Abstract
Physiological and ultrastructural evidence indicates that gap junctions link many classes of neurons in mammalian central nervous system (CNS), allowing direct electrical and metabolic communication. Among at least six gap junction-forming connexin proteins in adult rat brain, connexin- (Cx) 32, Cx36, and Cx43 have been reported to occur in neurons. However, no connexin has been documented at ultrastructurally defined neuronal gap junctions. To address this question directly, freeze-fracture replica immunogold labeling (FRIL) and immunofluorescence (IF) were used to visualize the subcellular and regional localization of Cx36 in rat brain and spinal cord. Three antibodies were generated against different sequences in Cx36. By Western blotting, these antibodies detected protein at 36 and 66 kDa, corresponding to Cx36 monomer and dimer forms, respectively. After double-labeling for Cx36 and Cx43 by FRIL, neuronal gap junctions in inferior olive, spinal cord, and retina were consistently immunogold-labeled for Cx36, but none were labeled for Cx43. Conversely, Cx43 but not Cx36 was detected in astrocyte and ependymocyte gap junctions. In >250 Cx32/Cx43 single- and double-labeled replicas from 10 CNS regions, no neuronal gap junctions were labeled for either Cx32 or Cx43. Instead, Cx32 and Cx43 were restricted to glial gap junctions. By IF, Cx36 labeling was widely distributed in neuropil, including along dendritic processes and within neuronal somata. On the basis of FRIL identification of Cx36 in neuronal gap junctions and IF imaging of Cx36 throughout rat brain and spinal cord, neuronal gap junctions containing Cx36 appear to occur in sufficient density to provide widespread electrical and metabolic coupling in adult CNS.
Gap junctions are composed of hexamers of “connexin” proteins that bond head-to-head across the extracellular space, thereby forming channels that permit intercellular exchange of ions and small molecules (1–3). In nervous tissue, gap junctions are a structural correlate of electrical synapses, and evidence has accumulated for the existence of such synapses throughout the mammalian central nervous system (CNS) (4–10). In addition, gap junctions too small to be seen by conventional thin section electron microscopy have been described at freeze-fractured “mixed” (chemical plus electrical) synapses in mammalian spinal cord (10). Indeed, on the basis of electrophysiological, dye-coupling, and ultrastructural analyses, virtually all major CNS structures from retina and olfactory bulb to spinal cord and primary sensory ganglia have been suggested to contain neurons linked by gap junctions (9). The functional significance of these electrical and mixed synapses in adult mammalian CNS has been difficult to assess because of the absence of practical methods for documenting, quantifying, or mapping the distribution of neuronal gap junctions. In addition, there has been substantial uncertainty concerning the identity of connexins that form neuronal gap junctions. For example, the consensus glial connexins Cx32 and Cx43 (11–19) also were reported to be expressed in neurons of adult rat or mouse (11, 20–25). However, neither of these connexins has been demonstrated to occur at ultrastructurally defined gap junctions in unambiguously identified neurons, which is essential for designating connexins as neuronal (13, 15).
Among six members of the connexin family identified in CNS tissues, Cx36 and its homologs Cx35 and Cx34.7 in fish have recently emerged as prime candidates for neuronal connexins (26–29). Cx36 was identified by reverse transcription–PCR of mRNA extracted from rat inferior olive (IO), then cloned from a mouse genomic library (26), and subsequently confirmed to be highly expressed in neural tissue, particularly retina (30). In situ hybridization revealed Cx36 mRNA in neuronal cell bodies in various regions of rat brain, and neuronal lesions of IO depleted it of Cx36, further suggesting a neuronal source of Cx36 mRNA (26).
To investigate Cx36 protein localization in neural tissue of adult rat, antibodies (Abs) against Cx36 were developed and used to localize Cx36 within individual gap junction plaques using freeze-fracture replica immunogold labeling (FRIL) (31–33). In addition, FRIL was used for single- and double-labeling of Cx32, Cx43, and Cx36 to determine the ultrastructural localization of the former two connexins in relation to that of Cx36. These three connexins are now documented to occur separately in each of three specific cell types and, in the case of Cx43, in two types of macroglial cells. Our data reveal that of these three connexins, only Cx36 is present in gap junctions of ultrastructurally defined neurons. An abstract of these results has been presented.**
Materials and Methods
Cx36 Abs and Western Blotting.
Three anti-Cx36 Abs were generated in rabbits immunized with keyhole limpet hemocyanin-conjugated synthetic peptides and then affinity-purified. Ab298 was against a peptide corresponding to amino acids 298–318 in Cx36. Ab51-6200 was against a cytoplasmic loop domain, and Ab51-6300 was against the carboxyl terminus. These latter two Abs are commercially available from Zymed. Brain tissues from male Sprague–Dawley rats (300–350 g) were homogenized in Hepes buffer [20 mM Hepes, pH 7.2/0.1 M NaCl/2 mM EDTA/1 mM phenylmethylsulfonyl fluoride (PMSF)] and centrifuged. Pellets were treated with 1% Nonidet P-40 (Igepal CA-630; Sigma), centrifuged, and resuspended in Hepes buffer. Total protein was determined with the Bio-Rad DC protein assay. Western blotting was conducted as described (12), with transblotted membranes processed by enhanced chemiluminescence. For analysis of specificity by peptide preadsorption, Ab (3 μg) was diluted in 100 μl of Tris-buffered saline, pH 7.4, alone or in combination with 100 μg of Cx36 peptide, and incubated overnight at 4°C before final dilution for use in primary solutions (1 μg/ml).
