Abstract
Pinus taeda L. (loblolly pine) and Arabidopsis thaliana differ greatly in form, ecological niche, evolutionary history, and genome size. Arabidopsis is a small, herbaceous, annual dicotyledon, whereas pines are large, long-lived, coniferous forest trees. Such diverse plants might be expected to differ in a large number of functional genes. We have obtained and analyzed 59,797 expressed sequence tags (ESTs) from wood-forming tissues of loblolly pine and compared them to the gene sequences inferred from the complete sequence of the Arabidopsis genome. Approximately 50% of pine ESTs have no apparent homologs in Arabidopsis or any other angiosperm in public databases. When evaluated by using contigs containing long, high-quality sequences, we find a higher level of apparent homology between the inferred genes of these two species. For those contigs 1,100 bp or longer, ≈90% have an apparent Arabidopsis homolog (E value < 10-10). Pines and Arabidopsis last shared a common ancestor ≈300 million years ago. Few genes would be expected to retain high sequence similarity for this time if they did not have essential functions. These observations suggest substantial conservation of gene sequence in seed plants.
All higher vascular plants are likely to be derived from a small, leafless, rootless ancestor ≈420 million years ago (Mya) during the Silurian (1). Gymnosperms and angiosperms are the two major taxa of seed plants, distinct since the end of the Carboniferous, ≈300 Mya (2). Angiosperms comprise the overwhelming diversity of woody and herbaceous plants with ≈250,000 species (3). Angiosperms include our major food crops and dominate many critical and diverse ecosystems such as our tropical forests. Extant gymnosperms represent a monophyletic clade and a sister lineage to the angiosperms (2–4). Gymnosperms dominate many temperate terrestrial ecosystems but include only ≈700–1,000 extant species. All known gymnosperms are woody plants.
Gymnosperms generally have significantly larger haploid DNA contents than angiosperms. The modal value for gymnosperms is 15,480 Mbp (5) compared with 588 Mbp for angiosperms. The haploid DNA content estimate of loblolly pine, a woody gymnosperm, is 20,000 Mbp (6), which is 160 times larger than the 125-Mbp genome of Arabidopsis thaliana (7) and 47 times larger than the rice genome (430 Mbp) (8, 9). The large size of the pine genome is not likely to be due to recent polyploidy. All pines have 12 chromosomes, and there is a narrow range of variation in the basic chromosome number (from 11 to 13) within the Pinaceae (10).
The significant differences in genome size, phenotypic diversity, and genetic distance raise the question of the extent to which gymnosperms and angiosperms share the same genes. A high level of gene sharing would suggest that gene function in plants and the great diversity observed in higher vascular plants evolves primarily through differential regulation of similar gene sets rather than through the evolution of new, function-specific genes. Strategies of genetic engineering often depend on functional homology of heterologous genes across taxa or on the presence of genes specific to taxa, for example, where pesticide or herbicide specificity is desired.
A major unresolved question in the evolution of higher plants is the extent to which they share a common functional genome. The past 400 million years provided many opportunities for the evolution of specific gene differences through gene loss, horizontal gene transfer, or rapid rates of sequence divergence within a lineage. For example, Allen (11) recently compared predicted gene content in three crop species, tomato, soybean, and medicago, with that of Arabidopsis. He found that 9.5%, 14.5%, and 13.3% of the crop EST contigs, respectively, failed to hit an Arabidopsis homolog (E-value cutoff of 10-3) and argued for gene loss as the most likely mechanism for this variation. More genomic and EST sequence, obtained from different angiosperms and gymnosperms, is needed to determine the extent and mechanisms of gene evolution in higher plants.
To contribute to this discussion, we compared a large number of expressed gene sequences from wood-forming tissues of loblolly pine with the inferred gene sequences of A. thaliana. This comparison allows us to examine sequence divergence over 300 million years, which is approximately three-fourths of the time since the emergence of the first higher plants. We generated and analyzed a total of 59,797 loblolly pine ESTs of high sequence quality (12, 13) from six partial cDNA libraries prepared from differentiating xylem harvested from trees of different ages and under different environmental conditions. We extended our results from previous studies (14, 15) by comparing these ESTs to the complete set of predicted expressed gene sequences from Arabidopsis (7). Such a comparison should highlight the differences in genes involved in wood or secondary cell-wall formation between an herbaceous angiosperm and a woody gymnosperm. We find a high degree of apparent sequence homology for loblolly pine cDNAs, particularly where sufficient length of high-quality sequence has been obtained.
