Abstract
Precursor protease vesicles are plant-specific compartments containing precursors of enzymes that are thought to participate in the degradation of cellular components in organs undergoing senescence. We report in vivo evidence that the precursor protease vesicle-localized vacuolar processing enzyme-γ (VPEγ) is critical for maturation of the plant vacuolar protease AtCPY. We also provide biochemical and functional evidence that VPEγ is involved in degradation of the vacuolar invertase AtFruct4 in aging tissues. Moreover, a proteomics-based approach identified various proteins found in the vacuoles of aging vpeγ mutants but not in WT plants, suggesting a unique role of VPEγ in protein processing and degradation in Arabidopsis.
Many vacuolar enzymes are synthesized as pro-proteins and need to be proteolytically processed to be active. Plant-specific, endoplasmic reticulum (ER)-derived compartments containing precursors of cysteine proteinases (PPVs) have been described in seed-storage tissues (1–4). Germination of the seeds induces the expression and processing of those proteases into the mature active forms, which are thought to participate in the degradation of cellular materials in senescing storage tissues, presumably to assist in the programmed cell death process and to provide nutrients to the growing embryo. However, direct evidence linking these compartments with the senescence process in plants is lacking, which in part may be because seed PPVs have been found in legumes, which are not very amenable to genetic studies. Recently, similar compartments also called the ER bodies have been described in vegetative tissues of Arabidopsis (2). In this article we refer to these compartments in Arabidopsis vegetative tissues as PPVs.
Two cysteine proteinases, vacuolar processing enzyme-γ (VPEγ) and RD21, are known to be present in Arabidopsis PPVs. For RD21, it has been shown that PPVs store the inactive precursor isoform. Senescence induces the expression of VPEγ and RD21 and the processing of RD21 into the mature active form (5), most likely in the acidic environment of the vacuole. In daylily, PPVs containing cysteine proteinase have been found in petals, which in this species undergo rapid senescence within 24 h of flower opening. These observations indicate that vegetative PPVs may also have a senescence-associated function, as has been suggested for seed PPVs. VPEγ belongs to the small gene family of VPEs (6) that are thought to process storage proteins in vacuoles of developing seeds. Processing activity of castor bean and pumpkin VPE on several seed-storage proteins has been demonstrated in vitro (7, 8). By analogy, the Arabidopsis seed-specific VPE isoform, VPEβ, is thought to process seed-storage proteins. In contrast, the substrates of VPEγ, which is expressed in vegetative tissues, are not known. However, through heterologous expression in yeast it has been shown that VPEγ processes the vacuolar pro-protein precursor of yeast carboxypeptidase Y (CPY), indicating that VPEγ has vacuolar processing activity.
We reasoned that mutations in VPEγ might result in the loss of activity of many downstream hydrolases processed by it and might therefore have an important and measurable effect on the senescence process. In this work, we report the identification and characterization of Arabidopsis null mutants of VPEγ. We show that vpeγ mutants are blocked in the maturation of an Arabidopsis homologue of yeast CPY (AtCPY) and that the defect in maturation can be complemented by transgenic expression of VPEγ. These results, which are in vivo evidence of the role of VPEs in the processing of vacuolar proteins, suggest that AtCPY is a direct substrate for the vegetative VPEγ.
VPEγ is induced in senescing organs of old plants and its protease activity is required for degradation of the vacuolar protein AtFruct4 in aging tissues. We propose that senescence induced by aging activates VPEγ, possibly by releasing the inactive precursor form from PPV into the acidic lumen of the vacuole, triggering the processing of downstream proteases for the degradation/recycling of proteins in dying cells.
Materials and Methods
Mutant Isolation. Primary screening of a collection of T-DNA insertional mutants was performed and the mutants were obtained from Mendel Biotechnology (Hayward, CA) by using a PCR-based methodology (9). We isolated the homozygous mutants and determined the insertion sites (data not shown). To complement the vpeγ mutations, we digested the Arabidopsis EST 208I21T7 with BamHI–PstI and cloned it directionally under the control of the 35S promoter into a derivative of the binary vector pCAMBIA1300 to produce plasmid pNVRE80. The plasmid pNVRE80 was introduced into Agrobacterium and transformed in vpeγ plants by infiltration as described (10).