Freeze-Fracture and Immunogold Labeling.
Twenty-six male and female Sprague–Dawley rats (145–585 g) were deeply anesthetized and fixed for 3–10 min via transcardiac perfusion with 1% or 0.1% formaldehyde in 0.15 M Sorenson's phosphate buffer. More than 250 Vibratome slices (150 μm thick) from seven regions of adult rat brain (IO, hippocampus, suprachiasmatic nucleus, supraoptic nucleus, paraventricular nucleus, cerebellum, and retina), and from spinal cord were freeze-fractured and replicated. A gold “index” grid was bonded to the frozen sample by using Lexan plastic dissolved in dichloroethane; the samples were thawed and “grid-mapped” by confocal microscopy (10, 34), and cellular material was removed by vigorous washing with SDS detergent (ref. 31; as modified in ref. 33).
For FRIL, residual connexin proteins adhering to the replica after SDS washing (Fig. 1) were single- or double-labeled using combinations of primary monoclonal and polyclonal anti-connexin Abs, followed by secondary labeling with anti-rabbit, anti-mouse, and anti-sheep Abs conjugated to 10 and 20 nm gold (33). For Cx36, only Ab298 was used for FRIL experiments. Monoclonal and rabbit polyclonal anti-Cx43 Abs were from Chemicon (Temecula, CA); mAbs to Cx32 were from Chemicon and Zymed; rabbit polyclonal Abs to Cx32 were from Chemicon, Sigma, and Zymed; and sheep anti-Cx32 was from Biogenesis (Poole, United Kingdom). Secondary Abs (10- and 20-nm immunogold goat anti-rabbit, goat anti-mouse, and donkey anti-sheep) were from Chemicon (now obtainable from Jackson Laboratories, Westgrove, PA.).
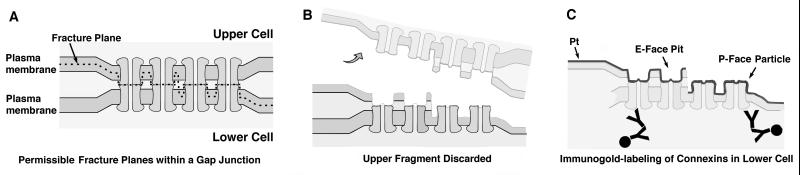
Figure 1.
Diagram of the SDS-FRIL technique. (A and B) Gap junction before A and after B freeze-fracturing, with dashed line indicating the two permissible fracture planes within the lipid bilayers. Regardless of whether the fracture plane splits the plasma membrane of the upper cell, the lower cell, or jumps from one cell to the other, all connexons of the upper gap junction plaque remain with the upper cell and all connexons of the lower plaque remain with the lower cell. (C) After replication of E- and P-faces, the samples are thawed and washed with SDS detergent, leaving a small amount of membrane proteins adhering beneath both E-face pits and P-face particles. The platinum (Pt) replica delineates the outer leaflet (membrane E-face) of the upper cell and the inner leaflet (P-face) of the lower cell. Residual connexin proteins are labeled by primary Abs (Y) and detected by labeling with gold-conjugated, species-specific secondary Abs (Y with ●). For reasons that are not yet clear, labeling is usually greater at the periphery of gap junctions (see Figs. 3 and 4).
Using a JEOL 2000 EX-II transmission electron microscope, we photographed replicas as stereoscopic pairs to distinguish specific labeling (occurring on tissue-side only) from nonspecific labeling [>90% on the nontissue/Lexan side (33)]. Freeze-fractured neurons, glial cells, and vascular endothelia were identified according to 24 criteria (see table 2 in ref. 35). Nerve terminals and dendrites were identified by the presence of >25 uniform-diameter synaptic vesicles in the cytoplasm, distinctive clusters of E- or P-face intramembrane particles (IMPs) that correspond in location to postsynaptic densities seen in conventional thin sections, specialized appositions (“active zones”) shared between dendrite and one or more nerve terminals, and absence of glial markers. Astrocytes were identified by aquaporin 4 square arrays in the plasma membranes, distinctive glial fibrillary acidic protein (GFAP) filaments in the cytoplasm, and high density of IMPs in their plasma membranes, particularly in E-faces (35–38). Oligodendrocytes were identified by low density of IMPs in both E- and P-faces, distinctive “reciprocal patches” of mixed IMPs and pits in both E- and P-faces, and unique “necklaces” of IMPs surrounding gap junction E-faces (35, 37). Ependymocytes lined the third ventricle and spinal central canal, and microvilli projected from their apical surfaces (35, 36).
