Abstract
Treatment of hyperthyroidism, a common clinical condition that can have serious manifestations in the elderly, has remained essentially unchanged for >30 years. Directly antagonizing the effect of the thyroid hormone at the receptor level may be a significant improvement for the treatment of hyperthyroid patients. We built a computer model of the thyroid hormone receptor (TR) ligand-binding domain in its predicted antagonist-bound conformation and used a virtual screening algorithm to select 100 TR antagonist candidates out of a library of >250,000 compounds. We were able to obtain 75 of the compounds selected in silico and studied their ability to act as antagonists by using cultured cells that express TR. Fourteen of these compounds were found to antagonize the effect of T3 on TR with IC50s ranging from 1.5 to 30 μM. A small virtual library of compounds, derived from the highest affinity antagonist (1-850) that could be rapidly synthesized, was generated. A second round of virtual screening identified new compounds with predicted increased antagonist activity. These second generation compounds were synthesized, and their ability to act as TR antagonists was confirmed by transfection and receptor binding experiments. The extreme structural diversity of the antagonist compounds shows how receptor-based virtual screening can identify diverse chemistries that comply with the structural rules of TR antagonism.
Overproduction of thyroid hormone (hyperthyroidism or thyrotoxicosis) is an extremely common clinical entity caused by a number of different pathological conditions of the thyroid gland. Approximately 0.5% of women will experience some clinical manifestation of hyperthyroidism in their lifetime (3–5 times more frequent than men), with potentially life-threatening effects on the cardiovascular system (e.g., cardiac arrhythmias, heart failure, angina, and myocardial infarction), particularly in the elderly (1–3).
The treatment of hyperthyroidism has essentially remained unchanged for the past 30 years and includes the use of radioactive [131I]iodine, surgery, or the use of antithyroid drugs, such as propylthiouracil, that inhibit thyroid hormone synthesis by blocking the iodination of thyroglobulin (1–3). Each approach has its own intrinsic limitations and/or side effects. Propylthiouracil and related drugs, which block thyroid hormone synthesis, act slowly and can take up to 6–8 weeks to fully deplete the thyroid gland and intrathyroidal stores of iodinated thyroglobulin, during which time hyperthyroidism can have severe consequences in certain individuals. Radiochemical destruction of thyroid tissues by 131I may require 4–6 months to be fully effective, whereas surgical thyroidectomy must be preceded with antithyroid drugs to prevent life-threatening complications such as thyroid storm.
The activity of the thyroid hormones, L-thyroxin (T4) and L-triiodothyronine (T3), is mediated by thyroid hormone nuclear receptors (TRs) (for a recent review see ref. 4). The TRs are members of the nuclear hormone receptor (NR) superfamily that also includes receptors for steroid hormones, retinoids, and 1,25-dihydroxyvitamin D3 (5–7). These receptors are transcription factors that can regulate expression of specific genes in various tissues and are targets for widely used drugs, such as tamoxifen, an estrogen receptor (ER) partial antagonist (8), flutamide, an antiandrogen (9), or rosiglitazone, a peroxisome proliferator-activated receptor-γ agonist (PPARγ) (10). TR is expressed as different isoforms (TRα1, TRβ1, and TRβ2) differentially expressed in various tissues (4). Gene knockout studies in mice indicate that TRβ plays a role in the development of the auditory system and in the negative feedback of thyroid stimulating hormone (TSH) by T3 in the pituitary (11, 12), whereas TRα modulates the effect of thyroid hormone on calorigenesis and on the cardiovascular system (13). The identification of TR antagonists could play an important role in the future treatment of hyperthyroidism. Such molecules would act rapidly by directly antagonizing the effect of thyroid hormone at the receptor level, a significant improvement for individuals with hyperthyroidism who require surgery, have cardiac disease, or are at risk for life-threatening thyrotoxic storm.
The crystal structure of the ligand-binding domain (LBD) of several NRs has been solved in the absence of ligand or in the presence of bound agonists or antagonists and has provided a detailed model for the structural mechanism of activation and inhibition of members of the NR family (14–19). Although most of the LBD remains relatively static, regardless of the activation state of the receptor, the most C-terminal helix (referred to as H12) is rather dynamic and can adopt a variety of conformations when no ligand is bound to the receptor. Binding of an agonist stabilizes a conformation of the receptor where H12 folds like a lid onto the agonist, contributing to the formation of a hydrophobic cavity at the surface of the receptor, involved in the binding of coactivator proteins. Antagonists, on the other hand, destabilize this coactivator-recruitment state by preventing H12 from folding on the ligand-binding pocket. The crystal structure of ERα bound to the partial antagonist raloxifene shows that H12 relocates onto the coactivator binding site (14). Alternatively, the pure antagonist ICI 164,384 presents an arm that extends out of the ligand-binding pocket and onto the coactivator binding site (18). The crystal structure of a PPARα/antagonist/corepressor peptide complex also recently showed that binding of corepressor proteins can further destabilize the active conformation of the receptor (19). In most cases, two important properties of antagonist molecules apply: (i) they bind within the ligand-binding pocket, making specific interactions with pocket-lining residues in a fashion similar to that for agonists; (ii) they present an extension that protrudes [slightly in the case of the PPARα antagonist GW6471 (19), more extensively in the case of ICI 164,384 (18)] out of the pocket and antagonizes the active conformation of H12. The recent crystal structure of the ERβ-selective antagonist (R,R)-5,11-cis-diethyl-5,6,11,12-tetrahydrochrysene-2,8-diol bound to the inactive form of the ERβ-LBD depicts an alternate mode of antagonism, where the ligand induces nonproductive conformations of binding pocket residues that destabilize the active state of the H12 helix (20). It is unclear whether this interesting “passive-antagonism” mechanism is an isolated case. On the other hand, the mechanism whereby a protruding extension directly induces relocation of helix H12 was experimentally observed in a variety of complexes (14, 18, 19, 21–23).