Antibody Production. AtCPY antibodies were raised against a fragment of the protein (amino acids 306–539) expressed in Escherichia coli as a His-fusion protein in the pET28b vector (Novagen). The protein was purified by nickel affinity chromatography and injected into two rabbits. Sera from both rabbits recognized the same bands as described in Fig. 2. Polyclonal AtFruct4 antibodies were raised by injecting a rabbit with the peptide LDIEAEFEINKESLDKIIGN coupled to keyhole limpet hemocyanin. The serum was affinity-purified as described (11) against a fragment of AtFruct4 expressed as a His-fusion protein in E. coli (12). Other AtFruct4 antibodies (12) gave identical results. All other antisera used in this study have been described.
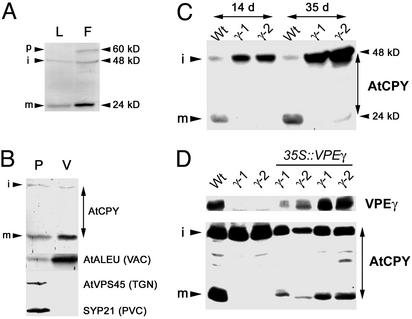
Fig. 2.
VPEγ is required for AtCPY maturation. (A) Total protein extracts from rosette leaves (L) and flowers (F) of WT plants were analyzed by Western blot with α-AtCPY antibodies. The positions of the precursor (p), intermediate (i), and mature (m) forms of AtCPY are indicated. (B) Total protein samples from protoplast (P) and vacuoles (V) from an Arabidopsis cell suspension culture were analyzed by Western blot for the presence of previously described Arabidopsis markers of different compartments of the endomembrane system. TGN, trans-Golgi network; PVC, prevacuolar compartment; VAC, vacuole. (C) Total protein extracts from rosette leaves of WT, vpeγ-1, and vpeγ-2 mutant plants 14 or 35 days after germination were analyzed by Western blot with α-AtCPY antibodies. (D) Total protein extracts from rosette leaves of WT, vpeγ-1, vpeγ-2, and four independent vpeγ mutant lines transformed with the VPEγ cDNA under the control of the 35S promoter were analyzed by Western blot with α-VPEγ and α-AtCPY antibodies.
Electron Microscopy. Roots from Arabidopsis plants grown in vitro were fixed in 1.5% formaldehyde, 0.5% glutaraldehyde, and 0.05 M sodium phosphate, pH 7.4, for 2 h at room temperature, postfixed in OsO4, and embedded in London Resin white (Polysciences). Single and double immunolabeling of ultrathin sections were performed as described (13).
Subcellular Fractionation Studies. Vacuoles were purified from Arabidopsis cell suspension cultures as described (14), and samples from protoplast and the vacuole-enriched fraction were analyzed by SDS/PAGE and immunoblotting. Sucrose gradients of Arabidopsis root cultures were performed as described (11). This procedure allows for efficient separation of vacuoles from other endomembrane compartments (15). Twenty-five fractions were collected, and the proteins in the fractions were precipitated with trichloroacetic acid. Protein samples from odd fractions were analyzed by SDS/PAGE and immunoblotting.