Immunofluorescence (IF).
Anesthetized rats were perfused transcardially with 50 ml of 50 mM sodium phosphate buffer (PB), pH 7.2, containing 0.9% saline (PBS), followed by perfusion with 4% formaldehyde in PB. Cryostat sections (3–15 μm) were collected on gelatin-coated slides and incubated for 16 h at 4°C with anti-Cx36 Abs diluted 1:500 in PBS containing 0.3% Triton X-100 (PBST). Sections were washed in PBST, incubated for 1.5 h with Cy3-conjugated donkey anti-rabbit IgG diluted 1:200 in PBST, washed, air-dried, and coverslipped.
Results
Ab Characterization.
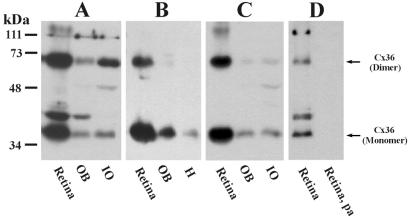
Western blots with three sequence-specific Abs to Cx36 are shown in Fig. 2. Ab298 (Fig. 2A), Ab51-6200 (Fig. 2B), and Ab51-6300 (Fig. 2C) detect monomeric Cx36 migrating at 36 kDa in homogenates of retina, olfactory bulb, and IO. In each tissue, the dimeric form of Cx36 migrating at 66 kDa was recognized by Ab298 and to various degrees by the other two Abs. An additional unidentified protein migrating at 40 kDa was recognized by Ab298 in retina and olfactory bulb, but was absent in IO, and was not recognized by the other Abs. After omission of primary Ab, blots probed with secondary Ab were devoid of all signal (not shown) except for a nonspecific band at 48 kDa in IO. Preadsorption of Ab298 (Fig. 2D) and Ab51-6300 (not shown) with synthetic peptide antigen eliminated detection of Cx36 as well as the band detected by Ab298 at 40 kDa.
Figure 2.
Western blots of tissues probed with anti-Cx36 Abs generated against three different sequences within Cx36. (A–C) Three blots containing homogenates of retina, olfactory bulb (OB), and IO probed with Ab298 (A), Ab51-6200 (B), and Ab51-6300 (C). The Abs detected Cx36 monomer migrating at 36 kDa in all three tissues. A Cx36 dimer migrating at 66 kDa also was detected to various degrees. The band at 48 kDa was seen after omission of primary Ab (not shown) and is nonspecific. Ab298 detected an unidentified protein at 40 kDa. (D) Recognition of Cx36 and the cross-reacting protein at 40 kDa (retina) was eliminated after preadsorption of Ab298 with synthetic peptide antigen (retina, pa).
Detection of Neuronal and Glial Connexins by FRIL.
Neurons and glia in selected areas of brain and spinal cord were examined in >165 replicas labeled for Cx43, 88 replicas labeled for Cx32, and 8 replicas double-labeled for Cx32/Cx43. Neurons and glia also were examined in 5 replicas of IO, 1 of spinal cord, and 4 of retina that were double-labeled for Cx36/Cx43, and in 1 sample of IO that was single-labeled for Cx36.
In Cx36/Cx43 double-labeled replicas, stereoscopic imaging revealed Cx36 immunogold beneath 24 of 26 neuronal gap junctions in IO (Fig. 3 A–D), beneath 10 of 10 in retina (Fig. 3E), and beneath 2 of 2 in spinal cord (Fig. 4A). No neuronal gap junctions were labeled for Cx43. In the same replicas, >90% of astrocyte gap junctions were labeled for Cx43 (Fig. 4B), but none were labeled for Cx36. The probabilistic nature of immunogold labeling of SDS-washed samples, combined with typical labeling efficiencies of approximately 1:40, occasionally resulted in unlabeled gap junctions (33), particularly those containing fewer than 50 connexons (see Fig. 4B for labeled and unlabeled junctions in the same astrocyte).
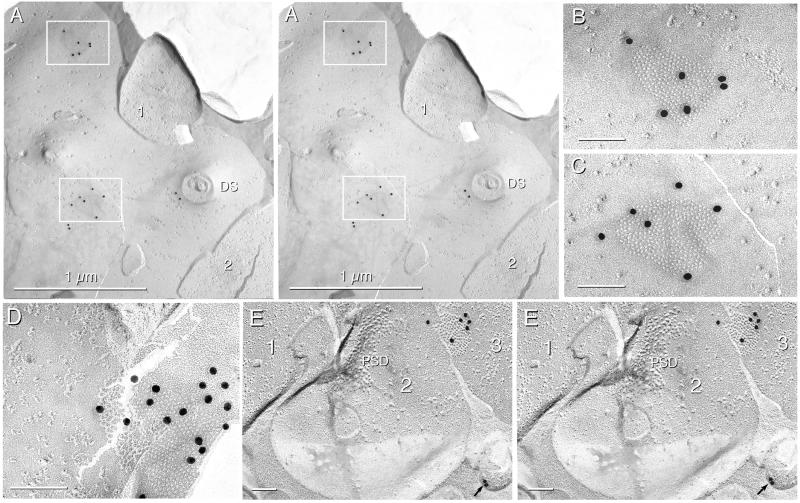
Figure 3.