Materials and Methods
cDNA Libraries. Six nonnormalized partial cDNA libraries were constructed from six different differentiating xylem tissues of loblolly pine. Xylem-forming tissues were harvested by using either a vegetable peeler (for primary xylem) or a block plane (for lignifying secondary xylem), frozen in liquid nitrogen in the field, and stored at -80°C until needed for mRNA isolation (14). The six types of tissues were (i) primary mature xylem from a 35-year-old tree harvested in the spring (normal mature wood library NXNV); (ii and iii) primary juvenile xylem from the side and underside of the bent segment of three 6-year-old trees of different genotypes inclined at a 45° angle for 40 days (side wood library NXSI and compression wood library NXCI); (iv) lignifying secondary transitional xylem from a 10-year-old tree (wood planings library NXPV); (ν) normal primary xylem collected late in the summer from a transitional area below the crown of a 20-year-old tree (late wood library NXLV); and (vi) normal primary xylem harvested from the root wood of a 12-year-old tree (root wood library NXRV).
For all six libraries, total RNA was extracted from 2–3 g of tissue (16) and evaluated on 2% agarose gels. mRNA was isolated from total RNA by using either the Promega Polyattract system IV kit (NXSI and NXCI) or the Stratagene poly(A) Quik mRNA isolation kit (NXNV, NXLV, NXRV, and NXPV). Each library was constructed by using 5 μg of mRNA and the ZAP-cDNA synthesis kit (Stratagene) to generate the cDNA, which was fractionated by using Size-Sep 400 spin columns (Amersham Pharmacia) to remove cDNAs <400 bp in size. The purified cDNA was unidirectionally cloned into the EcoRI site of the Uni-ZAP XR vector (Stratagene) and packaged by using Gigapack III Gold (Stratagene). pBluescript or pTriplEx (NXLV) plasmids containing cDNA inserts were mass-excised from the Uni-Zap XR vector by using Ex-Assist helper phage (Stratagene) and propagated in Escherichia coli strain XL1Blue or BM25.8 (NXLV). For more information on these libraries, see http://pinetree.ccgb.umn.edu.
DNA Sequencing. Plasmid-containing colonies were picked into 1.3 ml of Magnificent broth (McConnell Research, San Diego) containing ampicillin (0.1 mg/ml) and grown for 20 h at 37°C with shaking in deep 96-well blocks. Plasmids were purified from cells by using R.E.A.L. kits (Qiagen, Valencia, CA) and evaluated on 0.8% agarose gels. Plasmids were sequenced from the 5′ end of the cDNA insert by using the 5′-Tripl primer (pTriplEx) or T3 (pBluescript) and dRhodamine/BigDye terminator chemistry (Applied Biosystems) according to manufacturer instructions. Sequencing reactions were purified by using the Millipore multiscreen system and Sephadex G-50 Fine Fine (Sigma) and run on either ABI377XL-96 slab gel sequencers for 6–7 h (36/48 cm well to read distance) by using 5% GenePage Plus (Amresco, Euclid, OH) or ABI3700 capillary sequencers for 4 h by using POP-6. For the ABI377XL-96, samples were loaded with membrane combs (Gel Company, San Francisco).
Sequence Processing. ABI sequencing trace files were submitted to the University of Minnesota Center for Computational Genomics and Bioinformatics for batch processing. Raw sequence files were produced from the trace files by using the PHRED base-calling program (12, 13) with a PHRED quality threshold of 8. Bases with a PHRED quality score of <8 were converted to “N.” Vector and linker sequences were trimmed from each raw sequence. The subsequent quality checks on the remaining sequence were (i) determining the number of unknown or N base calls in a sequence and trimming leading and trailing high-N sections to obtain the best subsequence where the N content is ≤4%, and (ii) using an artifact filter to remove remaining E. coli, or vector sequences. The “usable” sequences (at least 100 bp of high-quality sequence) from all six libraries were clustered into contigs based on sequence overlap by using PHRAP (17) to generate a xylogenesis UniGene set of all resulting contigs and singlets. The PHRAP settings for generating the contigs were (i) a minimum length of matching word required to nucleate SWAT comparison of 50 bp, (ii) a minimum alignment score of 100, and (iii) a minimum base pair size of individual assembled sequences of 100. The contig sequences, images of the contig assemblies, and the BLAST targets for the entire contig set are at http://pinetree.ccgb.umn.edu. All ESTs used in this study were deposited into GenBank. All EST contigs and singlets were also compared with the entire GenBank nonredundant peptide sequence database (ftp://ftp.ncbi.nih.gov/blast/db). Sequences with high similarity to potential sources of contamination (bacterial or phage origin) were reanalyzed by using BLASTN (default parameters) for confirmation at the nucleotide level. The loblolly pine xylogenesis UniGene set was compared with the A. thaliana nuclear, chloroplast, and mitochondrial predicted gene sequences (ftp://ftpmips.gsf.de/cress/arabiprot-release07/26/2002) by using BLASTX (default parameters) (18). ORFs were identified by using DIOGENES (ref. 19 and www.cbc.umn.edu/diogenes), which is designed to identify ORFs in short sequences, based on organism-specific training sets.