Proteomics Analyses. Sample preparation and digestion. The vacuolar proteins were precipitated by the addition of ice-cold acetone at 4:1 followed by incubation at -80°C for 1 h and centrifugation at 20,000 × g at 4°C for 30 min. The pellets obtained were each rinsed twice with 1 ml of ice-cold 80% acetone and allowed to dry. The protein pellets were then dissolved in 100 μl of freshly prepared 100 mM ammonium bicarbonate/1 M urea/125 mM thiourea/5% acetonitrile and digested overnight at 37°C after the addition of 10 μg of trypsin. The next day, insoluble material was removed by centrifugation. Each digest was brought to 300 μl by using HPLC solvent A [5 mM phosphate buffer (pH 2.7), 25% acetonitrile] and acidified by using phosphoric acid to pH <3. Insoluble material was removed by centrifugation and 250 μl was loaded onto the strong cation exchange column that had been equilibrated in HPLC solvent A. HPLC separation on the strong cation exchange column was performed in two steps: first, a 5-min linear gradient from 0% to 30% HPLC solvent B (5 mM phosphate buffer (pH 2.7)/350 mM KCl/25% acetonitrile), and second, a 3-min gradient from 30% to 100% HPLC solvent B. The flow rate was 50 μl/min and fractions were collected starting at 5 min. Each fraction contained ≈37.5 μl. Fractions that contained a substantial amount of peptides as exhibited by absorbance at 214 nm were analyzed by liquid chromatography/tandem MS as described below.
Peptide identification by liquid chromatography/tandem MS. Nanoscale capillary liquid chromatography/tandem MS analysis of peptide samples was performed by using an autosampler/nanoscale HPLC system from LC Packings (San Francisco) coupled to a ThermoFinnigan (San Jose, CA) LCQ Deca XP ion trap mass spectrometer. Samples (6 μl) were applied to a reverse-phase peptide CapTrap (Michrom Bioresources, Auburn, CA) and the CapTrap was washed with ≈60 μl of 2% acetronitrile/0.1% trifluoroacetic acid. Trapped peptides were then fractionated by using a 75 μm × 5.1-cm prepacked PicoFrit Aquasil C-18 column with a 15-μm tip (New Objective, Woburn, MA) placed in-line with the trap. Solvents used for fractionation were 0.1% formic acid (solvent A) and 95% acetonitrile/0.1% formic acid (solvent B). A 70-min linear gradient from 5% to 20% solvent B followed by a 10-min gradient from 20% to 60% solvent B was used for fractionation at ≈200 nl/min. The mass spectrometer was operated in positive-ion electrospray mode and controlled with XCALIBUR (ThermoFinnigan). A precursor scan of m/z values from 400 to 1,200 was followed by data-dependent acquisition of tandem MS data of the four most intense ions. Tanden MS spectra were analyzed by using MASCOT (Matrix Science, London) to match the spectra to peptides in the National Center for Biotechnology Information nonredundant database and a database consisting of all predicted Arabidopsis proteins.
Results and Discussion
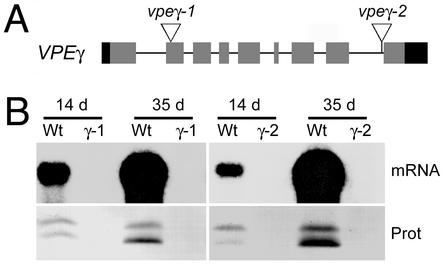
Isolation of vpeγ Insertional Mutants. The Arabidopsis genome contains three VPE genes (6). One of the isoforms, VPEβ, is expressed specifically in seeds, suggesting that it plays a role similar to that of seed-specific VPEs from other species. The other two isoforms, VPEγ and VPEα, are expressed in vegetative tissues and are induced during senescence and also by various stresses. Although VPEγ and VPEα have a similar expression pattern (16), the level of VPEγ expression is much higher than that of VPEα. GenBank database searches retrieved 40 ESTs corresponding to VPEγ in libraries made from vegetative tissues. In contrast, only one EST for VPEα was found corresponding to a cDNA library from developing seeds. Consistent with this result, we found that VPEγ steady-state transcript levels in Arabidopsis leaves are at least 100-fold higher than those of VPEα (data not shown). These results indicate that VPEγ is the major vacuolar processing enzyme expressed in vegetative tissues in Arabidopsis. To understand the role that PPV-contained cysteine proteinases have in plant senescence we searched for plants containing T-DNA insertions in the gene. We have identified two mutant alleles: the vpeγ-1 allele, which is a result of a T-DNA integration in the first exon of VPEγ, and the vpeγ-2 allele, which is a result of a T-DNA integration in the last intron of the gene (Fig. 1A). VPEγ mRNA levels are highly induced in senescing organs of WT plants, which accumulate large amounts of the active intermediate (43 kDa) and mature forms (40 kDa) of the VPEγ protein (Fig. 1B). In contrast, no VPEγ mRNA or protein is detected in the mutants, demonstrating that both alleles are null (Fig. 1B). In addition, no significant increase of VPEγ mRNA was detected (data not shown), indicating that the VPEα mRNA was not up-regulated in a compensatory manner in vpeγ mutants.