FRIL immunogold localization of Cx36 (20-nm gold) in rat IO and retina. (A) Stereoscopic image of two Cx36-labeled gap junctions in a large area of dendrite E-face. Two (of five) nerve terminals are numbered. Two nonspecific gold beads are at the base of a cross-fractured dendritic spine (DS). (B and C) Magnified images of gap junctions in inscribed areas of A. Six 20-nm gold beads localizing Cx36 are beneath both gap junction E-faces. (D) Gap junction in IO labeled for Cx36 by more than one dozen gold beads. (E) Stereoscopic image of three linked nerve terminals in retina, with two linked at a chemical synapse (1 and 2) and two connected by gap junctions (2 and 3). The nerve terminal on the left (1) is presynaptic to the middle terminal (2), as identified by the postsynaptic density (PSD). Two nonspecifically bound gold beads are on the Lexan side of the replica (arrow). In FRIL images, all calibration bars are 0.1 μm unless otherwise indicated.
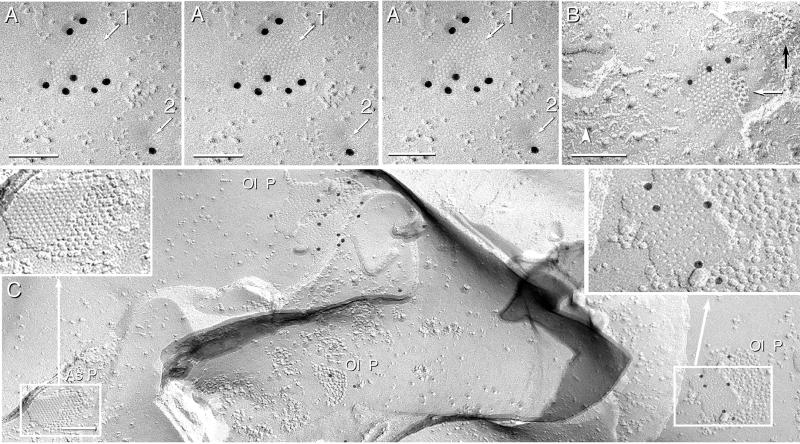
Figure 4.
Images from spinal cord labeled for Cx32 and Cx43 and from cerebellum labeled for Cx32. (A) Stereoscopic view (left and center images) and intaglio view (center and right images) of two Cx36-labeled neuronal gap junctions in spinal cord. The larger junction (arrow 1) and the smaller junction [four or five connexon imprints (arrow 2)] are labeled with 20-nm gold beads. The intaglio perspective reveals the “sidedness” of labeling and the topological deformation of membranes at gap junction close contacts. (B) Two astrocyte gap junctions in the same replica as A. One plaque has three 10-nm gold beads (white arrow), and one is unlabeled (black arrow). Aquaporin 4 square array (arrowhead) is a definitive marker for astrocytes (38). (C) Three gap junctions in oligodendrocyte P-faces (Ol P) in cerebellum are labeled for Cx32 by 12, 6, and 2 gold beads. One Cx32-labeled oligodendrocyte gap junction is enlarged (Right Inset). Nearby astrocyte gap junction (As P, and Left Inset) is not labeled for Cx32.
Two gap junctions were observed in each of three nerve terminals (two in IO and one in spinal cord), and five of those six gap junctions were labeled for Cx36 (Figs. 3 A–C and 4A), demonstrating that multiple gap junctions in the same neuronal processes contained Cx36. Although no protein is replicated within E-face pits, E-faces are labeled and often with higher efficiency than P-faces (Figs. 3A–D and 4A–C). The basis for this seeming contradiction is illustrated in Fig. 1. Overall, the 36 Cx36-labeled and two unlabeled neuronal gap junctions had a total of 259 gold beads beneath ≈9,800 connexons [average, 258 connexons and 7.2 gold beads per gap junction, or 1 gold bead per 38 connexons, which is approximately the same as reported for other connexins (31, 32, 38, 39)]. Interestingly, the 24 labeled and 2 unlabeled gap junctions in electrical synapses of IO had an average of 247 connexons, which is 5 times greater than the average number of connexons in gap junctions at mixed synapses in spinal cord (10).
To test Cx32 and Cx43 as possible neuronal vs. glial connexins, >200 single- and double-labeled replicas from throughout brain and spinal cord were examined. In these replicas, >3,000 astrocyte gap junctions (Fig. 4B) and >100 ependymocyte gap junctions were labeled for Cx43 (also see refs. 13, 15, and 33). However, no Cx43-labeled neuronal gap junctions were detected, even as “false positives” (defined and illustrated in ref. 33). Cx32 was found in >100 oligodendrocyte gap junctions (Fig. 4C) but was never detected in neuronal gap junctions (13, 15, 33) or in astrocyte (Fig. 4C) or ependymocyte gap junctions. However, more than one dozen unlabeled neuronal gap junctions were found in the same samples (negative data not shown).