Based on such observations, we built a low-energy model of the retinoic acid receptor-α (RARα) LBD where H12 adopts a conformation similar to raloxifene-bound ERα and showed that such a model can be used for structure-based discovery of novel RAR antagonists (24). This rationale was recently confirmed by another group which designed a TR antagonist by adding a steric moiety to T3 that would destabilize the transcriptionally active conformation of H12 (25). In this report, we demonstrate that, by deriving molecules from the structure of the receptor, rather than the structure of known agonists, a large array of TR antagonists can be identified, that present striking structural diversity. Such an approach could accelerate the discovery of new chemical entities for the treatment of hyperthyroidism.
Materials and Methods
Model Building of a TR Antagonist Structure. The crystal structure of raloxifene-bound ERα (14) was used as a template to derive from the structure of agonist-bound TRβ-LBD (26) a predicted model of the receptor bound to an antagonist. The conformation of the N-terminal domain of the TRβ-LBD was left unchanged from the agonist-bound form, up to residue Leu-440, whereas the C-terminal domain (residues His-441 to Asp-461) was remodeled in a two-step process, as described for RAR (24). This model is based on the hypothesis (derived from sequence alignment with the ER and the crystal structure of the ER/raloxifene complex) that the FXXVF pattern, at positions 455, 458, and 459 of helix H12, mimics, in the antagonist conformation, the LXXLL motif of the GRIP-1 coactivator observed in the crystal structure of agonist-bound TRβ (26). The isolated, flexible H12 helix was docked to a hydrophobic cavity at the surface of the receptor according to the internal coordinate mechanics (ICM) method (27, 37). The docked structure of helix H12 (residues 455–461) was then fixed, and the conformation of the C-terminal loop joining His-441 to Phe-455 was optimized by an extensive energy minimization Monte Carlo simulation in the internal coordinate space with ICM (28).
High-Throughput Docking. The ICM virtual library screening (VLS) module was used (Molsoft). A series of five grid potential representations of the receptor were automatically generated and superimposed that accounted for the hydrophobicity, carbon-based and hydrogen-based van der Waals boundaries, hydrogen-bonding profile, and electrostatic potential of the predefined ligand binding site. Each of the 250,000 flexible compounds of the Available Chemicals Directory database (MDL Information Systems, San Leandro, CA) that passed the Lipinski filters (35) was docked to the receptor grids by the ICM method (29) and assigned a score that reflects the quality of the complex and includes grid energy, continuum electrostatic, and entropy terms (refs. 30 and 31 for review). Docking took an average of 1 min per processor and per ligand. The whole process was conducted four times in parallel, and the lowest score assigned to each ligand was retained. Compounds scoring below (i.e., better than) the ICM VLS threshold used for RAR (24) and therefore most likely to bind the TR ligand binding pocket were then tested in silico for their predicted antagonist activity. Although all NR antagonists do not necessarily present a bulky moiety protruding out of the binding pocket (20), all known ligands that do present such moiety are antagonists (14, 18, 19, 21–23). Based on this observation, only the top 1,000 ligands that had a bound conformation incompatible with the active state of the receptor were retained (docked compounds that were further than 3 Å away from Phe-455 or Phe-459 of the active form of helix H12 were discarded). The structure of this top 1,000 selection was rapidly refined with both ligand and receptor side-chains flexible, according to the ICM local energy minimization method that uses internal coordinates and analytical derivatives (28, 29), and the refined structures presenting unacceptable receptor/ligand van der Waals repulsions were filtered out. Each of the remaining 300 top-scoring complexes was visually inspected: parameters taken into account were shape complementarity, hydrogen bond network and flexibility of the ligand. Based on this final subjective selection step, 100 compounds were retained to be characterized in vitro. It is difficult to evaluate the relevance and enrichment generated by the different automatic or visual filters added after the initial high-throughput docking; to better judge the efficiency of the first screening step, the score distribution of all compounds screened as well as that of the active compounds is provided (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org): of 14 active TR antagonists identified, 9 ranked in the top 600 of ≈250,000 compounds and 3 ranked in the top 300 [the source library included ≈250,000 ligands but only 190,000 passed the Lipinski filters (35) and were assigned a score].