Fig. 1.
vpeγ-1 and vpeγ-1 are null alleles. (A) Diagram of the VPEγ gene. Exons are shown as boxes, with gray indicating coding regions and black indicating the 5′ and 3′ UTR. The positions where the T-DNAs are inserted in the vpeγ-1 and vpeγ-2 alleles are shown. (B) Total mRNA and protein extracted from rosette leaves of WT, vpeγ-1, and vpeγ-2 mutant plants 14 or 35 days after germination were analyzed by Northern (mRNA) and Western blot (Prot) with a VPEγ probe or α-VPEγ antibodies, respectively. Positions of intermediate and mature forms of VPEγ are 43 and 40 kDa, respectively.
AtCPY Is a Direct Target for Vegetative VPEγ The in vitro processing activity of seed-specific VPEs on several storage proteins is well established. In contrast, the putative substrates of the vegetative VPEs are not known. It has been reported that the processing of RD21, the only other previously known cargo of Arabidopsis vegetative PPVs, does not depend on VPEγ or VPEα. However, the processing activity of the vegetative VPEγ on yeast carboxypeptidase Y (CPY) has been demonstrated by heterologous expression of VPEγ (16). To test whether VPEγ is required for maturation of Arabidopsis vacuolar carboxypeptidases, we searched the Arabidopsis genome for proteins homologous to yeast CPY. Several serine proteases homologous to yeast CPY can be found in the Arabidopsis genome. The carboxypeptidase with the highest homology score, At3g10410 (named AtCPY hereinafter), contains an N-terminal extension shared with CPY from yeast but is absent in other Arabidopsis homologues. The vacuolar sorting information for CPY is present in that domain, so we hypothesized that AtCPY may contain vacuolar-sorting signals in the N-terminal extension.
We raised rabbit polyclonal antibodies against the C-terminal half of AtCPY expressed as a His-fusion in E. coli. This region of the protein is the most divergent between the different carboxypeptidases from Arabidopsis. Sera from both rabbits recognized bands of 48 and 24 kDa in leaf extracts from WT plants (Fig. 2A), that were absent in immunoblots with preimmune sera (data not shown). Flower buds had the highest expression of AtCPY, and we detected with both sera an additional band of ≈60 kDa that is consistent with the molecular mass predicted for the precursor form of AtCPY. Proteins homologous to AtCPY, such as barley and wheat carboxypeptidase (17) and Arabidopsis glucose acyl transferase (18), are processed into two subunits. Presumably, the 48-kDa band is an intermediate form that is processed into an N-terminal subunit not recognized by our antibodies and a C-terminal subunit, which is the one detected by the antibodies as the 24-kDa band. AtCPY is present in highly purified preparations of isolated vacuoles (Fig. 2B), suggesting that it is localized in this organelle and may be a substrate of VPEγ processing. As shown in Fig. 2C, the mature C-terminal subunit of AtCPY is absent in vpeγ-1 and vpeγ-2 plants, indicating that VPEγ is involved in the processing of AtCPY. A concomitant increase in the abundance of the 48-kDa band is apparent in the mutants relative to WT plants, demonstrating that it constitutes a precursor of the mature 24-kDa subunit. The processing of AtCPY is recovered when the VPEγ cDNA is expressed under the 35S promoter in the mutant background (Fig. 2D), confirming that VPEγ is involved in AtCPY maturation. This in vivo evidence for a role of the VPE enzymes in the maturation of proteins suggests that AtCPY is a direct target for vegetative VPEs.