IF Labeling.
On the basis of the above results, light microscopy (LM) immunolabeling was used to examine neuronal localization of Cx36 protein and to investigate the extent of its distribution in adult brain and spinal cord. Regionally and qualitatively similar punctate IF staining was obtained with all three Cx36 Abs used in this initial study; however, only images obtained with Ab51-6200 are presented here.
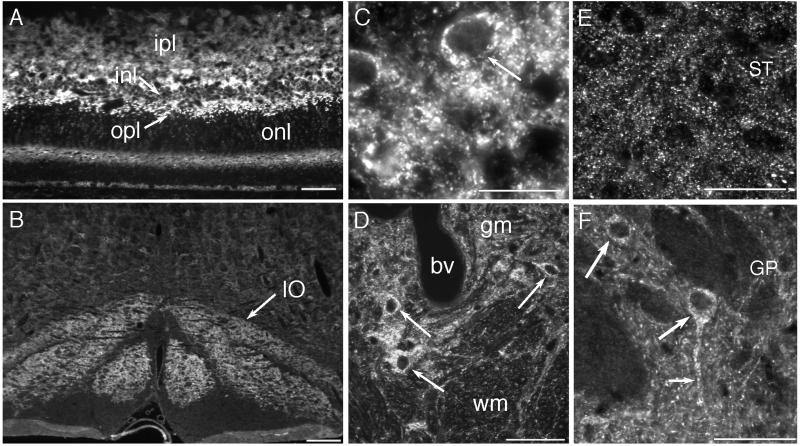
In the retina, Cx36-labeling was intense and stratified. Relatively large and brilliant neuronal puncta were seen within the outer plexiform layer (Fig. 5A), whereas a more extensive field of fine puncta typified labeling of the inner plexiform layer (Fig. 5A). Cx36 IF was densely distributed in the IO, which showed much higher Cx36-immunoreactivity than other brainstem regions at this level of section (Fig. 5B). Labeling in the IO consisted of puncta within the neuropil, as well as IF within neuronal somata (Fig. 5C), thereby providing data not obtainable by the membrane labeling methods of FRIL. In the spinal cord, punctate Cx36 IF was much more evident in gray matter than in white matter, and both the dorsal and ventral horn showed moderate immunoreactivity, paralleling observations of neuronal gap junctions in conventional freeze-fracture replicas (10). As had been noted in the IO, perikaryal staining was seen in large neurons within the ventral horn (Fig. 5D, arrows). In two areas not investigated by FRIL, the striatum and globus pallidus (Fig. 5 E and F), distinctly different subcellular distributions were seen, with a preponderance of punctate staining in the neuropil of the striatum vs. a preponderance of cytoplasmic labeling of neurons in globus pallidus. IF data showed a similarly striking regional and cellular heterogeneity in the distribution of Cx36 throughout many other brain regions, notably the hippocampus, thalamus, and brainstem.
Figure 5.
Distribution of Cx36 in rat retina, IO, and spinal cord by IF labeling. (A) The retina displays Cx36-IF in both the inner and outer plexiform layers (ipl and opl), with only moderate IF in the inner nuclear layer (inl) and very little IF in the outer nuclear layer (onl). Prominent IF is seen in the outer part of the opl (arrow). (B) The IO is delineated by more intense Cx36-IF than other brainstem regions (arrow). (C) The neuropil of the IO contains fine punctate immunolabeling and Cx36-IF within individual olivary neurons (arrow). (D) Cx36-IF is widely distributed in spinal cord gray matter (gm) and very sparse within white matter (wm). Cx36-IF also is seen within individual ventral horn neurons (arrows). bv, Blood vessel. (E) The striatum (ST) exhibits a greater density of Cx36-IF than the globus pallidus (GP). (F) In globus pallidus, Cx36-IF is seen within neuronal somata (large arrows) and along dendrites (small arrow). (Calibration bars: A, D, E, and F = 50 μm; B = 200 μm; and C = 20 μm.)
Discussion
FRIL analysis of three regions of adult rat CNS revealed that Cx36 is present in gap junctions linking neurons. In contrast, neither Cx32 nor Cx43 was detectable in neuronal gap junctions in any of seven regions of brain or three regions of spinal cord. These results indicate that: (i) neuronal gap junctions contain Cx36, but not Cx32 or Cx43; (ii) astrocyte gap junctions contain Cx43, but not Cx36 or Cx32; and (iii) oligodendrocyte gap junctions contain Cx32, but not Cx36 or Cx43. The present FRIL data are consistent with our previous estimates of >1,000-fold more glial than neuronal gap junctions in CNS tissue (15, 35). In comparison with freeze-fractured astrocyte junctions, where 10–50 gap junctions were commonly seen in single grid openings, neuronal gap junctions were encountered, on average, in ≈1/100 grid openings (10, 15). However, in the broader views afforded by LM, widespread IF labeling for Cx36 was seen in neuropil, in neuronal cell bodies, and along dendrites.