Results
Establishment and Analysis of the Loblolly Pine Xylogenesis UniGene Set. ESTs (59,797) with at least 100 bp of high-quality sequence from six nonnormalized partial cDNA libraries were assembled, by using PHRAP, into a xylogenesis UniGene set of contigs and singlets totaling 20,377 (Table 1). The woody tissues used for these libraries differ in age, location in the tree, tissue source, season of collection, and extent of mechanical stress. The individual trees sampled represent seven different normal genotypes. Contigs and singlets were classified according to the level of similarity (BLASTX E value) to A. thaliana. The term “similarity” is used for sequence matches and “homology” for a relationship by descent. Convergent evolution of protein sequence is rare; therefore high BLASTX similarity scores infer but do not prove relationship by descent. Our analysis primarily is based on BLASTX E values, which estimate the probability of sequence similarity due to chance. E values provide statistical support for inferences of sequence similarity based on identity and similarity of amino acids and the length of sequences that contain similarity. Thus, a pair of long sequences with a low percentage of amino acid identity and a pair of short sequences with a high percentage of amino acid identity can have similar E values. For the loblolly pine xylogenesis UniGene set, 50% of the sequences showed a significant sequence similarity to Arabidopsis at a BLASTX E value of 10-5, with 56% of the contigs and 38% of the singlets having “hits.”
Table 1.
Xylogenesis EST statistics and phrap assembly results for six pine cDNA libraries and the xylogenesis unigene set
| Library | No. of ESTs | Average length, bp | No. of phrap contigs | No. of phrap singlets | Library or combined unigene set* | EST redundancy†, % | Contig redundancy‡ |
|---|---|---|---|---|---|---|---|
| NXNV early | 8,490 | 312 | 1,387 | 3,982 | 5,369 | 53 | 3.3 |
| NXCI bent | 9,333 | 311 | 1,670 | 2,580 | 4,250 | 72 | 4.0 |
| NXSI side | 11,904 | 387 | 2,063 | 3,652 | 5,715 | 69 | 4.0 |
| NXPV planings | 9,642 | 380 | 1,768 | 2,187 | 3,955 | 77 | 4.2 |
| NXLV late | 10,244 | 345 | 1,216 | 4,320 | 5,536 | 58 | 4.9 |
| NXRV root | 10,184 | 436 | 1,878 | 3,043 | 4,921 | 70 | 3.8 |
| Combined | 59,797 | 364 | 8,070 | 12,307 | 20,377 | 79 | 5.9 |
Contigs + singlets
{[(No. of ESTs per library) - (no. of phrap singlets per library)]/(no. of ESTs per library)} × 100
[(No. of ESTs per library) - (no. of phrap singlets per library)]/(no. of phrap contigs per library)
Many Loblolly Pine Sequences Lack Similarity to A. thaliana. Approximately 50% of the total number of contigs and singlets in our xylogenesis UniGene set show no apparent homologs in Arabidopsis even at the moderate E-value cutoff of 10-5. Searching all of GenBank for additional homologs in different species at this E-value cutoff does not decrease the percentage of “no hits” by >2%. These no-hit sequences could include genes unique to pines, conifers, gymnosperms, or woody plants as well as homologs that are unrecognized because of the limitations of EST analysis. Alternatively, many of these no-hit sequences may simply represent unrecognized contaminants from (i) loblolly pine genomic DNA, (ii) DNA from other microbes or organisms not well represented in genome databases, or (iii) a variety of transcriptional artifacts (20, 21) particularly relevant to singlets, which include alternative splice variants, unspliced/incorrectly spliced mRNA fragments, and transcription initiation from multiple 3′-poly(A) tracts common to the same gene.
Pine Retrotransposon Sequences and ESTs. Some no-hit ESTs might be due to loblolly pine nuclear DNA contamination in our cDNAs. We searched our ESTs for a pine retrotransposon sequence family called IFG (22), named after the Institute of Forest Genetics (Placerville, CA). IFG accounts for ≈1% of the total pine DNA. None of these ESTs, selected from the loblolly pine xylogenesis libraries, contain the IFG sequence. Six clones of IFG were identified in loblolly pine partial cDNA libraries from loblolly pine shoot tips and pollen cones used in previous studies (15). If our no-hit category of ESTs, representing a total of 10,250 clones, were due to loblolly pine nuclear DNA, then based on the amount of IFG in total pine DNA we would expect ≈100 IFG clones. We found none, indicating that our xylogenesis UniGene set is essentially clear of contamination from loblolly pine nuclear DNA.
