nonintegrin (DC-SIGN) (13, 14). One limitation with these studies is that they measure only HCV gp–cell attachment and not virus-mediated cell fusion. To overcome the lack of a conventional cell culture system for the propagation of infectious HCV particles, pseudotype viruses expressing the HCV envelope gps have been generated. Several laboratories have reported on the infectivity of vesicular stomatitis virus (VSV) pseudotypes expressing chimeric HCV E1E2 gps encoding the transmembrane domain and cytoplasmic tail of VSV G gps, but with conflicting results (15–17).
HIV readily forms pseudotypes with the envelope proteins of many different viruses. In this article, we present data showing that HIV pseudotypes bearing native HCV E1 and E2 gps are infectious for the human hepatoma cell lines Huh-7 and PLC/PR5. Significantly, infectivity is pH-dependent and can be neutralized by a number of E2-specific mAbs. These pseudotype viruses will be invaluable for further investigation into the mechanism of HCV entry and the identification of cellular receptor(s) mediating virus attachment and fusion. This system will also allow us to address the role of the humoral immune response in HCV infection and to evaluate therapeutics targeting the HCV gp–cell interaction.
Materials and Methods
Cells. Hos.CD4.R5 were obtained from the National Institutes of Health AIDS Reagent Program and propagated in DMEM with 10% FBS and 1 μg/ml puromycin. Huh-7 (gift of R. Lanford, Southwest Foundation of Biomedical Research, San Antonio, TX), Huh-7.5 (18), and HeLa cells were propagated in DMEM/10% FBS. HepG2 cells were propagated on collagen type 1-coated tissue culture plastic in DMEM/10% FBS (gift of Y. Matsuura, Osaka University, Osaka). PLC/PR5 cells were propagated in DMEM/10% FBS (gift of J. Garson, University College London, London). THLE cells (gift of S. Feinstone, Food and Drug Administration, Washington, DC) were propagated as described (19). RBL cells stably expressing human CD81 were propagated in DMEM/10% FBS with 400 μg/ml G418 (gift of P. Monk, University of Sheffield, Sheffield, U.K.) (8). U937 cells expressing human CD81 (gift of S. Levy, Stanford University, Stanford, CA) and THP cells expressing DC-SIGN (gift of R. Doms, University of Pennsylvania, Philadelphia) were propagated in RPMI/10% FBS. All cells were grown at 37°C/5% CO2.
Plasmids. Plasmids encoding E1 (pE1; polyprotein residues 171–383), E2 (pE2; polyprotein residues 364–746), and E1 plus E2 (pE1E2; polyprotein residues 171–746) were constructed by PCR amplification from template pBRTM/HCV1–3011 as reported (20, 21). The plasmid-encoding strain Con1 E1E2 was similarly generated by PCR amplification of the E1E2 ORF from Con1/FL (18) and ligated into pCAGGS/MCS. Plasmids expressing the HCV Sindbis virus (SIN) and VSV G chimeric constructs, VSV G and SF162 gp160, have been described (13, 22). The plasmid encoding amphotropic murine leukemia virus envelope was a gift of S. Goff (Columbia University, New York).
Antibodies. Murine mAbs specific for CD81 (5A6; a gift of S. Levy), DC-SIGN (m507; R & D Systems), LDLR (Ab-1; Oncogene Research Products, San Diego), SR-BI (25, BD Biosciences), NB 400 101 (Novus Biologicals, Littleton, CO), PDI (SPA-891; Stress-Gen Biotechnologies, Victoria, Canada), HIV gp120 (B4a1; National Institutes of Health AIDS Reagent Program), and HCV E2 (H52; J. Dubuisson, Institute Pasteur, Lille, France) were used. Human mAbs specific for HCV E1 (H111) and E2 (CBH5) were a gift of S. Foung (Stanford University). Rat mAbs specific for HCV E2 have been described (8).
Flow Cytometric Analysis. Expression of CD81, LDLR, SR-B1, DC-SIGN, and HCV gps was quantified as described (18). All cells were incubated with an irrelevant isotype-matched IgG, and the fluorescence signal was used to establish threshold values of detection for the test mAbs. Analyses were performed by using a FACSCalibur flow cytometer and CELLQUEST software (Becton Dickinson).
Pseudotype Production, Characterization, and Infection. HIV pseudotypes were generated by cotransfection of 293-T cells with equal amounts of expression plasmids expressing the viral gps or an empty vector and the envelope-defective pNL4.3.Luc.R-E- proviral genome as described (13, 23). The supernatants were collected 48 h posttransfection, and HIV p24 antigen content was assessed by using a commercially available EIA (Beckman Coulter). Virus particles were purified by velocity centrifugation through a 5–20% sucrose gradient in PBS at 35,000 rpm at 4°C in a Beckman SW41 rotor for 30 min. Fractions were collected, and aliquots were precipitated with trichloroacetic acid for immunoblotting or diluted in 3% FBS/DMEM for infectivity studies. To investigate pseudotype virus infectivity, target cells were seeded into 96-well plates (8 × 103 cells per well) 24 h before infection. Equal volumes of p24 antigen-normalized viral supernatants were diluted in 3% FBS/DMEM plus 4 μg/ml polybrene and 100 μl was added per well. Cells were incubated at 37°C for 72 h, washed in PBS once, and lysed with 40 μl per well of cell lysis buffer (Promega). Twenty microliters of lysate was tested for luciferase activity by the addition of 50 μl of luciferase substrate and was measured for 10 s in a luminometer (Lumat LB 9507, Berthold Technologies, Oak Ridge, TN). To evaluate the reversibility of pH-dependent infectivity, pseudotype viruses were treated with low-pH buffer (Mes, pH 5.0) for 10 min at room temperature, the pH was neutralized to pH 7.2, and the viruses were tested for their ability to infect Huh-7.5 cells as described above.