Our Western blotting results extend earlier reports (26, 30) of widespread Cx36 mRNA expression in rodent neural tissue by demonstrating Cx36 protein in homogenates from several CNS regions. Detection of similar protein bands with three Abs directed against different sites in Cx36, in combination with FRIL localization of one of those Abs to ultrastructurally defined neuronal gap junction plaques, leaves little possibility of protein misidentification. Moreover, the high levels of Cx36 protein expression in retina are consistent with the abundance of Cx36 mRNA reported in that tissue (30). More generally, detection of Cx36 on Western blots of several brain regions confirms and extends data obtained for Cx36 expression by in situ hybridization (26).
Previous studies, using LM methods, have reported expression of Cx32 and Cx43 protein and/or mRNA in neurons (11, 16, 20, 21, 24, 25, 40–45). However, inherent limits of resolution of LM preclude assignment to structures smaller than 0.3 μm. Astrocyte and oligodendrocyte processes, which are well established to contain Cx43 and Cx32 (13, 16–19, 24), often are thinner than 0.2 μm. Where those thin processes follow the contours of neurons, it would be difficult to distinguish whether connexin IF is localized to oligodendrocyte, astrocyte, or neuronal plasma membranes.
In this study, no evidence for Cx32 or Cx43 in neuronal gap junctions was found in >250 single- and double-labeled FRIL replicas. To date we have evaluated >3,000 labeled glial gap junctions (this study), >1,000 unlabeled glial gap junctions (35), >100 unlabeled neuronal gap junctions (10, 15), and 36 (of 38) labeled neuronal gap junctions (this study) in 10 regions of brain and spinal cord. In this sampling, no evidence was found for gap junctions between glial cells and neurons. These results and our previous estimates of 1,000-fold greater number of glial than neuronal gap junctions contrast with reports that 17.9% of all gap junctions in adult rat cortex represent neuronal/glial junctions and that neuronal/glial gap junctions contain Cx32 and Cx43 (21). Thus, it is possible that either gap junctions or cells linked by gap junctions were misidentified in previous reports, or alternatively, that putative neuronal/glial gap junctions were not recognized in our FRIL replicas, and if they exist, that they use connexins other than Cx32, Cx36, or Cx43.
With increasing evidence for multiple connexins in individual gap junction plaques in diverse cell types (19, 39), our data do not exclude the possibility that neuronal gap junctions contain additional as-yet-unidentified connexins that may participate in the formation of novel gap junctions. Also requiring further analyses are the subcellular sites to which neuronally expressed connexins are targeted. These may be localized at, e.g., typical electrical synapses that have been described in mammalian brain (reviewed in ref. 9) or at mixed synapses in spinal cord (10, 15). FRIL methods should be of value in assessing each of those issues. In FRIL, each gold bead acts as an independently targeted probe. Thus, multiple gold beads on the same gap junction plaque provide multiple independent confirmations of target connexins in individual gap junction plaques of identified cells (33). Despite its relatively low labeling efficiency (≈1 gold bead per 40 connexins), FRIL is a highly specific technique for “flagging” even the smallest gap junctions, identifying the constituent connexins, and identifying the coupled cells (15, 31, 33).
Connexin immunostaining by LM usually appears punctate in most tissue (9, 13), and those puncta have been presumed to reflect gap junctions. Although the degree to which Cx36-positive puncta correspond to neuronal gap junctions will require correlative LM-transmission electron microscopy analysis, confirmation of such a correspondence would indicate that neurons in diverse areas of mammalian brain are relatively richly invested with gap junctions. Intracellular labeling for Cx36 also was seen, and this may be fortunate as it will enable LM correlation of Cx36-positive neurons with appropriate transmitter markers, just as FRIL will allow colocalization of Cx36 and neurotransmitter receptors within individual mixed synapses (unpublished results). As with other proteins expressed by neurons, only a portion of neurons expressing Cx36 may show recognizable levels of the protein in the perikaryon. For example, both Cx36-positive puncta and neuronal somata were seen in the globus pallidus, whereas Cx36-positive somata were not seen in the striatum, which nevertheless contained a high density of puncta. As one possibility, these cytoplasmic differences may reflect high vs. low rates of connexin turnover through cytomembrane pathways.
The present FRIL and IF data, as well as in situ hybridization results (26) suggest that adult mammalian CNS contains sufficient neuronal gap junctions to provide structural pathways for widespread electrical and/or metabolic integration between neurons. Our identification of Cx36 in ultrastructurally defined neuronal gap junction plaques provides the basis for obtaining detailed immunohistochemical maps of the entire CNS to establish the regional and cellular densities of neuronal Cx36. Such maps will be of value in illuminating areas in which to focus functional studies of the roles of neuronal gap junctions in synaptic integration and metabolic coupling in the CNS.