Effect of Antagonists on T3-Stimulation of Gene Expression and T3-Association with NRs in Vivo and in Vitro. For functional chloramphenicol acetyltransferase (CAT) assays, HeLa cells were inoculated at 50,000 cells per well in 24-well plates in DMEM containing 10% calf serum. The cells were transfected 5 h later by calcium phosphate precipitation with 450 ng of the T3-responsive ΔMTV-IR-CAT reporter and 250 ng of a vector expressing TRα. At the time of transfection, the cells also received 6 nM T3 and the different concentrations of the antagonist candidates. Cells were harvested 40 h after transfection and assayed for protein content and CAT activity as described (24). Results are expressed as the extent of inhibition of the T3-stimulation of CAT activity observed in the presence of the antagonist candidates. Each data point reflects the average of triplicate samples that showed <10% variation.
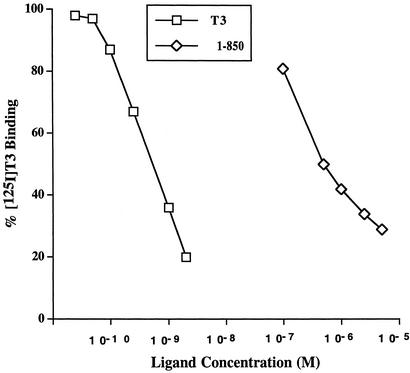
Inhibition of binding of [125I]T3 to TRs in intact GH4 cells by the antagonist candidate 1-850 was carried out as described (32) with slight modifications. GH4 rat pituitary cells, which contain endogenous TRs (TRα1, TRβ1, and TRβ2), were grown in monolayer culture in DMEM containing 10% calf serum. Cells were dispersed by incubation in a buffered solution of EDTA and incubated at 37°C for 60 min in serum-free DMEM to lower endogenous levels of thyroid hormones. Aliquots containing ≈1.5 million cells were centrifuged at 1,000 × g for 10 min and then suspended in 1 ml of serum-free medium containing 0.1 nM [125I] T3 and the indicated concentrations of unlabeled T3 or the antagonist candidate, 1-850. After incubation at 37°C for 60 min, the cells were chilled in ice and then centrifuged at 4°C at 1,000 × g for 10 min. The samples were washed twice by resuspension and vortexing with 1 ml of 50 mM Tris·HCl, pH 7.85, containing 1 mM MgCl2 and 0.5% Triton X-100 and centrifugation at 1,000 × g for 10 min to isolate the nuclear fraction of the cells. The amount of [125I]T3 retained in the nuclei was determined by using a Packard spectrometer. The results are plotted as percent of [125I]T3 retained in washed nuclei from cells incubated with 0.1 nM [125I]T3 in the presence of unlabeled T3 or the 1-850 antagonist compared with cells incubated with 0.1 nM [125I]T3 alone. Each point represents the average of duplicates, which varied by <5%.
Effect of Antagonist 1-850 and Derivatives on the T3-Mediated Binding of TR to the Coactivator NRC in Vitro. Full-length TRα was synthesized in the presence of L-[35S]methionine by using TNT reticulocyte lysates (Promega). Approximately 2.5–5 × 104 cpm of 35S-labeled TRα (20 fmol) in 2 μl of lysate was incubated with 500 ng of GST fused to the receptor interaction region of the coactivator NRC (NRC15) immobilized on glutathione-agarose beads (33). The samples were incubated at room temperature for 15 min in 500 μl of binding buffer [20 mM Hepes (pH = 7.8)/1 mM MgCl2/100 mM KCl/0.05% Triton X-100/1mMDTT/10% (vol/vol) glycerol/100 μg/ml ovalbumin/0.1 μM ZnCl2] with the indicated concentrations of 1-850 and the 1-850 derivatives D1 and D4. The samples were then chilled on ice and incubated with 1 nM T3 for an additional 60 min at 4°C. Control samples contained no T3 or antagonists or received only T3. The beads were collected by centrifugation (≈500 × g) at 4°C for 5 min and washed three times with 1 ml of binding buffer. The bound [35S]TRα was electrophoresed in an SDS/10% polyacrylamide gel followed by analysis and quantitation of the amount of [35S]TRα bound by using a Molecular Dynamics PhosphorImager and IMAGEQUANT software. The percent inhibition of T3-mediated binding of [35S]TRα to GST-NRC15 by 1-850, D1, and D4 was determined after subtracting the amount of [35S]TRα bound to GST-NRC15 in the absence of T3.