Huh-7.5 cells were incubated with anti-CD81, anti-LDLR, and anti-SR-B1 at 5 μg/ml (100 μl per well) for 30 min on ice, and 100 μl of pseudotyped virus, diluted in 3% FBS/DMEM plus 4 μg/ml polybrene, was added. Cultures were incubated at 37°C for 72 h, and luciferase activity was measured. mAbs specific for E2 (final concentration 5 μg/ml) and soluble CD81 (various concentrations) were incubated with pseudotype virus for 30 min at 37°C, and virus/ligand mixtures were tested for infectivity of Huh-7.5 cells.
Huh-7.5 cells were infected with pseudotype viruses in the presence or absence of 10 mM ammonium chloride or 25 nM concanamycin for 8 h at 37°C. Treatment of cells had no effect on proliferation over the time course of the experiment. Virus was removed by aspiration, and cells were washed once with DMEM and cultured in 10% FBS/DMEM for 72 h. Cells were lysed as described above and luciferase activity was determined.
Results
HIV Pseudotypes Expressing Native HCV gps Are Infectious. Because HIV assembles at the plasma membrane and HCV gps are retained in the ER, we hypothesized that it would be necessary to express the gps at the plasma membrane for efficient pseudotype formation. We (13, 20) and others (15–17) have reported that truncated forms of HCV E1 and E2 fused to the transmembrane domain and cytoplasmic tail of VSV G (E1/G and E2/G, respectively) and SIN gps (E1/SIN and E2/SIN) are efficiently expressed at the cell surface.
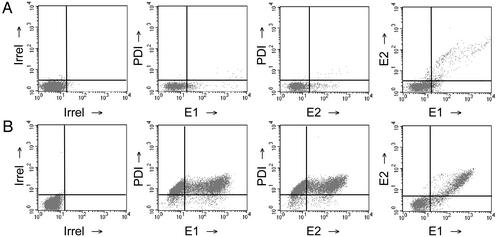
To produce virus pseudotypes, 293-T cells were cotransfected with plasmids encoding an envelope-defective HIV-1 proviral genome expressing a luciferase reporter gene (NL4.3.Luc.R-E-) (23), full-length strain H (H E1E2), and chimeric HCV E1E2 gps (E1E2-G and E1E2-SIN). As controls, plasmids encoding HIV SF162 gp160 and VSV G were cotransfected to generate pseudotype viruses with known entry characteristics. To assess whether full-length E1E2 gps were expressed at the cell surface, transfected cells were quantified for their level of total and cell-surface E1E2 antigen by flow cytometry. Membrane damage and subsequent exposure of internal cell membrane proteins to the staining mAbs would falsely suggest plasma membrane localization. To assess this, cells were fixed and dually stained for expression of protein disulfide isomerase (PDI), an ER resident protein, and E1E2 gps. Both gps were detected at the surface of cells transfected with pE1E2 and NL4.3.Luc.R-E-, which failed to stain for PDI (E1E2+/PDI- cells, 12%; E1E2+/PDI+ cells, 0.8%; Fig. 1A), suggesting transport of the HCV gps to the cell surface. Detection of E1E2 after cell permeabilization suggested that the majority of the gps were retained within the cell (Fig. 1B). Dual staining of transfected cells for both E1 and E2 confirmed that both gps were present at the cell surface (Fig. 1A). Similar results were obtained for Con1 E1E2 gps (data not shown). Expression of full-length E1E2 at the cell surface did not depend on NL4.3 cotransfection (data not shown). Chimeric gps E1E2-G and E1E2-SIN were efficiently expressed at the cell surface (42% and 36% of nonpermeabilized transfected 293-T cells stained for expression of E1E2-G and E1E2-SIN, respectively).
Fig. 1.
HCV E1E2 gp expression. Transfected 293-T cells were tested for expression of HCV E1 and E2 gps. Cells transfected with pNL4.3.Luc.R-E- and pE1E2 were tested for cell surface and total expression of E1 (mAb H111), E2 (mAbs CBH5 or H52), and PDI (SPA-891). Cells were fixed and analyzed for antigen expression with (B) or without (A) permeabilization. Regions were established based on irrelevant isotype-matched IgG staining of the cells.