Acknowledgments
We thank K. Mandelic and B. Tinner for outstanding technical assistance. This work was supported by National Institutes of Health Grants NS-31027 and NS-39040 (to J.E.R.) and MH-59995 (to F. E. Dudek, Colorado State University), and by grants from the Medical Research Council of Canada (to J.I.N. and to W.A.S.). Additional higher-resolution images related to this report are available at: http://microscopy.cvmbs.colostate.edu/imagefiles/rash/.
Abbreviations
- Cx
connexin
- IF
immunofluorescence
- CNS
central nervous system
- FRIL
freeze-fracture replica immunogold labeling
- Ab
antibody
- IO
inferior olive
- IMP
intramembrane particle
- LM
light microscopy
Footnotes
Rash, J. E., Yasumura, T., Staines, W. A., Patel, D. & Nagy, J. I. (1999) Mol. Biol. Cell, 10, 404a.
References
- 1.Goodenough D A, Goliger J A, Paul D L. Annu Rev Biochem. 1996;65:475–502. doi: 10.1146/annurev.bi.65.070196.002355. [DOI] [PubMed] [Google Scholar]
- 2.Gilula N B, Reeves O R, Steinbach A. Nature (London) 1972;235:262–265. doi: 10.1038/235262a0. [DOI] [PubMed] [Google Scholar]
- 3.Bruzzone R, White T W, Paul D L. Eur J Biochem. 1996;238:1–27. doi: 10.1111/j.1432-1033.1996.0001q.x. [DOI] [PubMed] [Google Scholar]
- 4.Bennett M V L. In: Electrical Transmission: A Functional Analysis and Comparison with Chemical Transmission. Kandel E R, editor. Bethesda, MD: Am. Physiol. Soc.; 1977. pp. 357–416. [Google Scholar]
- 5.Bennett M V L, Goodenough D A. Neurosci Res Prog Bull. 1978;16:373–486. [PubMed] [Google Scholar]
- 6.Bennett M V L. J Neurocytol. 1997;26:349–366. doi: 10.1023/a:1018560803261. [DOI] [PubMed] [Google Scholar]
- 7.Llinas R, Baker R, Sotelo C. J Neurophysiol. 1974;37:560–571. doi: 10.1152/jn.1974.37.3.560. [DOI] [PubMed] [Google Scholar]
- 8.Sotelo C, Korn H. Int Rev Cytol. 1978;55:67–107. doi: 10.1016/s0074-7696(08)61887-2. [DOI] [PubMed] [Google Scholar]
- 9.Nagy J I, Dermietzel R. In: Advances in Molecular and Cell Biology. Hertzberg E L, editor. Vol. 30. New York: JAI Press; 2000. pp. 323–396. [Google Scholar]
- 10.Rash J E, Dillman R K, Bilhartz B L, Duffy H S, Whalen L R, Yasumura T. Proc Natl Acad Sci USA. 1996;93:4235–4239. doi: 10.1073/pnas.93.9.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dermietzel R, Traub O, Hwang T K, Beyer E, Bennett M V L, Spray D C, Willecke K. Proc Natl Acad Sci USA. 1989;86:10148–10152. doi: 10.1073/pnas.86.24.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Hertzberg E L, Nagy J I. J Comp Neurol. 1997;379:571–591. doi: 10.1002/(sici)1096-9861(19970324)379:4<571::aid-cne8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Nagy J I, Rash J E. Brain Res Rev. 2000;32:29–44. doi: 10.1016/s0165-0173(99)00066-1. [DOI] [PubMed] [Google Scholar]
- 14.Ochalski P A Y, Frankenstein U N, Hertzberg E L, Nagy J I. Neuroscience. 1997;76:931–945. doi: 10.1016/s0306-4522(96)00394-6. [DOI] [PubMed] [Google Scholar]
- 15.Rash J E, Yasumura T, Dudek F E. Cell Biol Int. 1998;22:731–749. doi: 10.1006/cbir.1998.0392. [DOI] [PubMed] [Google Scholar]
- 16.Dermietzel R, Spray D C. Trends Neurosci. 1993;16:186–192. doi: 10.1016/0166-2236(93)90151-b. [DOI] [PubMed] [Google Scholar]
- 17.Dermietzel R, Farooq M, Kessler J A, Hertzberg E L, Spray D C. Glia. 1997;20:101–114. [PubMed] [Google Scholar]
- 18.Yamamoto T, Ochalski A, Hertzberg E L, Nagy J I. J Comp Neurol. 1990;302:853–883. doi: 10.1002/cne.903020414. [DOI] [PubMed] [Google Scholar]
- 19.Nagy J I, Patel D, Ochalski P A Y, Stelmack G L. Neuroscience. 1999;88:447–468. doi: 10.1016/s0306-4522(98)00191-2. [DOI] [PubMed] [Google Scholar]
- 20.Micevych P E, Abelson L. J Comp Neurol. 1991;305:96–118. doi: 10.1002/cne.903050110. [DOI] [PubMed] [Google Scholar]