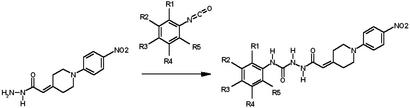
Generation of a Virtual Focused Library. A virtual library of derivatives of the antagonist 1-850 (the best hit derived from firstgeneration VLS) was generated, based on the chemical synthesis scheme described below, so that all molecules generated in silico could be easily and rapidly synthesized. All phenylisocyanates commercially available from Sigma–Aldrich were extracted from virtual structure libraries with the ICM “find molecule” command (27) and attached to the nonvariable moiety of 1-850. Each molecule was then assigned ECEPP3 potentials and mmff partial charges, energy-minimized, and stored in a focused library of 101 compounds. All chemo-informatics procedures were carried out with ICM (27).
Preparation of 1-850 Active Analogs D1–D57. Selected compounds from the series of 1-850 analogs D1–D101 were prepared by coupling of compound 4 (see Supporting Text, which is published as supporting information on the PNAS web site, for more details) with commercially available phenylisocyanates (Fig. 3). The general procedure involved adding 0.5 mmol of phenylisocyanate to a solution of 0.13 g of compound 4 (0.5 mmol) in 1 ml of dry CH2Cl2, stirring at room temperature for 2 h, and then separation of the final product by filtration. The purity of the all library compounds was determined by LC-MS. The most active compounds D1–D4 were characterized more fully by 1H NMR and MS.
Fig. 3.
Chemical synthesis of 1-850 derivatives was conducted by preparing a nonvariable core structure and rapidly linking commercially available polysubstituted phenylisocyanate building blocks in a simple step of parallel synthesis (see Materials and Methods for details).
Results
Modeling of the Antagonist-Bound Conformation of TR. A low-energy model of the antagonist structure of the TRβ-LBD was built by homology to agonist-bound TRβ-LBD (26) and raloxifene-bound ERα (14). Following a strategy devised with RAR (14), we docked the C-terminal helix H12 of the TRβ LBD to the coactivator recruitment site as described (34), and remodeled the 20 C-terminal residues of the receptor by an extensive global energy minimization (27, 38).
Virtual Screening. A virtual database of 250,000 commercially available compounds was rapidly screened in silico. All continuously flexible ligands were docked to a grid representation of the receptor and assigned a score reflecting the quality of the complex, according to the ICM method (Molsoft). Briefly, a series of grid potentials representing the shape, hydrophobicity, hydrogen bonding profile, and electrostatic potential of the receptor was generated. Each ligand was then docked to the grid representation of the receptor by a Monte Carlo simulation in the internal coordinates space, and the complex was optimized with flexible receptor side chains (see Materials and Methods). Putative ligands that did not extend out of the binding pocket in the docked conformation were automatically filtered out. After visual inspection of the 300 best scoring compounds, 100 were selected as potential TR antagonists, 75 of which were still commercially available (chemicals listed in the Available Chemicals Directory are often not resynthesized once they run out of stock).
Discovery of 14 Novel TR Antagonists. The ability of the 75 commercially available compounds to inhibit the functional activity of TR in mammalian cells was assayed. The compounds were dissolved in DMSO, and the effect of various concentrations of the compounds to inhibit T3-stimulation of a reporter gene by TRα expressed in HeLa cells was studied. Fourteen of the 75 compounds showed antagonist activity; their structure and order of potency are shown in Table 1. Compound 1-850 showed the greatest antagonist activity. To verify that 1-850 inhibited the action of T3 by competition at the TR level, the ability of 1-850 to inhibit the binding of [125I]T3 to endogenous TRs in GH4C1 cells was compared with nonradioactive T3 (Fig. 1). Based on this study, 1-850 has an ≈1,000-fold lower affinity for the receptor in intact cells compared with T3. Although the results of Table 1 were obtained with TRα, similar results for these compounds were also found by using TRβ. All of the compounds that exhibited antagonist activity in the presence of T3 showed no partial agonist activity in the absence of T3. In addition, these compounds had no effect on the activity of RARα (data not shown), suggesting specificity for TR.
Table 1. Name, structure, and relative activity of the 14 TR antagonists identified after the first round of virtual screening.
*Compound 2-1060 was toxic at 20 μM.
Fig. 1.
Inhibition of [125I]T3 binding to TRs by 1-850 in intact cells. The GH4C1 pituitary cell line, which contains endogenous TRs (TRα, TRβ1, and TRβ2), was incubated with 0.1 nM [125I]T3 alone and with the indicated concentrations of unlabeled T3 and 1-850 as described in Materials and Methods. After incubation for 60 min at 37°C, the cells were chilled and washed, and the nuclei were isolated as described in Materials and Methods. The results indicate the inhibition of binding of [125I]T3 by T3 and 1-850.