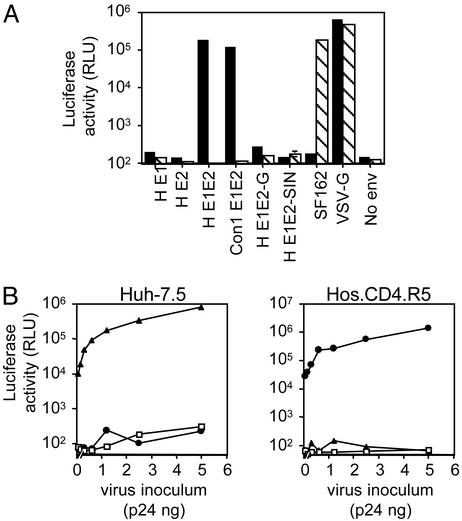
Extracellular supernatants were tested for their ability to infect Hos cells stably expressing the HIV receptors CD4 and CCR5 (Hos.CD4.R5) and the human hepatoma cell line Huh-7.5. HIV produced from cells expressing HCV E1E2 infected Huh-7.5 cells, whereas virus from cells expressing the chimeric HCV gps did not (Fig. 2A). Infectious pseudotypes were produced only from cells expressing both HCV gps either from a single plasmid (pE1E2) or by cotransfection of two separate plasmids (pE1 and pE2) (data not shown). As expected, HIV pseudotypes bearing the SF162 gp160 infected only Hos.CD4.R5 cells and HIV-VSV G infected both cell types (Fig. 2A). Particles lacking any virus gp (no envelope) failed to infect either cell type. Luciferase activity (relative light units, RLUs) was proportional to the amount of virus used for the infection, demonstrating a dose-dependent relationship (Fig. 2B).
Fig. 2.
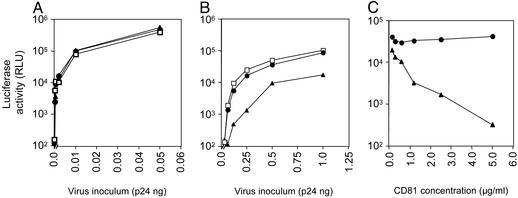
HIV pseudotype infectivity. (A) HIV pseudotypes bearing strain H native E1 and E2 gps alone or together (H E1, H E2, or H E1E2), strain Con1 E1E2 gps (Con1 E1E2), E1 and E2 ectodomains fused to the transmembrane domain and cytoplasmic tail of VSV G (H E1E2-G), SIN E2 and E1 gps (H E1E2-SIN), HIV–SF162 gp160 (SF162), VSV-G, or no envelope (no env) were tested for their ability to infect Huh-7.5 (filled bars) or Hos.CD4.R5 cells (hatched bars). All virus stocks were normalized for p24 HIV core antigen and infected at 1 ng per well, with the exception of VSV G, which was used at 0.1 ng per well. All infections were performed in triplicate, and the mean luciferase activity (RLU) is shown. The coefficient of variance was <10% in all cases. (B) Increasing concentrations of pseudotypes bearing H E1E2 (▴), SF162 (•), and no envelope (□) were tested for infection of Huh-7.5 and Hos.CD4.R5 cells.
HIV pseudotypes bearing an independent strain of HCV E1E2 (Con 1, genotype 1b) showed levels of infectivity similar to HIV-H E1E2 for Huh-7.5 cells (Fig. 2A). Sucrose density gradient centrifugation of HIV–HCV pseudotype preparations demonstrated that infectivity and HCV E2 were detected in fractions containing particulate HIV p24 antigen, consistent with HIV incorporation of E2 (data not shown). However, the majority of E2 present in the extracellular supernatant was not incorporated into sedimenting particles and was located in fractions at the top of the gradient. In conclusion, cotransfection of plasmids encoding HCV E1E2 and HIV produced pseudotypes that incorporated HCV gps and were infectious for Huh-7.5 cells.
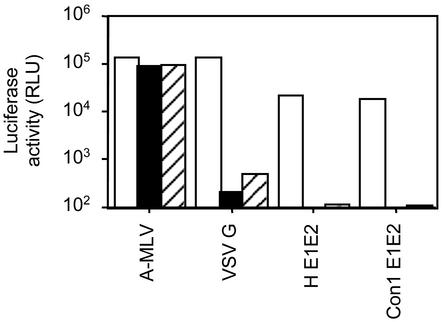
HCV Pseudotype Entry Is pH-Dependent. Enveloped viruses enter cells through two main pathways: direct fusion at the plasma membrane and receptor-mediated endocytosis. In the latter case, the fusion of the viral envelope protein(s) is triggered by low pH within the endosome. Inhibitors of vacuolar acidification, such as concanamycin and ammonium chloride, have been used to demonstrate the pH sensitivity of virus entry. We therefore tested the infectivity of pseudotypes after treatment of target cells with ammonium chloride and concanamycin. As controls, HIV bearing VSV G and amphotropic murine leukemia virus gps, demonstrating pH-dependent and -independent routes of entry, respectively, were tested. HIV–HCV pseudotypes demonstrated a >90% reduction in infectivity in the presence of ammonium chloride and concanamycin (Fig. 3), suggesting a pH-sensitive route of virus entry. As expected, infectivity of HIV–VSV G was reduced in treated cells, whereas HIV amphotropic murine leukemia virus showed no change (Fig. 3). Because pH-dependent changes in VSV G conformation are reversible (24), we were interested to determine whether low-pH treatment of HCV pseudotypes would affect their infectivity. HCV pseudotype infectivity (luciferase activity) was reduced by up to 70% after a 10-min incubation at pH 5.0, whereas infectivity of HIV–VSV G was not affected. These data suggest that low-pH-induced changes in the HCV gps are not readily reversible and may play a critical role in the HCV-gp fusion pathway.