VPEγ Is Essential for Degradation of Vacuolar Invertase in Aging Tissues. Our results indicate that VPEγ may regulate the hydrolytic activity of vacuoles by processing proteases such as AtCPY into their mature forms. Accordingly, one-dimensional electrophoresis analysis of vacuolar contents reveals proteins that accumulate at higher amounts in vpeγ plants (data not shown), which may reflect a higher rate of proteolysis in WT vacuoles. This finding is consistent with the role proposed for other cysteine proteinases from seed PPVs in the turnover of proteins accumulated in storage organs. On the basis of its induced expression in aging or stressed tissues and its vacuolar localization, it has been proposed that VPEγ is involved in the recycling of vacuolar proteins in tissues that undergo senescence. However, no evidence for regulation of protein turnover by VPEγ has been presented yet.
We searched the literature to find putative candidates for VPEγ-dependent turnover. The search criteria were that the protein should be localized to the vacuole and that its expression should be down-regulated in aging organs. The members of a subset of Arabidopsis invertases have been classified as vacuolar invertases based on sequence similarity with those of other species, although their localization had not been determined previously. Importantly, in maize, a lower vacuolar invertase activity in mature leaves relative to young leaves has been reported (19).
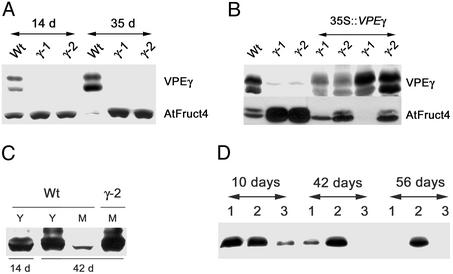
We raised antibodies against AtFruct4 (At1g12240), a putative Arabidopsis vacuolar invertase, to determine whether its accumulation is regulated by VPEγ. We used antibodies raised against a fragment of AtFruct4 expressed as a fusion protein in E. coli or against an AtFruct4 peptide, which gave identical results in Western blot and immunoelectron microscopy experiments. As shown in Fig. 3A, the levels of AtFruct4 decline in senescing leaves of WT plants, but not in vpeγ mutants, although the mRNA levels are similar in mutant and WT plants (data not shown), suggesting that senescence induces the VPEγ-dependent breakdown of AtFruct4. The overexpression of the VPEγ cDNA in the mutant background complements the defect in AtFruct4 turnover (Fig. 3B), demonstrating the role of VPEγ in AtFruct4 breakdown. To confirm that the degradation of AtFruct4 was related to senescence and not to the developmental stage of the plant, we analyzed the protein levels in young and senescing leaves of the same plant. As shown in Fig. 3C, AtFruct4 is degraded in the senescing leaves of WT but not in mutant plants or in the young leaves of aging WT plants, indicating that AtFruct4 degradation is regulated by senescence. We also tested whether AtFruct4 is degraded in senescing tissues other than leaves. As shown in Fig. 3D, AtFruct4 is also degraded in roots from aging plants, and the degradation depends on VPEγ activity.
Fig. 3.
Senescence-induced degradation of AtFruct4 requires VPEγ. (A) Total protein was extracted from WT, vpeγ-1, and vpeγ-2 rosette leaves from in vitro-grown plants at the indicated days after germination and analyzed by Western blot with α-VPEγ and α-AtFruct4 antibodies. (B) Transformation with the VPEγ cDNA complements the defect in AtFruct4 degradation. Total protein extracts from senescing rosette leaves of WT, vpeγ-1, vpeγ-2, and four independent vpeγ lines transformed with the VPEγ cDNA under the 35S promoter were analyzed by Western blot with α-VPEγ and α-AtFruct4 antibodies. Positions of intermediate and mature forms of VPEγ are 43 (A) and 40 (B) kDa, respectively. (C) Total protein was extracted from young (Y) or mature (M) leaves of WT and vpeγ-2 plants grown in soil for 14 and 42 days and analyzed by Western blot with α-AtFruct4 antibodies. (D) Total protein was extracted from roots from in vitro-grown WT (lane 1), vpeγ-2 (lane 2), and vpeγ-2 plants expressing the VPEγ cDNA under the 35S promoter (lane 3) at the indicated days after germination and was analyzed by Western blot with α-AtFruct4 antibodies.