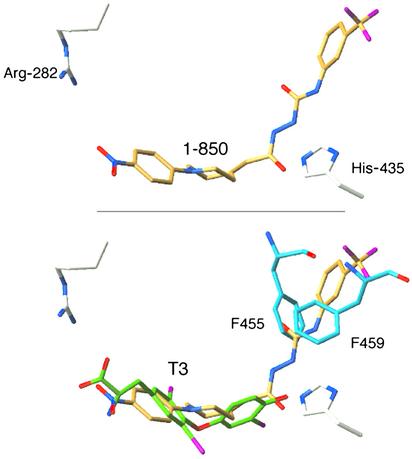
Structure-Based Optimization of Compound 1-850. The docking of 1-850, the most active antagonist identified by our first round of virtual screening, suggests that the antagonist makes mostly hydrophobic interactions with residues lining the ligand-binding pocket. A carbonyl oxygen of 1-850 also forms a hydrogen bond with His-435 in the docked conformation, whereas the nitro group is relatively close to Arg-282 (4 Å between the nitro oxygen and the closest arginine nitrogen), deep inside the receptor pocket (Fig. 2 Upper). As expected, the antagonist superimposes well with the crystal structure of bound T3 and presents an additional extension that would clash with the active conformation of H12 (Fig. 2 Lower).
Fig. 2.
Predicted conformation of 1-850 (gold) bound to the TRβ ligand-binding pocket. (Upper) A hydrogen bond between His-435 and a carbonyl oxygen of 1-850 and possibly between Arg-282 and a nitro oxygen of 1-850 constitute the only polar interactions. All other contacts are hydrophobic (not shown for clarity). (Lower)1-850 superimposes with the crystal structure of T3 (green), bound to active TR and clashes with the active conformation of helix H12 (cyan). Color code is red for oxygen, blue for nitrogen, and magenta for fluoride/iodide in 1-850/T3, respectively.
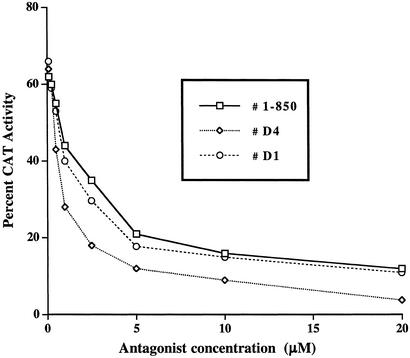
Based on this model, we synthesized derivatives of 1-850 that might display higher affinity for the receptor. 1-850 was divided into one core unit that was synthesized separately and one variable extremity, where polysubstituted phenylisocyanates could be used as building blocks to easily derivatize the hydrophobic, trifluoromethylated moiety of 1-850 (Fig. 3 and Supporting Text). A virtual library of all such building blocks available from the Sigma–Aldrich catalog was generated, and all corresponding derivatives of 1-850 were constructed in silico with ICM. The resulting chemical library of 101 compounds was docked to the receptor as described above, and each 1-850 derivative was assigned a score reflecting its predicted fit with the receptor. To address the relevance of this virtual automatic parallel synthesis and docking strategy, we synthesized 8 of the 57 top-ranking 1-850 analogs and compared calculated scores with observed activities. A dose–response inhibition of T3-stimulation comparing 1-850 with the best two antagonist derivatives (D4 and D1) is shown in Fig. 4, and the results of the 8 compounds are shown in Table 2. None of the 1-850 derivatives exhibited partial agonist activity in the absence of T3. The best two inhibitors (D4 and D1), were among the top four scoring compounds whereas the two less active molecules were the lowest scoring ones (D37 and D57). One derivative (D4) reached IC50 in the nanomolar range (0.75 μM), and all compounds inhibited TR from 10% to 84% at 5 μM (Table 2). According to our docking simulation, the methyl and isopropyl groups of D4's hydrophobic end would make more extensive hydrophobic interactions with the receptor than the trifluoromethyl of 1-850 (Fig. 7, which is published as supporting information on the PNAS web site.) To document that D4 and D1 acted through inhibition of T3 binding to receptor, we compared their activity to 1-850 using a T3-dependent in vitro coactivator binding assay (Fig. 5). In this study, we incubated rabbit reticulocyte labeled [35S]TRα with GST-NRC15 immobilized on glutathione-agarose beads. NRC15 is a region of the nuclear receptor coactivator NRC that interacts with agonist-bound TR through the NRC LXXLL-1 motif (33). GST-NRC15 was incubated with [35S]TRα with the indicated concentrations of D4, D1, and 1-850 followed by incubation with 1 nM T3 (a concentration that leads to a maximal effect in the absence of antagonist). After incubation and washing as indicated in Materials and Methods, the samples were electrophoresed in an SDS/10%-polyacryalimide gel, followed by imaging and quantitation by using a PhosphorImager. The results shown in Fig. 5 indicate that the antagonists block the T3-mediated interaction of TRα with NRC, and the extent of inhibition generally parallels the relative ability of these compounds to inhibit T3-mediated stimulation of gene expression in cells (D4 > D1 > 1-850; Fig. 4).