Functional Categories of Loblolly Pine ESTs. Sequences with significant similarity (BLASTX E value < 10-5) to the A. thaliana genome were assigned cellular functional categories based on the annotation of the Arabidopsis Genome Initiative (7) generated by the Munich Information Center for Protein Sequences. This comparison provides a general overview of the cellular functional categories for genes expressed during xylogenesis. Arabidopsis is a useful common standard, although the current annotation is limited (23), because Arabidopsis is the plant with the genome that has been sequenced most completely.
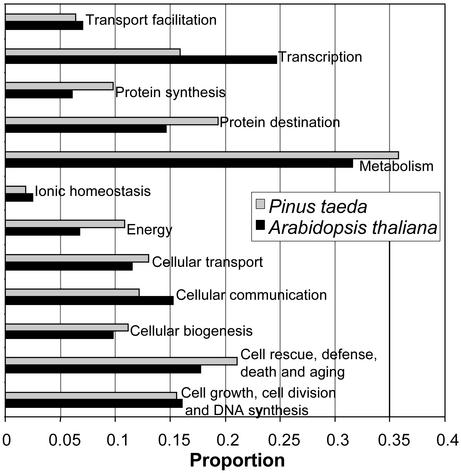
Of the 20,377 total contigs and singlets in our UniGene set, 49.7% have a predicted BLASTX homolog at E < 10-5. The diversity of loblolly pine sequences is high, and the relative frequency of inferred genes in each category for loblolly pine is similar to the spectrum of inferred genes for Arabidopsis (Fig. 1). A relatively smaller fraction of pine ESTs is assigned to transcription and cellular communication, but relatively more are assigned to protein synthesis and targeting. Few of our loblolly pine xylogenesis ESTs are assigned to photosynthesis (as expected for xylem-forming tissues).
Fig. 1.
Loblolly pine xylogenesis UniGene set, classified by cellular functional categories, compared with A. thaliana. The proportion of Arabidopsis genes in each functional category is relative to the 12,922 total predicted genes that were assigned by the Arabidopsis Genome Initiative (7) to 1 of 12 major categories. The proportion of predicted loblolly pine genes in each functional category is relative to the total number of contigs and singlets (xylogenesis UniGene set) for which homology was found to an Arabidopsis gene (blastx E value < 10-5) that was assigned to at least one functional category.
Properties of Contigs as a Function of Length. Contig consensus sequences are more informative than singlets, because their construction from multiple, overlapping ESTs increases both the quality and length of the sequence. Consequently, the sequence is more accurate, and the longer contigs usually contain more, and often complete, coding sequence. Analyzing contigs rather than singlets also minimizes the deleterious effect of transcriptional artifacts. We surveyed our contigs for ORFs using DIO-GENES and for full-length coding sequences using BLASTX (Table 2) to analyze contig properties as a function of sequence length.
Table 2. Properties of contigs based on length of sequence.
| Contig length, bp | No. of contigs | No. of ORFs | No. of full-length coding sequences |
|---|---|---|---|
| 100-500 | 2,921 | 1,936 | 55 |
| 501-900 | 4,306 | 3,772 | 630 |
| 901-1,300 | 623 | 618 | 289 |
| 1,301-1,700 | 159 | 159 | 109 |
| >1,700 | 61 | 61 | 45 |
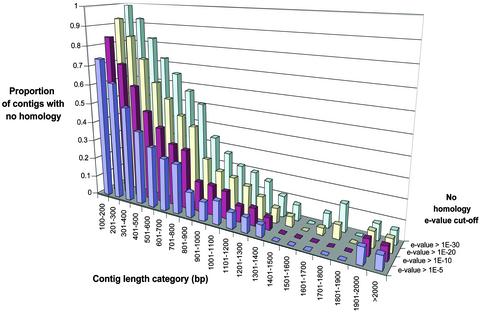
Apparent Homology of Contigs Increases with Increased Sequence Length and Quality. No-hit contig sequences could be short segments of genes that would be recognized as homologs if more sequence were available. These segments could be from 3′ or 5′ ends or regions of proteins sufficiently diverged to escape our screening criteria for similarity. Therefore, we examined the relationship of homolog identification as a function of length of contigs composed of high-quality sequence (Fig. 2). Above 900 bases, 92% of the contigs show homology to A. thaliana by using a BLASTX E-value cutoff of 10-5. A similar proportion (93%) is reached for sequences above 1,300 bases by using a far more stringent BLASTX E-value cutoff of 10-30. If we select contigs predicted by DIOGENES to contain ORFs, we find a very similar distribution (data not shown).
Fig. 2.
Proportion of P. taeda (loblolly pine) contigs with no homology to A. thaliana predicted gene sequences (y axis) relative to the contig length category (x axis). Four blastx E-value thresholds (>10-30, >10-20, >10-10, and >10-5) were used to indicate no homology (z axis).