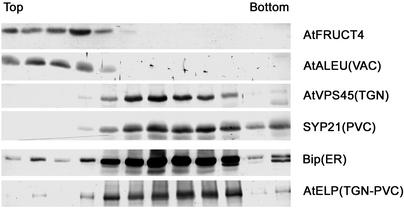
VPEγ is synthesized as an inactive precursor, and it is autocatalytically converted into the active forms under acidic conditions (20). It has been suggested that the inactive precursor of VPEγ accumulates in PPVs and that it is selfactivated by delivery into the acidic environment of the vacuole. On the basis of that suggestion, we predicted that the VPEγ-dependent degradation of AtFruct4 occurred in the vacuole. To discern the subcellular localization of AtFruct4, we fractionated Arabidopsis root culture extracts in sucrose density gradients, and the distribution of AtFruct4 was compared with previously characterized markers of different compartments of the Arabidopsis endomembrane system. AtFruct4 fractionates at the top of the gradient, together with AtALEU, a vacuolar marker (Fig. 4), and segregated from markers from the ER, trans-Golgi network, and prevacuolar compartment, which accumulate in denser fractions of the gradients. This fractionation pattern suggests that AtFruct4 is localized in the vacuole and is consistent with its VPEγ-dependent degradation occurring in this organelle.
Fig. 4.
AtFruct4 colocalizes with a vacuolar marker in subcellular fractionation studies. (A) A postnuclear supernatant of an extract from a 4-week-old Arabidopsis root culture was analyzed by fractionation on a sucrose density gradient. Protein samples from odd fractions were analyzed by SDS/PAGE and immunoblotting with the indicated antibodies against previously described Arabidopsis markers of different compartments of the endomembrane system. TGN, trans-Golgi network; PVC, prevacuolar compartment; VAC, vacuole.
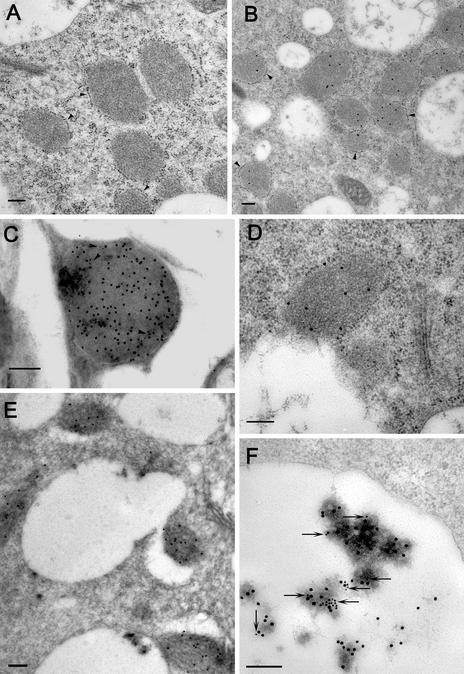
To unequivocally determine the subcellular localization of AtFruct4, we performed immunoelectron microscopy on Arabidopsis plants. Remarkably, in young seedlings AtFruct4 colocalizes with VPEγ in PPVs (Fig. 5C), indicating that vegetative PPVs store not only precursors of proteases but also other hydrolases such as invertases. AtFruct4 and VPEγ accumulate in the spindle-shape compartments, with geometry identical with that described in ref. 2. Their unique shape and size were completely different from other ER-derived compartments, including precursor-accumulating vesicles. In older plants, we could detect the incorporation of PPVs into vacuoles (Fig. 5E) and the presence of AtFruct4 in the vacuolar lumen, where it was colocalized (Fig. 5F) with the vacuolar marker barley lectin (14). This age-induced change in localization coincides with the onset of AtFruct4 degradation in roots (Fig. 3D). A similar incorporation of PPVs into vacuoles has been reported in salt-stressed Arabidopsis seedlings (2). In that work, it was proposed that this incorporation leads to the conversion of the precursor of VPEγ into the active, mature form that assists in the cell death of the epidermal cells damaged by the salt stress. Our results provide evidence for the activation of VPEγ-dependent proteolysis of AtFruct4 in vacuoles of senescing organs, supporting the model of VPEγ activation by release from PPVs into the acidic vacuolar lumen. However, mature VPEγ is also present in nonsenescing tissues, where it is involved in AtCPY maturation (Figs. 1B and 2C). This activity indicates that some VPEγ reaches the vacuole in nonsenescing tissues, by PPVs or Golgi-derived vesicles. The accumulation of AtFruct4 in PPVs, together with the VPEγ zymogen, may therefore constitute an additional mechanism to protect AtFruct4 from degradation by the residual activity of VPEγ in vacuoles of nonsenescing tissues. The localization of an invertase isoform in PPVs is without precedence and raises many questions about the function of the enzyme in that compartment.