- 21.Nadarajah B, Thomaidou D, Evans W H, Parnavelas J G. J Comp Neurol. 1996;376:326–342. doi: 10.1002/(SICI)1096-9861(19961209)376:2<326::AID-CNE13>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 22.Micevych P E, Popper P, Hatton G I. Neuroendocrinology. 1996;63:39–45. doi: 10.1159/000126933. [DOI] [PubMed] [Google Scholar]
- 23.Bennett M V L, Barrio L C, Bargiello T A, Spray D C, Hertzberg E L, Saez J Z. Neuron. 1991;6:305–320. doi: 10.1016/0896-6273(91)90241-q. [DOI] [PubMed] [Google Scholar]
- 24.Dermietzel R. Cell Biol Int. 1998;22:719–730. doi: 10.1006/cbir.1999.0393. [DOI] [PubMed] [Google Scholar]
- 25.Dermietzel R, Spray D C. Glia. 1998;24:1–7. doi: 10.1002/(sici)1098-1136(199809)24:1<1::aid-glia1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 26.Condorelli D F, Parenti R, Spinella F, Salinaro A T, Belluardo N, Cardile V, Cicirata F. Eur J Neurosci. 1998;10:1202–1208. doi: 10.1046/j.1460-9568.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- 27.O'Brien J, Al-Ubaidi M R, Ripps H. Mol Biol Cell. 1996;7:233–243. doi: 10.1091/mbc.7.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Brien J, White T W, Al-Ubaidi M R, Ripps H. J Neurosci. 1998;18:7625–7637. doi: 10.1523/JNEUROSCI.18-19-07625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srinivas M, Rozental R, Kojima T, Dermietzel R, Mehler M, Condorelli D F, Kessler J A, Spray D C. J Neurosci. 1999;19:9848–9855. doi: 10.1523/JNEUROSCI.19-22-09848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sohl G, Degen J, Teubner B, Willecke K. FEBS Lett. 1998;428:27–31. doi: 10.1016/s0014-5793(98)00479-7. [DOI] [PubMed] [Google Scholar]
- 31.Fujimoto K. J Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- 32.Fujimoto K. Histochem Cell Biol. 1997;107:87–96. doi: 10.1007/s004180050092. [DOI] [PubMed] [Google Scholar]
- 33.Rash J E, Yasumura T. Cell Tissue Res. 1999;296:307–321. doi: 10.1007/s004410051291. [DOI] [PubMed] [Google Scholar]
- 34.Rash J E, Dillman R K, Morita M, Whalen L R, Guthrie P B, Fay-Guthrie D, Wheeler D W. In: Rapid Freezing, Freeze Fracture, and Deep Etching. Severs N J, Shotton D M, editors. New York: Wiley; 1995. pp. 127–150. [Google Scholar]
- 35.Rash J E, Duffy H S, Dudek F E, Bilhartz B L, Whalen L R, Yasumura T. J Comp Neurol. 1997;388:265–292. doi: 10.1002/(sici)1096-9861(19971117)388:2<265::aid-cne6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 36.Peters A, Palay S L, Webster H D. The Fine Structure of the Nervous System, Neurons and Their Supporting Cells. New York: Oxford Univ. Press; 1991. [Google Scholar]
- 37.Massa P T, Mugnaini E. Neuroscience. 1982;7:523–538. doi: 10.1016/0306-4522(82)90285-8. [DOI] [PubMed] [Google Scholar]
- 38.Rash J E, Yasumura T, Hudson C S, Agre P, Nielsen S. Proc Natl Acad Sci USA. 1998;95:11981–11986. doi: 10.1073/pnas.95.20.11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Severs N J. Novartis Found Symp. 1999;219:188–206. doi: 10.1002/9780470515587.ch12. [DOI] [PubMed] [Google Scholar]
- 40.Chang Q, Gonzalez M, Pinter M J, Balice-Gordon R J. J Neurosci. 1999;19:10813–10828. doi: 10.1523/JNEUROSCI.19-24-10813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nadarajah B, Parnavelas J G. Novartis Found Symp. 1999;219:157–170. doi: 10.1002/9780470515587.ch10. [DOI] [PubMed] [Google Scholar]
- 42.Nadarajah B, Jones A M, Evans W H, Parnavelas J G. J Neurosci. 1998;17:3096–3111. doi: 10.1523/JNEUROSCI.17-09-03096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto A, Arai Y, Urano A, Hyodo S. Neurosci Res. 1992;14:133–144. doi: 10.1016/0168-0102(92)90089-u. [DOI] [PubMed] [Google Scholar]
- 44.Fisher R S, Micevych P E. In: Progress in Cell Research: Gap Junctions. Hall J E, Zampighi G A, Davis R M, editors. Vol. 3. New York: Elsevier; 1993. pp. 141–148. [Google Scholar]
- 45.Simburger E, Stang A, Kremer M, Dermietzel R. Histochem Cell Biol. 1997;107:127–137. doi: 10.1007/s004180050097. [DOI] [PubMed] [Google Scholar]