Although the number of contigs in the length range above 900 bases represents a relatively small fraction (843 contigs) of the total number of contigs in our xylogenesis UniGene set (8,070 contigs) (Table 2), there is a clear trend toward higher sequence similarity with predicted A. thaliana genes as contig length increases. Many proteins have hydrophobic cores, which tend to be more conserved across major taxa than the N- and C-terminal regions (24). If untranslated regions (UTRs) and 5′ and 3′ ends of protein-coding sequences are less conserved relative to the middle of the sequences, the longer sequences are simply more likely to extend into regions of recognizable similarity. DIOGENES could identify no ORF for more than half of the contigs in the size range of 100–201 bases as well as for a significant proportion (>30%) of the contigs with <500 bases (Table 2).
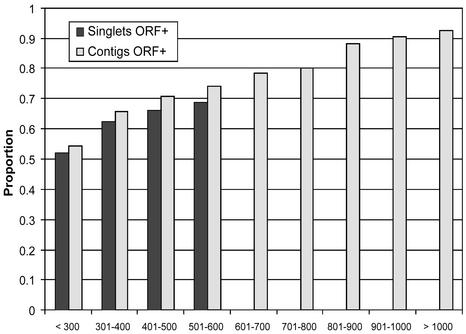
Is There Bias Resulting from the Preference to Clone-Abundant Sequences? The relationship of similarity and sequence length could be biased toward the longer sequences if more-abundantly expressed genes are more highly conserved in plants. Contigs should represent the more-abundant sequences, and singlets should represent the less-abundant ones. We have compared the relationship of sequence length and Arabidopsis similarity for both singlets and contigs with identifiable ORFs. If abundant sequences are more conserved, and therefore sequence similarity to Arabidopsis more readily identified, we should see a difference in the distribution of singlets showing similarity to Arabidopsis as a function of length compared with contigs when both are selected only for those that have ORFs. We find little difference in the two distributions (Fig. 3). Our results for singlets extend up to a length of 600 bp, which is our practical limit for PHRED 20 quality single-sequence reads on both the ABI377 and ABI3700. There is a small (0.06) difference in the proportions for the 501- to 600-length class (significant at a P = 0.01 level by using a two-sided t test), supporting our conclusion that there is little bias toward conservation as length increases.
Fig. 3.
Relationship of length of ORF-containing contigs and length of ORF-containing singlets to percentage of apparent Arabidopsis homologs.
Estimation of Length and Location of UTRs for the Loblolly Pine Xylogenesis UniGene Set. There are no estimates of the length of 5′ or 3′ UTRs for a significant number of genes from any gymnosperm. UTRs are known to play important regulatory roles in posttranscriptional processing of mRNAs (25). We selected 751 contigs that have both very high similarity (BLASTX E value < 10-30) to Arabidopsis and where each contig extends over the entire length of the Arabidopsis homolog. The shortest contig was a 60S ribosomal L38-like protein (207 bp in Arabidopsis), and the longest contig was a subtilisin-like serine protease ARA 12 (2,271 bp in Arabidopsis). We estimated the length of 5′ UTRs by determining the number of base pairs between the 5′ end of the contig and the predicted AUG start codon common to both Arabidopsis and loblolly pine. This approximation of the average length of 5′ UTRs in loblolly pine is 132 bp (median = 106), with the largest region being 1,490 bp (contig 8,061). A similar approximation can be made for 3′ ends based on the inferred C terminus (the amber, ochre, or opal stop codon). The approximation of the average length of 3′ UTRs is 256 bp (median = 254), with a maximum of 696 bp (contig 7,670). This method provides a minimum estimate, because many contigs may not be complete. Corresponding loblolly pine transcript and genomic sequences for these contigs are needed to better define the 5′ and 3′ UTRs (26).
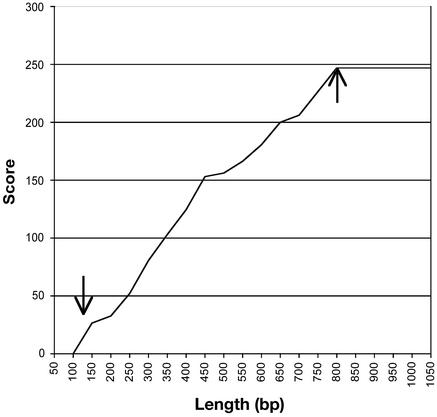
Sequence Similarity Is Typically Distributed over the Length of the Contigs. It is important to know how similarity is distributed within the loblolly pine contig sequences. Similarity could reside only in limited, conserved domains, but it could also be distributed across the entire expressed sequence. When similarity extends over a large fraction of an expressed gene, functional conservation is more likely. We have used a “gene-scan” method where we divide a sequence into regions of 50 bp and scan the sequence from the 5′ end, looking at the cumulative BLASTX scores for 50-bp intervals (e.g., for the first 50 bp, then the first 100 bp, the first 150 bp, etc., through the entire sequence) to determine how the similarity is distributed. Typically, a scan of a contig consensus sequence shows a lag followed by an upward slope followed by a plateau (Fig. 4). The sequence alignments from the BLASTX reports reveal that the inflection points (see arrows) are close to the junctions of the putative 5′ and 3′ UTRs, and the upward slope represents regions of similarity extending over the length of the inferred gene sequence. Most contigs show a similar pattern, although slopes differ, suggesting different times since divergence of ancient paralogs or different rates of sequence evolution. Two-thirds of the loblolly pine contigs spanning the full coding sequence have regions of very high similarity (BLASTX E value < 10-30) extending over 90% of the sequence (Table 3). Almost all (98%) of these contigs have very high similarity over >50% of the inferred coding sequence. Regions of similarity are not restricted to one or two short domains but are typically distributed over a long stretch of the coding sequence.