Fig. 3.
pH dependency of HIV–HCV pseudotype infectivity. Huh-7.5 cells were infected with HIV pseudotypes bearing amphotropic murine leukemia virus (A-MLV), VSV-G, H E1E2, and Con1 E1E2 in medium alone (open bars) or containing 10 mM ammonium chloride (filled bars) or 25 nM concanamycin A (hatched bars). All infections were performed in triplicate, and the mean luciferase activity (RLU) is shown. The coefficient of variance was <15% in all cases.
The Role of Cell-Surface Molecules in HIV–HCV Pseudotype Virus Entry. To examine the relevance of the cellular molecules identified as putative receptors for HCV, we monitored the expression of these molecules on a variety of cell types, which were also tested for their ability to support pseudotype virus infection. mAbs specific for these cell-surface molecules were also tested for their ability to inhibit pseudotype virus infection of Huh-7.5 cells. The human cell lines Hos.CD4.R5, Huh-7, Huh-7.5, HepG2, PLC/PR5, THLE, HeLa, THP-DC-SIGN, and SW13 were tested for their ability to support HIV pseudotype virus infection and their expression of CD81, LDLR, and SR-B1 (Table 1). All viruses were tested at two infecting doses (5 ng per well and 1 ng per well, p24), with the exception of HIV–VSV G, for which a single concentration (0.1 ng per well), shown to give saturable RLU signals on infection of Huh-7.5, was used. HIV–HCV pseudotypes were able to infect Huh-7, Huh-7.5, and PLC/PR5 cells (Table 1). Both HIV–HCV pseudotype stocks (H and Con1) infected PLC/PR5 with a lower titer than Huh-7.5 cells (end-point titer of HIV–H E1E2 for Huh-7.5 and PLC/PR5 cells was 5 × 104 and 1 × 103 tissue culture 50% infective dose per ng of p24, respectively). HIV–SF162 only infected Hos.CD4.R5 cells, whereas HIV–VSV G infected all cell types tested (Table 1). All human cell types, with the exception of HepG2, expressed CD81 (Table 1). THP cells expressing DC-SIGN were unable to support HIV–H E1E2 or HIV–SF162 pseudotype virus infection, suggesting that DC-SIGN does not directly support HCV pseudotype infection in the context of a monocytic cell background (Table 1). The majority of cells expressed LDLR and SR-B1 and yet were refractory to HIV–HCV pseudotype infection.
Table 1. Cell phenotype and susceptibility to HIV pseudotype infection.
| Infectivity of pseudotypes expressing env gps*
|
Cell-surface expression†
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cell line | Cell type | No env | SF162 | VSV G | H E1E2 | Con1 E1E2 | CD81 | LDLR | SR-B1 |
| Hos.CD4.R5 | Human osteosarcoma | - | ++ | +++ | - | - | +++ | + | + |
| Huh-7.5 | Human hepatoma | - | - | +++ | ++ | ++ | ++ | + | + |
| Huh-7 | Human hepatoma | - | - | +++ | ++ | ++ | ++ | + | + |
| HepG2 | Human hepatoma | - | - | +++ | - | - | - | + | + |
| PLC/PR5 | Human hepatoma | - | - | ++ | + | + | ++ | + | - |
| THLE | Human liver epithelial | - | - | +++ | - | - | +++ | - | - |
| Hela | Human epithelial | - | - | ++ | - | - | +++ | - | + |
| SW13 | Human colorectal adenocarcinoma | - | - | +++ | - | - | +++ | + | + |
| U937 | Human monocytoid | - | - | + | - | - | - | - | - |
| U937-humCD81 | Human monocytoid | - | - | + | - | - | +++ | - | - |
| THP | Human monocytoid | - | - | ++ | - | - | +++ | - | ++ |
| THP-DC-SIGN | Human monocytoid | - | - | ++ | - | - | +++ | - | ++ |
| RBL | Rat basophil | - | - | ++ | - | - | - | - | - |
| RBL-humCD81 | Rat basophil | - | - | ++ | - | - | +++ | - | - |
Virus pseudotypes with no env, H E1E2, Con1 E1E2, and SF162 were tested for infection of the various cell lines at 5 and 1 ng per well (data not shown) and VSV G at 0.1 ng per well
Infectivity is measured as RLU where: -, <300; +, 10,000-20,000; ++, >20,000-100,000; and +++, >100,000
Cell-surface expression of antigens, where an irrelevant isotype-matched IgG gave mean fluorescence intensity (MFI) in the range of 4-6 and specific mAbs gave staining intensities of +, 10-30; ++, >30-100; and +++, >100
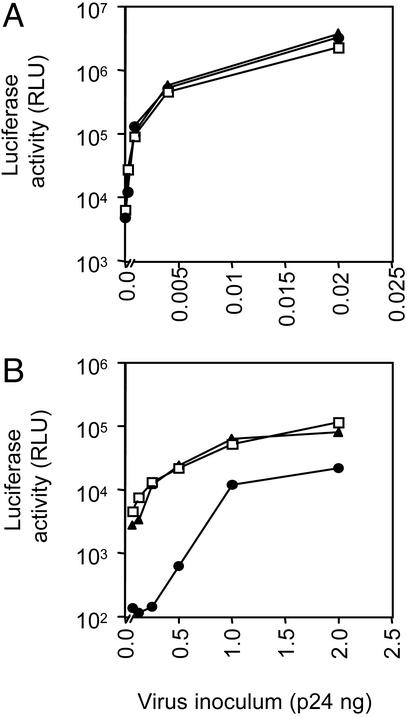
To elucidate the role of these cellular molecules in HCV entry, Huh-7.5 cells were incubated with mAbs specific for CD81, SR-B1, or LDLR at saturating concentrations (data not shown) and tested for their susceptibility to HIV–H E1E2 and HIV–VSV G pseudotype virus infection. HIV–H E1E2 virus infection was inhibited by the anti-CD81 mAb but not by the SR-B1 and LDLR Abs (Fig. 4 and data not shown). At lower concentrations of input HIV–H E1E2 virus, the anti-CD81 mAb blocked infection by >90% (Fig. 4B). Similarly, only the anti-CD81 mAb was able to inhibit HIV–Con1 E1E2 virus infection of Huh-7.5 cells (data not shown). HIV–HCV pseudotype (strain H and Con1) infection of PLC/PR5 cells was also inhibited by the anti-CD81 mAb (data not shown). None of the mAbs affected infection by HIV–SF162 (data not shown) or HIV–VSV G (Fig. 4A).