Fig. 4.
Comparison of the antagonist activity of 1-850 and two compounds (D1 and D4) identified through a VLS of derivatives of 1-850.
Table 2. Comparison of the biological activity of the 1-850 derivatives actually synthesized with the predicted ranking derived from the calculated score of the receptor/ligand complex.
Of the 101 analogs of 1-850 docked to the receptor, 8 were synthesized and tested in vitro. The calculated score, corresponding rank (of 101), structure, and activity for each compound are listed.
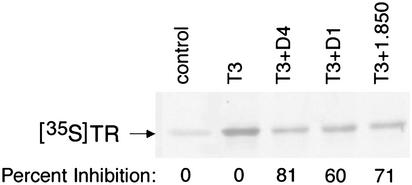
Fig. 5.
Inhibition of T3-mediated coactivator recruitment to TR by D1, D4, and 1-850 in vitro. Approximately 2.5–5 × 104 cpm of 35S-labeled TRα (20 fmol) in 2 μl of lysate was incubated with 500 ng of GST fused to the receptor-interaction region of the coactivator NRC (NRC15) immobilized on glutathione-agarose beads. The samples were also incubated for 15 min at room temperature with 2 μM D1 or D4 or 5 μM 1-850 in binding buffer as indicated in Materials and Methods. The samples were then chilled on ice and incubated with 1 nM T3 for an additional 60 min at 4°C. Control samples contained no T3 or antagonists or received only T3. The beads were washed and the bound [35S]TRα was electrophoresed in a an SDS/10% polyacrylamide gel followed by analysis and quantitation of the amount of [35S]TRα bound by using a Molecular Dynamics PhosphorImager and imagequant software. The percent inhibition of T3-mediated binding of [35S]TRα to GST-NRC15 by 1-850, D1, and D4 was determined after subtracting the amount of [35S]TRα bound to GST-NRC15 in the absence of T3.
Discussion
In this study, we used the crystal structure of agonist-bound TRβ and raloxifene-bound ERα to construct a computer model of the predicted antagonist form of TRβ and used this model as a template to discover in silico small molecule inhibitors of TR. Using the most active initial hit, we generated a small virtual library that can be easily and rapidly synthesized with commercially available building blocks. High-throughput docking of this library with TR identified a derivative with increased activity.
First, this work confirms a conceptually important observation that we made with RAR (24), documenting that it is possible to rationally design NR antagonists by building a computer model of the predicted antagonist-bound form of the receptor. This result reinforces the hypothesis that the structural mechanism for NR inhibition initially depicted for ER (14) can be generalized to other members of the family, as was shown for RAR (23) or PPAR (19). Although this strategy appears successful at rapidly identifying TR antagonists, it is likely that it also misses active molecules for the following reasons. (i) Our implementation of receptor modeling and selection of antagonist candidates was directed toward the identification of compounds that do fit in the TR ligand binding pocket but present a bulky moiety that would clash with the active conformation of helix H12. However, smaller compounds may induce nonproductive conformations of binding pocket residues that could destabilize the active state of the H12 helix, as was recently shown for the ER (20). Such active antagonists would be missed by our selection strategy. (ii) The VLS algorithm is mainly designed to filter out nonbinders and enrich significantly in active molecules smaller focused libraries. However, it also misses true positives. For instance, the only TR antagonist known before this work, recently described by Baxter et al. (25), was assigned a VLS score that did not pass our selection threshold. Possible reasons are that the receptor adopts an alternative conformation when bound to this ligand, or that the interactions with this moderate binder are not strong enough to be detected.
A second observation is the extraordinary structural diversity of the 14 TR antagonists discovered after our first round of virtual screening (Table 1). In a recent study, Baxter et al. (25) showed that adding a hydrophobic moiety to 3,5-dibromo-4-(3′-isopropyl-4′-hydroxyphenoxy)benzoic acid, a known TR agonist, resulted in TR antagonist activity. Here, we demonstrate that a diverse array of compounds, representing extremely heterogeneous chemistries, can comply with the structural rules that govern TR inhibition.
Underlying this surprising observation is the strategy used to discover TR antagonists, where small molecule modulators for a specific target are derived from the structure of the receptor rather than from existing ligands. Such an approach can open up entire domains of the chemistry space that would be overlooked by ligand-based pharmacophore approaches. This work demonstrates that high-throughput docking is a relevant approach to screen very large and diverse chemical libraries that best represent the infinite diversity of potential lead molecules. Additionally, this powerful strategy can be applied to orphan receptors, for which no ligand is known.
Our work also suggests that receptor-based virtual screening coupled to parallel synthesis can significantly accelerate the lead optimization process. We generated a virtual library focused on one of the initial hits (compound 1-850) by branching 101 commercially available building blocks to a central core, according to a single step of parallel synthesis. We showed that rapid docking of this virtual library efficiently ranked the derivatives according to their activity, with the best two antagonists tested in vitro corresponding to the fourth and first best scoring analogs of 101 compounds. Such lead optimization strategy can dramatically reduce the number of compounds to synthesize and test, thereby allowing better characterization of more relevant molecules.