Fig. 4.
Sequence similarity (blastx score on y axis) of contig 6,593 at increasing lengths (from 5′ to 3′) compared with the Arabidopsis probable homolog. The left and right arrows indicate the beginning and end of the Arabidopsis gene coding sequence, respectively.
Table 3. Extent of coding sequence similarity for contigs containing full-length homologs.
| Extent of similarity, % | >50 | >60 | >70 | >80 | >90 |
| Proportion | 0.99 | 0.96 | 0.90 | 0.83 | 0.69 |
Two Percent of Pine Contigs That Do Not Have Homologs in Arabidopsis Have Similarity to Other Entries in GenBank. One hundred and sixty-one loblolly pine xylogenesis EST contigs showed no homology to the Munich Information Center for Protein Sequences database for Arabidopsis but did show homology (BLASTX E value < 10-5) to other entries in GenBank. For example, a cytochrome P450 (loblolly pine contig 7,997) has a homolog in tobacco but not in Arabidopsis. Similarly, two contigs have very high homology to chitinases found in a mollusk, the cone shell (Conus tulipa L.). Sixty-six contigs have been found only in pine or spruce. Eleven sequences are found in both monocots and other dicots, whereas 18 are found only in monocots and 6 only in dicots. Thirteen are found in animals, 4 in fungi, and 10 in bacteria.
These contigs are biased toward specific categories of sequences. Twenty-eight contigs are found for arabinogalactan-like proteins, known to be highly diverse in sequence but with specific protein (27–29). Twenty-one contigs are homologous to proteins induced by abscisic acid, or water stress. Five are late embryo-abundant proteins of conifers. Six resemble leucine-rich repeat sequences found in disease-resistance genes. Seventeen are hypothetical proteins in a diversity of organisms.
Discussion
Our results support the use of Arabidopsis for comparative genomics not only for angiosperms but for gymnosperms as well. The major question we address is the relationship of the expressed genes in different vascular plant genomes using loblolly pine and Arabidopsis as representatives of the two major taxa of seed plants. Allen (11) compared ESTs from three dicotyledon crop species to Arabidopsis and found that ≈10% of expressed genes failed to hit homologs in Arabidopsis. Gymnosperms and angiosperms diverged ≈300 Mya, and the major division of the angiosperms into monocotyledons and dicotyledons occurred ≈200 Mya (30). The divergence of the lineages of tomato (Solanales) from Arabidopsis (Brassicales) is estimated at 100–150 Mya (11). Allen's results suggest that one should find an even higher number of differences between pine and Arabidopsis. We find a somewhat higher frequency of homologs. Among the no-hit to Arabidopsis category, we found many proteins recognizable as arabinogalactan proteins, which may evolve under selection for structural motifs. Therefore, the number of differences between taxa can be more or less under different criteria for homology.
Our study focused on pine EST contigs containing long high-quality sequences to maximize the possibility that they would extend into coding regions. For <10% of these contigs above 1,000 bp, no Arabidopsis homolog was found at a BLASTX E-value cutoff of 10-10. For the other 90%, the regions of similarity are typically distributed over long regions of coding sequence, providing confidence in these homologies. However, even if most loblolly pine genes have a homolog in Arabidopsis, it does not mean that loblolly pine and Arabidopsis have nearly the same number of genes. One or the other species could have very different numbers of genes within homologous gene families. For the tissues we sampled, we did not observe a higher level of gene-content diversity within families for loblolly pine, despite the 160-fold difference in genome size. Certainly, many loblolly pine genes have not been found yet. The samples of pine tissues used for this study were limited to wood-forming tissues specialized for secondary wall biosynthesis and programmed cell death.