Fig. 4.
Receptor dependency of HIV–HCV pseudotype infectivity. Huh-7.5 cells were incubated with mAbs specific for CD81 (5A6, •), SR-B1 (clone 25, ▴), and an irrelevant isotype-matched IgG (□) at 5 μg/ml and were infected with HIV pseudotypes bearing VSV G (A), H E1E2 (B), and no envelope (data not shown) at varying concentrations of p24 antigen. Infection with HIV-no envelope gave a maximal mean RLU count of 420 ± 50 at 2 ng per well. The coefficient of variance was <10% in all cases.
Soluble recombinant forms of the human CD81 large extracellular loop (LEL) (GST-humCD81) are able to bind HCV E2 gp and inhibit its interaction with cellular CD81 (8). We therefore tested the ability of GST-humCD81 to inhibit HIV–H E1E2 and HIV–VSV G infection of Huh-7.5 and PLC/PR5 cells. A recombinant protein encoding the African green monkey CD81 LEL sequence (GST-agmCD81) is unable to interact with E2 and was used as a specificity control (25). GST-humCD81 inhibited HIV–H E1E2 infection of both cell types but had no effect on HIV–VSV G infectivity (Fig. 5 and data not shown). GST-humCD81 showed a dose-dependent neutralization of infectivity, inhibiting HIV–H E1E2 infection of Huh-7.5 cells by 50% at 2.5 μg/ml (Fig. 5). To further investigate the role of CD81 in HIV–HCV pseudotype entry, we tested the ability of a human monocytoid cell line, U937, and a rat basophilic cell line, RBL, both transfected to stably express human CD81, to support virus infection. Neither cell line, independent of CD81 expression, was infected by HIV–H E1E2, whereas both were infected by HIV–VSV G (Table 1). In conclusion, these data suggest that expression of CD81, LDLR, or SR-B1 alone or together is not sufficient to permit infection of a cell by HIV–HCV pseudotypes. The ability of anti-CD81 and soluble forms of CD81 to block HIV–HCV pseudotype infectivity suggests that CD81 is a component of the receptor complex. However, the inability of CD81-expressing U937 or RBL cells to support HIV–HCV pseudotype infection suggests that CD81 needs to be expressed in the context of additional cellular factors to modulate productive HCV entry.
Fig. 5.
Soluble CD81 neutralization of HIV–HCV pseudotype infectivity. (A and B) HIV pseudotypes bearing VSV G (A) and H E1E2 (B) at varying concentrations of p24 antigen were incubated with recombinant soluble forms of CD81, GST-humCD81 (▴), and GST-agmCD81 (•)ata final concentration of 1 μg/ml or medium alone (□) for 1 h at 37°C. Virus–CD81 mixtures were tested for infectivity of Huh-7.5 cells. (C) A single dose of HIV-H E1E2 pseudotype (1 ng per well) was incubated with varying concentrations of GST-humCD81 (▴) and GST-agmCD81 (•)for 1 h at 37°C and tested for infection of Huh-7.5 cells. All infections were performed in triplicate, and the mean luciferase activity (RLU) is shown. The coefficient of variance was <10% in all cases.
Pseudotype Virus Infection Is Neutralized by E2-Specifc mAbs. To confirm the specificity of HIV–H E1E2 virus infection, a panel of well characterized E2-specific mAbs were tested for their ability to neutralize infection of Huh-7.5 cells. A neutralizing mAb specific for HIV gp120 (B4a1) was also tested. All of the HCV-specific mAbs were previously shown to interact with a soluble truncated form of E2 with varying affinities (Table 2; ref. 8). All mAbs were incubated with HIV–H E1E2 or HIV–SF162 for 1 h at 37°C, and the virus–mAb mixtures were tested for infection of Huh-7.5 and Hos.CD4.R5 cells, respectively (Table 2). Four of the five mAbs specific for the E2 hypervariable region (HVR), a proposed neutralization epitope, had no effect on the infectivity of either pseudotype virus (Table 2). mAb 9/27, specific for amino acids 396–407 within the HVR, specifically neutralized HIV–H E1E2 (Table 2). mAbs specific for E2 amino acids 412–423 (3/11), 432–443 (2/69a), and 436–447 (7/16b and 11/20) neutralized HIV–H E1E2, but not HIV–SF162, infectivity, suggesting that this region of the gp contains several neutralization epitopes. In contrast, mAbs specific for epitopes within the C-terminal region of E2 (6/1a, 6/41a, and 6/53) had no effect on HIV–H E1E2 infectivity. Several mAbs were evaluated for their ability to recognize E2 expressed at the surface of 293-T cells transfected with NL4.3 and pE1E2 plasmids, as characterized in Fig. 2. There was no correlation between mAb reactivity with cell-surface-expressed E2 and neutralization of pseudotype infectivity (Table 2).