Although we built an antagonist-bound conformation of TR where the helix H12 occupies the coactivator recruitment site, based on the crystal structure of the ER/raloxifene complex (14), none of the docked compounds contact receptor residues C-terminal to helix H11 (even though they all would clash with the active conformation of H12). Carrying out the VLS with H12 in an alternative, extended conformation, as observed in apo-RXR (36), or with a representation of the receptor truncated at the end of H11 may also have selected the active hits identified in this work. The recent crystal structure of ERβ-LBD bound to ICI 164,384 showed that, unlike partial ER antagonists such as tamoxifen or raloxifene, this pure antagonist presents an extension long enough to reach the coactivator recruitment site of the receptor and prevent interaction between helix H12 and the core of the receptor (18). If a similar mechanism applied to other NRs, such as TR or androgen receptor, performing a high-throughput docking with a receptor truncated at the end of helix H11 might result in the identification of pure antagonists, with potentially interesting therapeutic properties.
Conclusion
In this work, we showed that high-throughput docking can be used to rapidly identify lead TR antagonists that differ from known ligands and to prioritize subsequent lead optimization. The extreme structural diversity of the antagonist molecules discovered in this work illustrates the power of receptor-based virtual screening and demonstrates that diverse chemistry can comply with strict structural rules. Further optimization of TR antagonists could open the way for improved modalities for the treatment of hyperthyroidism.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant DK16636 (to H.H.S.) and Small Business Innovation Research/Small Business Technology Transfer Grant DK59041 (to Molsoft) for collaborative research with New York University.
Abbreviations: ER, estrogen receptor; ICM, internal coordinate mechanics; LBD, ligand-binding domain; NR, nuclear hormone receptor; PPARγ, peroxisome proliferator-activated receptor-γ agonist; RAR, retinoic acid receptor; T3, L-triiodothyronine; TR, thyroid hormone receptor; VLS, virtual library screening.
See commentary on page 6902.
References
- 1.DeGroot, L. J. (2001) Endocrinology (Saunders, Philadelphia), 4th Ed.
- 2.Braverman, L. E., Utiger, R. D., Ingbar, S. H. & Werner, S. C., eds. (2000) Werner and Ingbar's the Thyroid: A Fundamental and Clinical Text (Lippincott Williams & Wilkins, Baltimore), 8th Ed.
- 3.Wilson J. D., ed. (1998) Textbook of Endocrinology (Saunders, Philadelphia).
- 4.Yen, P. M. (2001) Physiol. Rev. 81, 1097-1142. [DOI] [PubMed] [Google Scholar]
- 5.Evans, R. M. (1988) Science 240, 889-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carson-Jurica, M. A., Schrader, W. T. & O'Malley, B. W. (1990) Endocr. Rev. 11, 201-220. [DOI] [PubMed] [Google Scholar]
- 7.Chambon, P. (1993) Gene 135, 223-228. [DOI] [PubMed] [Google Scholar]
- 8.Dees, E. C. & Kennedy, M. J. (1998) Curr. Opin. Oncol. 10, 517-522. [DOI] [PubMed] [Google Scholar]
- 9.Labrie, F. (1993) Cancer 72, 3816-3827. [DOI] [PubMed] [Google Scholar]
- 10.Olefsky, J. M.& Saltiel, A. R. (2000) Trends Endocrinol. Metab. 11, 362-368. [DOI] [PubMed] [Google Scholar]
- 11.Forrest, D., Erway, L. C., Ng, L., Altschuler, R. & Curran, T. (1996) Nat. Genet. 13, 354-357. [DOI] [PubMed] [Google Scholar]
- 12.Weiss, R. E., Forrest, D., Pohlenz, J., Cua, K., Curran, T. & Refetoff, S. (1997) Endocrinology 138, 3624-3629. [DOI] [PubMed] [Google Scholar]
- 13.Wikstrom, L., Johansson, C., Salto, C., Barlow, C., Campos Barros, A., Baas, F., Forrest, D., Thoren, P. & Vennstrom, B. (1998) EMBO J. 17, 455-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brzozowski, A. M., Pike, A. C., Dauter, Z., Hubbard, R. E., Bonn, T., Engstrom, O., Ohman, L., Greene, G. L., Gustafsson, J. A. & Carlquist, M. (1997) Nature 389, 753-758. [DOI] [PubMed] [Google Scholar]