In microbes and eukaryotes, many genes retain regions of sequence conservation over 1 billion years of evolution, whereas other genes show no apparent relationship. Many of the differences between closely related taxa may turn out to be simply artifacts of annotation or other methodological limitations of either genomic or EST sequencing. For example, rice (Oryza sativa L.) and Arabidopsis last shared a common ancestor ≈200 Mya (30). In the comparison of the rice draft-genome sequence (8, 9) with the genome sequence of Arabidopsis, 81% of the predicted Arabidopsis genes had an apparent homolog in rice, but only 49% of the inferred rice genes had a homolog in Arabidopsis. Most of the 51% “novel” genes in rice were not found in EST databases, and although they could be very rarely expressed sequences, Bennetzen (31) suggests that they are more likely to be artifacts of annotation of the rice genome. Some differences may also be due to the rice genome sequence not yet being in “base-perfect” form.
Given that the apparent homology of EST contigs increases with increased sequence length and quality, more full-length, high-quality cDNA sequences are clearly needed to find meaningful sequence similarity between distant taxa. Evaluation of distant functional relationships also requires full-length cDNA sequences including 5′ and 3′ UTRs. UTRs may have important posttranscriptional regulatory functions that are highly conserved (25, 32, 33).
Low-redundancy genomic sequencing of the first tree species, Populus (34), is now in progress. Obtaining the full genome sequence of loblolly pine, or any of its close relatives, is currently not feasible because of their very large genome sizes. A comparison of pine cDNAs with the genome of a woody angiosperm should provide additional insight into the conservation of gene content and gene regulation in gymnosperms and angiosperms. A comparison of loblolly pine, Populus, and Arabidopsis is also likely to improve our current understanding of the genetic basis of wood formation.
Based on the evolution of the woody and herbaceous growth habits, it is plausible that the genes for wood formation are functionally conserved. Wood is a primitive character, and the herbaceous habit is a derived state for angiosperms (35). With daily inflorescence pruning, Arabidopsis can grow rosettes up to 7 inches in diameter and produce woody inflorescence stems and roots with detailed anatomy similar to woody dicotyledons (36–39). Some woody plants in island floras are derived recently from more-herbaceous founders (40, 41). The evolution of wood formation per se, or the herbaceous habit, may simply involve the differential regulation of sets of similar genes rather than the evolution of new gene functions. Thus, a common set of genes for woodiness could exist for all seed plants. Although it is plausible that genes involved in wood formation are conserved between plant species, this may not be the case for genes involved in other plant traits, such as flower and cone formation, or in the formation of the many diverse plant secondary products. A more-specific and comprehensive survey of pine genes involved in flowering and other tissues therefore is needed.
Supplementary Material
Acknowledgments
We thank Gisele Gurgel and Reenah Schaffer for efforts in the early stages of library construction and sequencing; Dr. David M. O'Malley for involvement in the early planning and organization of this project; Andrea Brawner, Kihlon Golden, Lesley Ireland, Sheila Maxwell, Jamal Mitchell, Sabrina Piercy, Brian Smith, and Amy Stambaugh for high-throughput EST sequencing; and John Freeman, Suzanne Grindle, Lee Parsons, Shalini Raghavan, and Martina Stromvik for the construction and maintenance of our EST databases and EST pipeline, assembly of ESTs into contigs, and assistance with EST analysis. Emily Honeycutt provided assistance with programming. We also thank Dr. Bryon Sosinski and Limei He of the North Carolina State Genome Research Laboratory for assistance with the ABI3700 capillary sequencer. This work was supported by National Science Foundation Grant DBI 9975806, a North Carolina State University Genomics program fellowship (to M.K.), and the North Carolina State University Forest Biotechnology Industrial Research Consortium.
Abbreviation: Mya, million years ago.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. can be found in Table 4, which is published as supporting information on the PNAS web site, www.pnas.org).