Table 2. Neutralization of HIV pseudotype infectivity.
| % neutralization of§
|
||||||
|---|---|---|---|---|---|---|
| mAb | E2 epitope | Half-maximal binding, μg/ml* | Inhibition of E2-CD81† | Reactivity with 293-T expressed HCV E1E2‡ | HIV-H E1E2 | HIV SF162 |
| 7/59 | 384-391 | 0.01 | - | NT | <5 | <5 |
| 6/82a | 384-395 | 2.0 | - | NT | <5 | <5 |
| 6/16 | 384-395 | 0.02 | - | +/+ | <5 | <5 |
| 9/27 | 396-407 | 0.04 | - | +/+ | ≥99 | <5 |
| 9/86a | cHVR¶ | 1.0 | - | NT | <5 | <5 |
| 3/11 | 412-423 | 0.02 | + | NT | 70 | <5 |
| 2/69a | 432-443 | NS | + | +/+ | ≥99 | <5 |
| 1/39 | 432-443 | 0.15 | + | NT | 20 | <5 |
| 7/16b | 436-447 | 0.10 | + | +/+ | ≥99 | <5 |
| 11/20 | 436-447 | 0.02 | + | NT | ≥99 | <5 |
| 6/1a | 464-471 | 1.20 | - | NT | <5 | <5 |
| 6/41a | 480-493 | NS | + | NT | <5 | <5 |
| 2/64a | 524-531 | 0.08 | + | NT | 40 | <5 |
| 9/75 | 524-531 | 0.01 | + | +/+ | <5 | <5 |
| 6/53 | 544-551 | 0.80 | - | +/+ | <5 | <5 |
| B4a1 | HIV gp120 | NA | NA | NA | <5 | ≥99 |
NA, not applicable; NT, not tested
mAb recognition of truncated soluble E2, relative affinities are shown as the concentration (μg/ml) required to give half-maximal binding in a capture ELISA (8)
mAb (10 μg/ml) inhibition of soluble E2 binding to GST-humCD81 (8)
mAb (10 μg/ml) reactivity with nonpermeabilized and permeabilized 293-T cells transfected with NL4.3.Luc.R-E- and pE1E2 (depicted in Fig. 1) (represented as +/+ non-perm/perm cell recognition)
mAb (10 μg/ml) neutralization of HIV-H E1E2 and HIV SF 162 (1 ng per well inoculum) infection of Huh-7.5 and Hos.CD4.R5 cells, respectively, as determined by inhibition of luciferase activity
cHVR, conformation-dependent epitope within the HVR
Discussion
In this study, we demonstrate that HIV pseudotypes bearing HCV E1E2 gps are infectious for the human hepatoma cell lines Huh-7 and PLC/PR5. In contrast, pseudotypes bearing chimeric HCV gps (E1E2-G and E1E2-SIN) failed to infect any of the cell types tested. HIV–HCV pseudotype infectivity depended on expression of both gps and was blocked by several anti-E2 mAbs. This result was surprising, because HIV is reported to assemble at the plasma membrane, whereas HCV gps are retained within the ER (8). However, a small, but significant, number of transfected 293-T cells expressed E1E2 at the cell surface independent of HIV protein expression. Because signals for protein retention within the ER or Golgi complex are rarely 100% efficient, high-level E1E2 expression in 293-T cells appears to be leaky, and a fraction of the gps is transported to the plasma membrane and incorporated into HIV particles. This observation is consistent with that reported recently by Bartosch et al. (26). The noninfectious nature of the pseudotypes carrying the chimeric proteins may be attributed to the defective nature of these proteins, because they lack regions of the transmembrane domains important for E1E2 oligomerization and membrane insertion (27). VSV pseudotypes expressing chimeric HCV E1E2–VSV G gps have been reported to be infectious for a wide variety of cell types (15, 16). However, Buonocore et al. (17) recently reported that VSV pseudotypes expressing high levels of HCV–VSV G chimeric proteins demonstrated a low level of infectivity that was neutralized by anti-VSV G. The authors concluded that trace amounts of VSV G present after genesis of pseudotyped viruses through BHK cells accounted for this low level of infectivity. These conflicting reports cast doubt over the functional nature of such chimeric HCV gps.