Fig. 5.
Immunolocalization of AtFruct4. (A–E) Sequence of events in the delivery of AtFruct4 from PPVs into vacuoles. Immunoelectron microscopy was performed in ultrathin sections of roots from seedlings grown in vitro for 10 days (A–D) and 30 days (E and F) after germination. Preimmune sera (A) or affinity-purified α-AtFruct4 sera (B–F) were used as primary antibodies, followed by detection with protein A coupled to gold particles. (A and B) Isolated PPVs surrounded by ribosomes (arrowheads). (C) AtFruct4 colocalizing in PPVs with VPEγ. VPEγ-bound antibodies were visualized with 10-nm gold particles (indicated by arrows) and AtFruct4 antibodies were visualized with 15-nm gold particles. (D) Demonstration of direct contact between the tonoplast and the PPV membrane free of attached ribosomes. (E) PPVs are being delivered into the vacuoles. (F) AtFruct4 colocalizing in vacuoles with the vacuolar marker barley lectin (14) in roots of transgenic Arabidopsis. Barley lectin-bound antibodies were visualized with 15-nm gold particles and AtFruct4 antibodies were visualized with 10-nm gold particles (indicated by arrows). (Scale bars = 200 nm.)
To test the hypothesis that a set of proteins exists that are specifically processed/degraded in plants with a functional copy of VPEγ, vacuolar proteins from vpeγ-2 and WT plants were subjected to tandem MS analysis as described in Materials and Methods. Vacuoles were of high purity and had undetectable contamination from other endomemebranes (15). Thus far, we have identified various proteins that were detected only in mature leaves of vpeγ-2 plants, suggesting that these proteins may be processed/degraded in a VPEγ-dependent manner in WT plants (data not shown). Among these unique proteins, we have found four glycosidases, including a putative β-glucosidase (At1g52400), two α-mannosidases (At3g26720 and At5g13980), and an α-galactosidase (At3g56310). Their primary sequences are predicted to contain signal peptides on the N termini, thus these proteins are likely to be entering an Arabidopsis endomembrane system. It will be of great importance to identify vacuolar-targeting pathways and natural substrates of these proteins because of the striking similarity in their function and likely subsequent degradation in aging tissues. It is conceivable that these proteins may be targeted to the vacuole through the PPV-mediated pathway as we have found for AtFruct4. Recently, a β-glucosidase PYK10 was localized to PPVs (21), but its physiological role and processing/degradation remain unknown.
In conclusion, we have identified VPEγ as a key component of protein processing and degradation machinery in Arabidopsis. We have also demonstrated the uniqueness of the VPEγ-mediated pathway, on the basis of various biochemical studies in both WT and vpeγ mutant plants. The comparative proteomics approach allows for the identification of other potential targets of VPEγ, which would be overlooked if the proteomics were applied only to WT plants. To investigate the role of VPEγ in Arabidopsis vegetative tissues further, we have initiated characterization of the vpeγ mutant phenotype under various conditions that may trigger vacuolar remodeling.