References
- 1.Edwards, D., Davies, K. L. & Axe, L. (1992) Nature 357, 683-685. [Google Scholar]
- 2.Bowe, L. M., Coat, G. & de Pamphilis, C. W. (2000) Proc. Natl. Acad. Sci. USA 97, 4092-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuzoff, R. K. & Gasser, C. S. (2002) Trends Plant Sci. 5, 330-336. [DOI] [PubMed] [Google Scholar]
- 4.Chaw, S.-M., Parkinson, C. L., Cheng, Y., Vincent, T. M. & Palmer, J. D. (2000) Proc. Natl. Acad. Sci. USA 97, 4086-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leitch, I. J., Hanson, L., Winfield, M., Parker, J. & Bennett, M. D. (2001) Ann. Bot. (London) 88, 843-849. [Google Scholar]
- 6.Wakamiya, I., Newton, R. J., Johnston, J. S. & Price, H. J. (1993) Am. J. Bot. 80, 1235-1241. [Google Scholar]
- 7.The Arabidopsis Genome Initiative (2000) Nature 408, 791-826.11130708 [Google Scholar]
- 8.Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. (2002) Science 296, 92-100. [DOI] [PubMed] [Google Scholar]
- 9.Yu, J., Hu, S., Wang, J., Wong, G. K., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. (2002) Science 296, 79-92. [DOI] [PubMed] [Google Scholar]
- 10.Sax, K. & Sax, H. J. (1933) J. Arnold Arbor. 14, 356-389. [Google Scholar]
- 11.Allen, K. (2002) Proc. Natl. Acad. Sci. USA 99, 9568-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 13.Ewing, B. & Green, P. (1998) Genome Res. 8, 186-194. [PubMed] [Google Scholar]
- 14.Allona, I., Quinn, M., Shoop, E., Swope, K., St. Cyr, S., Carlis, J., Riedl, J., Retzel, E., Campbell, M. M., Sederoff, R. & Whetten, R. W. (1998) Proc. Natl. Acad. Sci. USA 95, 9693-9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whetten, R., Sun, Y.-H., Zhang, Y. & Sederoff, R. (2001) Plant Mol. Biol. 47, 275-291. [PubMed] [Google Scholar]
- 16.Chang, S., Puryear, J. & Cairney, J. (1993) Plant Mol. Biol. Rep. 11, 113-116. [Google Scholar]
- 17.Ewing, B. & Green, P. (2000) Nat. Genet. 25, 232-234. [DOI] [PubMed] [Google Scholar]
- 18.Altschul, S., Madden, T., Schaffer, A., Zhang, J. H., Zhang, Z., Miller, W. & Lipman, D. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crow, J. F. & Retzel, E. F. (2001) diogenes (Univ. of Minnesota, Minneapolis), Version 2.1.2.
- 20.Liang, F., Holt, I., Pertea, G., Karamycheva, S., Salzberg, S. & Quackenbush, J. (2000) Nat. Genet. 25, 239-240. [DOI] [PubMed] [Google Scholar]
- 21.Makalowski, W. & Boguski, M. S. (1998) Proc. Natl. Acad. Sci. USA 95, 9407-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kossack, D. S. & Kinlaw, C. S. (1999) Plant Mol. Biol. 39, 417-426. [DOI] [PubMed] [Google Scholar]
- 23.Bork, P. (2000) Genome Res. 10, 398-400. [DOI] [PubMed] [Google Scholar]
- 24.Brandon, C. & Tooze, J. (1999) Introduction to Protein Structure (Garland, New York), 2nd Ed., p. 410.
- 25.Jackson, R. J. (1993) Cell 74, 9-14. [DOI] [PubMed] [Google Scholar]
- 26.Kan, Z., Gish, W., Rouchka, E., Glasscock, J. & States, D. (2000) Proc. Int. Conf. Intell. Syst. Mol. Biol. 8, 218-227. [PubMed] [Google Scholar]
- 27.Zhang, Y., Sederoff, R. R. & Allona, I. (2000) Tree Physiol. 20, 457-466. [DOI] [PubMed] [Google Scholar]
- 28.Schultz, C. J., Johnson, K. L., Currie, G. & Bacic, A. (2000) Plant Cell 12, 1751-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, Y., Brown, G., Whetten, R., Loopstra, C. A., Neale, D., Kileiszewski, M. J. & Sederoff, R. R. (2003) Plant Mol. Biol., in press. [DOI] [PubMed]
- 30.Wolfe, K. H., Gouy, M. L., Yang, Y. W., Sharp, P. M. & Li, W. H. (1989) Proc. Natl. Acad. Sci. USA 86, 6201-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennetzen, J. (2002) Science 296, 60-63. [DOI] [PubMed] [Google Scholar]
- 32.Duret, L., Dorkeld, F. & Gautier, C. (1993) Nucleic Acids Res. 21, 2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jareborg, N., Birney, E. & Durbin, R. (1999) Genome Res. 9, 815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mann, C. C. & Plummer, M. L. (2002) Science 295, 1626-1629. [DOI] [PubMed] [Google Scholar]
- 35.Sporne, K. R. (1980) New Phytol. 85, 419-445. [Google Scholar]
- 36.Lev-Yadun, S. (1994) J. Exp. Bot. 45, 1845-1849. [Google Scholar]
- 37.Lev-Yadun, S. (1997) Ann. Bot. (London) 80, 125-129. [Google Scholar]
- 38.Zhao, C. S., Johnson, B. J., Kositsup, B. & Beers, E. P. (2000) Plant Physiol. 123, 1185-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lev-Yadun, S. & Flaishman, M. A. (2001) IAWA J. 22, 159-169. [Google Scholar]
- 40.Bohle, U.-R., Hilger, H. & Martin, W. F. (1996) Proc. Natl. Acad. Sci. USA 93, 11740-11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Givnish, T. J. & Sytsma, K. J. (2000) Molecular Evolution and Adaptive Radiation (Cambridge Univ. Press, Cambridge, U.K.), p. 621.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.