The pH dependence of infection by the HIV–HCV pseudotype viruses supports a receptor-mediated endocytic route of virus entry. Viruses that enter cells via pH-dependent pathways generally synthesize their viral fusion gps in an inactive form, to prevent premature fusion of the internal cellular membranes (28). Within the Flaviviridae family, the best-studied viral gp is the envelope protein E of tick-borne encephalitis (TBE) virus. The TBE E protein exists as a homodimer in the virus particle, which is converted to a trimer at the pH of fusion. Because infectivity of the HIV–HCV pseudotypes depends on coexpression of both E1 and E2, these particles will enable us to characterize the oligomeric status of functionally active HCV gps and begin to address the mechanism(s) of low-pH gp activation.
The cellular tropism of HCV is an important and much debated issue. The principal site of replication is thought to be the liver, leading to the hypothesis that liver-specific molecules act as viral receptors. Experiments to address whether any of the candidate cellular receptors are involved in HIV–HCV infectivity suggest that none of the molecules expressed alone, or in combination, define permissivity to infection. Several cell lines, including Hos and SW13, expressed CD81, LDLR, and SR-B1 and yet were refractory to infection by the HIV–HCV pseudotypes (Table 1). The ability of the anti-CD81 mAb, 5A6, and soluble GST-humCD81 to specifically inhibit infection of Huh-7.5 cells by the HIV–HCV pseudotypes suggests that CD81 may be a component of a receptor complex. However, the inability of CD81 to confer pseudotype infectivity to U937 and RBL cells suggests that CD81 may need to be expressed in concert with one or more specific molecules that are expressed in Huh-7 and PLC/PR5 cells to exert its effect.
Because HCV particles purified from human plasma have been reported to interact with LDLR via an indirect association with LDL or very-low-density lipoprotein, our inability to inhibit pseudotype infection with the anti-LDLR mAb may be attributed to the different lipid composition of in vitro-generated particles (4, 6). In a similar HCV pseudotyping system, Bartosch et al. (26) reported partial neutralization of HCV pseudotypes with anti-apolipoprotein E mAbs, suggesting that virus-associated lipids may contribute to the infection process. At the present time, it is unclear how HCV associates with plasma lipids. Whether this represents a passive interaction after release from the cell or utilization of the LDL synthesis and secretion pathway by HCV to exit from the cell is unknown. Future experiments will be required to address the issue of whether particles synthesized in a cell culture system are representative of “infectious” virus in plasma.
The ability of several mAbs raised against soluble E2 to neutralize pseudotype infection is encouraging for future vaccine design. The most potently neutralizing mAbs tested mapped to a 15-aa region in E2 (amino acids 432–447), which displays some variability between different strains. The HVR has been proposed to be a neutralization epitope (29). It is interesting to note that four mAbs (7/59, 6/82a, 6/16, and 9/86a) to epitopes within the HVR failed to neutralize infectivity, whereas mAb 9/27, specific for amino acids 396–407 within the C-terminal region of the HVR, did neutralize infectivity. No association was found between the ability of a mAb to neutralize HIV–HCV pseudotype infectivity and block soluble E2–CD81 interaction, suggesting that the mechanism of neutralization may be independent of CD81 interaction(s). However, conformational differences may exist between soluble forms of E2 and that present on pseudotype particles. Both of the HVR-specific mAbs, 6/16 and 9/27, were able to recognize 293-T cell-surface-expressed E2 and yet demonstrated different neutralization profiles (Table 2). Future experiments should address the antigenic conformation of the gps on infectious particles compared with soluble E2.
In summary, HIV pseudotypes expressing native HCV gps have allowed us to identify cell types that can support HCV gp-dependent virus entry. Future experiments should aim to identify cell types within the liver that can support HCV entry and the cellular molecules mediating such entry. The potential to measure neutralizing Abs will allow researchers to address the role of the humoral immune response in HCV infection. Understanding this process will be important in the development of new antiviral therapeutics targeting these early steps in the HCV life cycle.
Acknowledgments
We thank Peter Balfe, Lynn Dustin, Brett Lindenbach, Ivo Lorenz, Peggy MacDonald, Darius Moradpour, and Tim Tellinghuissen for reading the manuscript and for their helpful comments; Shoshana Levy and Peter Monk for reagents; and David Ho for interest and support. J.Z., M.F., C.L., C.M.R., and J.A.M. are supported by the Greenberg Medical Research Institute and Public Health Service Grants CA57973 and AI40034. C.C.-M. is supported by Public Health Service Grant CA72822. M.H. is a recipient of an American Foundation for AIDS Research fellowship.
Abbreviations: HCV, hepatitis C virus; gp, glycoprotein; ER, endoplasmic reticulum; LDLR, low-density lipoprotein receptor; SR-B1, scavenger receptor class B type 1; DC-SIGN, dendritic cell-specific intercellular adhesion molecule 3 grabbing nonintegrin; VSV, vesicular stomatitis virus; SIN, Sindbis virus; RLU, relative light unit; HVR, hypervariable region.