- 15.Moras, D. & Gronemeyer, H. (1998) Curr. Opin. Cell Biol. 10, 384-391. [DOI] [PubMed] [Google Scholar]
- 16.Weatherman, R. V., Fletterick, R. J. & Scanlan, T. S. (1999) Annu. Rev. Biochem. 68, 559-581. [DOI] [PubMed] [Google Scholar]
- 17.Bourguet, W., Germain, P. & Gronemeyer, H. (2000) Trends Pharmacol. Sci. 21, 381-388. [DOI] [PubMed] [Google Scholar]
- 18.Pike, A. C., Brzozowski, A. M., Walton, J., Hubbard, R. E., Thorsell, A. G., Li, Y. L., Gustafsson, J. A. & Carlquist, M. (2001) Structure (Cambridge, Mass.) 9, 145-153. [DOI] [PubMed] [Google Scholar]
- 19.Xu, H. E., Stanley, T. B., Montana, V. G., Lambert, M. H., Shearer, B. G., Cobb, J. E., McKee, D. D., Galardi, C. M., Plunket, K. D., Nolte, R. T., et al. (2002) Nature 415, 813-817. [DOI] [PubMed] [Google Scholar]
- 20.Shiau, A. K., Barstad, D., Radek, J. T., Meyers, M. J., Nettles, K. W., Katzenellenbogen, B. S., Katzenellenbogen, J. A., Agard, D. A. & Greene, G. L. (2002) Nat. Struct. Biol. 9, 359-364. [DOI] [PubMed] [Google Scholar]
- 21.Shiau, A. K., Barstad, D., Loria, P. M., Cheng, L., Kushner, P. J., Agard, D. A. & Greene, G. L. (1998) Cell 95, 927-937. [DOI] [PubMed] [Google Scholar]
- 22.Pike, A. C., Brzozowski, A. M., Hubbard, R. E., Bonn, T., Thorsell, A. G., Engstrom, O., Ljunggren, J., Gustafsson, J. A. & Carlquist, M. (1999) EMBO J. 18, 4608-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bourguet, W., Vivat, V., Wurtz, J. M., Chambon, P., Gronemeyer, H. & Moras, D. (2000) Mol. Cell 5, 289-298. [DOI] [PubMed] [Google Scholar]
- 24.Schapira, M., Raaka, B. M., Samuels, H. H. & Abagyan, R. (2000) Proc. Natl. Acad. Sci. USA 97, 1008-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baxter, J. D., Goede, P., Apriletti, J. W., West, B. L., Feng, W., Mellstrom, K., Fletterick, R. J., Wagner, R. L., Kushner, P. J., Ribeiro, R. C., et al. (2002) Endocrinology 143, 517-524. [DOI] [PubMed] [Google Scholar]
- 26.Darimont, B. D., Wagner, R. L., Apriletti, J. W., Stallcup, M. R., Kushner, P. J., Baxter, J. D., Fletterick, R. J. & Yamamoto, K. R. (1998) Genes Dev. 12, 3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molsoft (2002) ICM 2.8 Program Manual (Molsoft, San Diego).
- 28.Abagyan, R. & Totrov, M. (1994) J. Mol. Biol. 235, 983-1002. [DOI] [PubMed] [Google Scholar]
- 29.Totrov, M. & Abagyan, R. (1997) Proteins, Suppl. 1, 215-220. [DOI] [PubMed]
- 30.Totrov, M. & Abagyan, R. (2001) in Drug-Receptor Thermodynamics: Introduction and Applications, ed. Raffa, R. B. (Wiley, New York), pp. 603-624.
- 31.Abagyan, R. & Totrov, M. (2001) Curr. Opin. Chem. Biol. 5, 375-382. [DOI] [PubMed] [Google Scholar]
- 32.Raaka, B. M. & Samuels, H. H. (1983) J. Biol. Chem. 258, 417-425. [PubMed] [Google Scholar]
- 33.Mahajan, M. A. & Samuels, H. H. (2000) Mol. Cell. Biol. 20, 5048-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, D., Desai-Yajnik, V., Lo, E., Schapira, M., Abagyan, R. & Samuels, H. H. (1999) Mol. Cell. Biol. 19, 7191-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipinski, C. A., Lombard, F., Dominy B. W. & Feeney, P. J. (1997) Adv. Drug Delivery Rev. 23, 3-25. [DOI] [PubMed] [Google Scholar]
- 36.Bourguet, W., Ruff, M., Chambon, P., Gronemeyer, H. & Moras, D. (1995) Nature 375, 377-382. [DOI] [PubMed] [Google Scholar]
- 37.Strynadka, N. C., Eisenstein, M., Katchalski-Katzir, E., Shoichet, B. K., Kuntz, I. D., Abagyan, R., Totrov, M., Janin, J., Cherfils, J., Zimmerman, F., et al. (1996) Nat. Struct. Biol. 3, 233-239. [DOI] [PubMed] [Google Scholar]
- 38.Abagyan, R., Batalov, S., Cardozo, T., Totrov, M., Webber, J. & Zhou, Y. (1997) Proteins, Suppl. 1, 29-37. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.