Acknowledgments
We thank Dr. José J. Sánchez Serrano for helpful discussions. We are also grateful to members of the Raikhel laboratory for comments, suggestions, and support and Ms. Jocelyn Brimo for helping with the artwork and formatting the manuscript. We also thank the Michigan State Mass Spectrometry Facility for mass spectrometric sequencing. This work was supported by funds from the National Science Foundation (MCB-0296060; to N.V.R.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CPY, carboxypeptidase Y; PPV, precursor protease vesicle; VPE, vacuolar processing enzyme; ER, endoplasmic reticulum.
References
- 1.Chrispeels, M. J. & Herman, E. M. (2000) Plant Physiol. 123, 1227-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayashi, Y., Yamada, K., Shimada, T., Matsushima, R., Nishizawa, N. K., Nishimura, M. & Hara-Nishimura, I. (2001) Plant Cell Physiol. 42, 894-899. [DOI] [PubMed] [Google Scholar]
- 3.Schmid, M., Simpson, D. J., Sarioglu, H., Lottspeich, F. & Gietl, C. (2001) Proc. Natl. Acad. Sci. USA 98, 5353-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toyooka, K., Okamoto, T. & Minamikawa, T. (2000) J. Cell Biol. 148, 453-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada, K., Matsushima, R., Nishimura, M. & Hara-Nishimura, I. (2001) Plant Physiol. 127, 1626-1634. [PMC free article] [PubMed] [Google Scholar]
- 6.Hara-Nishimura, I., Kinoshita, T., Hiraiwa, N. & Nishimura, M. (1998) J. Plant Physiol. 152, 668-674. [Google Scholar]
- 7.Yamada, K., Shimada, T., Kondo, M., Nishimura, M. & Hara-Nishimura, I. (1999) J. Biol. Chem. 274, 2563-2570. [DOI] [PubMed] [Google Scholar]
- 8.Hara-Nishimura, I., Takeuchi, Y. & Nishimura, M. (1993) Plant Cell 5, 1651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krysan, P. J., Young, J. C. & Sussman, M. R. (1999) Plant Cell 11, 2283-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-774. [DOI] [PubMed] [Google Scholar]
- 11.Bassham, D. C. & Raikhel, N. V. (1998) Plant Physiol. 117, 407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rojo, E., Gillmor, C. S., Kovaleva, V., Somerville, C. R. & Raikhel, N. V. (2001) Dev. Cell 1, 303-310. [DOI] [PubMed] [Google Scholar]
- 13.Sanderfoot, A. A., Ahmed, S. A., Marty-Mazars, D., Rapoport, I., Kirchhausen, T., Marty, F. & Raikhel N. V. (1998) Proc. Natl. Acad. Sci. USA 95, 9920-9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed, S. U., Rojo, E., Kovaleva, V., Venkataraman, S., Dombrowski, J. E., Matsuoka, K. & Raikhel, N. V. (2000) J. Cell Biol. 149, 1335-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rojo, E., Zouhar, J., Kovaleva, V., Hong, S. & Raikhel, N. V. (2003) Mol. Biol. Cell 14, 361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinoshita, T., Yamada, K., Hiraiwa, N., Kondo, M., Nishimura, M. & Hara-Nishimura, I. (1999) Plant J. 19, 43-53. [DOI] [PubMed] [Google Scholar]
- 17.Degan, F. D., Rocher, A., Cameron-Mills, V. & von Wettstein, D. (1994) Proc. Natl. Acad. Sci. USA 91, 8209-8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, A. X. & Steffens, J. C. (2000) Proc. Natl. Acad. Sci. USA 97, 6902-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, J.-Y., Mahe, A., Brangeon, J. & Prioul, J.-L. (2000) Plant Physiol. 124, 71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuroyanagi, M., Nishimura, M. & Hara-Nishimura, I. (2002) Plant Cell Physiol. 43, 143-151. [DOI] [PubMed] [Google Scholar]
- 21.Matsushima, R., Kondo, M., Nishimura, M. & Hara-Nishimura, I. (2003) Plant J. 33, 493-502. [DOI] [PubMed] [Google Scholar]