References
- 1.Lerat, H., Rumin, S., Habersetzer, F., Berby, F., Trabaud, M. A., Trepo, C. & Inchauspe, G. (1998) Blood 91, 3841-3849. [PubMed] [Google Scholar]
- 2.Sung, V. M., Shimodaira, S., Doughty, A. L., Picchio, G. R., Can, H., Yen, T. S., Lindsay, K. L., Levine, A. M. & Lai, M. M. (2003) J. Virol. 77, 2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Op De Beeck, A., Cocquerel, L. & Dubuisson, J. (2001) J. Gen. Virol. 82, 2589-2595. [DOI] [PubMed] [Google Scholar]
- 4.Agnello, V., Abel, G., Elfahal, M., Knight, G. B. & Zhang, Q. X. (1999) Proc. Natl. Acad. Sci. USA 96, 12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andre, P., Komurian-Pradel, F., Deforges, S., Perret, M., Berland, J. L., Sodoyer, M., Pol, S., Brechot, C., Paranhos-Baccala, G. & Lotteau, V. (2002) J. Virol. 76, 6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wunschmann, S., Medh, J. D., Klinzmann, D., Schmidt, W. N. & Stapleton, J. T. (2000) J. Virol. 74, 10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pileri, P., Uematsu, Y., Compagnoli, S., Galli, G., Falugi, F., Petracca, R., Weiner, A. J., Houghton, M., Rosa, D., Grandi, G. & Abrignani, S. (1998) Science 282, 938-941. [DOI] [PubMed] [Google Scholar]
- 8.Flint, M., Maidens, C., Loomis-Price, L. D., Shotton, C., Dubuisson, J., Monk, P., Higginbottom, A., Levy, S. & McKeating, J. A. (1999) J. Virol. 73, 6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambot, M., Fretier, S., Op De Beeck, A., Quatannens, B., Lestavel, S., Clavey, V. & Dubuisson, J. (2002) J. Biol. Chem. 277, 20625-20630. [DOI] [PubMed] [Google Scholar]
- 10.Wellnitz, S., Klumpp, B., Barth, H., Ito, S., Depla, E., Dubuisson, J., Blum, H. E. & Baumert, T. F. (2002) J. Virol. 76, 1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clayton, R. F., Owsianka, A., Aitken, J., Graham, S., Bhella, D. & Patel, A. H. (2002) J. Virol. 76, 7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scarselli, E., Ansuini, H., Cerino, R., Roccasecca, R. M., Acali, S., Filocamo, G., Traboni, C., Nicosia, A., Cortese, R. & Vitelli, A. (2002) EMBO J. 21, 5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pohlmann, S., Zhang, J., Baribaud, F., Chen, Z., Leslie, G. J., Lin, G., Granelli-Piperno, A., Doms, R. W., Rice, C. M. & McKeating, J. A. (2003) J. Virol. 77, 4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lozach, P. Y., Lortat-Jacob, H., De Lacroix De Lavalette, A., Staropoli, I., Foung, S., Amara, A., Houles, C., Fieschi, F., Schwartz, O., Virelizier, J. L., et al. (2003) J. Biol. Chem., in press. [DOI] [PubMed]
- 15.Lagging, L. M., Meyer, K., Owens, R. J. & Ray, R. (1998) J. Virol. 72, 3539-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuura, Y., Tani, H., Suzuki, K., Kimura-Someya, T., Suzuki, R., Aizaki, H., Ishii, K., Moriishi, K., Robison, C. S., Whitt, M. A. & Miyamura, T. (2001) Virology 286, 263-275. [DOI] [PubMed] [Google Scholar]
- 17.Buonocore, L., Blight, K. J., Rice, C. M. & Rose, J. K. (2002) J. Virol. 76, 6865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blight, K. J., McKeating, J. A. & Rice, C. M. (2002) J. Virol. 76, 13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeifer, A. M., Cole, K. E., Smoot, D. T., Weston, A., Groopman, J. D., Shields, P. G., Vignaud, J. M., Juillerat, M., Lipsky, M. M., Trump, B. F., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 5123-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flint, M., Thomas, J. M., Maidens, C. M., Shotton, C., Levy, S., Barclay, W. S. & McKeating, J. A. (1999) J. Virol. 73, 6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn, E. R., Chan, C. H., Hadlock, K. G., Foung, S. K., Flint, M. & Levy, S. (2001) Blood 98, 3745-3749. [DOI] [PubMed] [Google Scholar]
- 22.Tan, R. C., Harouse, J. M., Gettie, A. & Cheng-Mayer, C. (1999) J. Med. Primatol. 28, 164-168. [DOI] [PubMed] [Google Scholar]
- 23.Connor, R. I., Chen, B. K., Choe, S. & Landau, N. R. (1995) Virology 206, 935-944. [DOI] [PubMed] [Google Scholar]
- 24.Roberts, P. C., Kipperman, T. & Compans, R. W. (1999) J. Virol. 73, 10447-10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higginbottom, A., Quinn, E. R., Kuo, C. C., Flint, M., Wilson, L. H., Bianchi, E., Nicosia, A., Monk, P. N., McKeating, J. A. & Levy, S. (2000) J. Virol. 74, 3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartosch, B., Dubuisson, J. & Cosset, F. L. (2003) J. Exp. Med. 197, 633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubuisson, J. (2000) Curr. Top. Microbiol. Immunol. 242, 135-148. [DOI] [PubMed] [Google Scholar]
- 28.Skehel, J. J. & Wiley, D. C. (1998) Cell 95, 871-874. [DOI] [PubMed] [Google Scholar]
- 29.Farci, P., Shimoda, A., Wong, D., Cabezon, T., De Gioannis, D., Strazzera, A., Shimizu, Y., Shapiro, M., Alter, H. J. & Purcell, R. H. (1996) Proc. Natl. Acad. Sci. USA 93, 15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]