Abstract
The constitutive activation of G-protein-coupled receptors is a major new approach to investigating their physiopathology and pharmacology. A large number of spontaneous and site-directed mutations resulting in constitutive activity have been identified, but systematic mapping of the amino acids involved for a given receptor would be extremely useful for complete elucidation of the molecular mechanisms underlying its activation. We carried out such mapping for the angiotensin II type 1A (AT1A) receptor by screening a randomly mutated cDNA library after expressing the mutated clones in eukaryotic cells. To test the AT1A mutants generated, we developed an original, specific, and highly sensitive assay based on the properties of CGP42112A. This classical AT2 agonist is a weak partial agonist of the wild-type AT1A receptor and becomes a full agonist for constitutively active AT1A mutants, as shown experimentally and in allostery-based theoretical models. Activation of the mutated receptors by CGP42112A was monitored by using the bioluminescent protein æquorin, a very sensitive and specific sensor of intracellular calcium mobilization. The screening of 4,800 clones, providing an exhaustive coverage of all of the mutations generated, led to the identification of 16 mutations in sequences encoding the transmembrane domains that were responsible for high sensitivity to CGP42112A. The constitutive activity was confirmed by agonist-independent production of inositol phosphates, which showed that at least half of the clones had significantly increased basal activity. These data demonstrate that this new type of approach is very efficient for the systematic identification of constitutively active mutants of G-protein-coupled receptors.
The seven transmembrane receptors (7TMRs) belong to a large superfamily of several thousand proteins and are involved in most major physiological processes. They share a common structure, seven hydrophobic putative transmembrane domains (TMs), and a common function, the transduction of an extracellular signal to intracellular heterotrimeric G-proteins. Despite extensive mutagenesis studies and molecular modeling based on the three-dimensional structures of bacteriorhodopsin and bovine rhodopsins, the precise molecular description of their activation process is still unclear. How does an extracellular interaction with a ligand, that may be as small as a calcium ion, lead to a conformational change resulting in intracellular protein activation? One approach to answering this question is to identify constitutively active mutant (CAM) receptors, which are active even in the absence of agonist, and continuously stimulate the signaling pathways. Such receptors are stuck in an active conformation and can therefore be used for the modeling of molecular activation process (1). They were first observed in site-directed mutagenesis experiments, in which CAM adrenergic receptors have been particularly studied (2–5). Constitutive activity has later been shown to be associated with the physiopathology of 7TMRs. CAMs have been shown to be responsible for developmental (6–8) and tumoral (9–12) human diseases and have been used to produce transgenic mice serving as unique experimental models (13, 14). Studies of CAM receptors have also enriched our understanding of 7TMR pharmacology. Several models based on allostery suggest that there are at least two interconverting conformations of the receptor, one active and one inactive, for which a given ligand has different affinities (1, 4, 15). Thus, the affinity and efficacy of the ligand depend on the number of receptors in the active state and are not intrinsic properties of the ligand. A particularly striking consequence is that apparent affinity and efficacy of agonists are better for CAM than for wild-type (WT) receptor (4, 16, 17). CAM receptors may therefore be useful for the screening of agonists by the pharmaceutical industry. They are also useful for the screening of inverse agonists, ligands that stabilize the inactive conformation (18).
Complete mapping of the amino acids responsible for constitutive activation of a given 7TMR would be extremely useful for elucidation of the molecular mechanisms underlying its activation. If a priori reasoning about the positions of amino acids is to be avoided, this requires the screening of a library of randomly generated mutants of the entire cDNA. In this study, we used a systematic approach of this type involving an original, specific, and very sensitive bioassay based on the greater sensitivity of CAM receptors to partial agonists. This strategy was successfully applied to the angiotensin II (AngII) type 1A (AT1A) receptor, a 7TMR responsible for most, if not all, of the cardiovascular effects of the vasoactive peptide AngII. It is coupled to a Gαq/11 protein responsible for inositol phosphate (IP) production and intracellular calcium mobilization. Little is known about CAM AT1 receptors. No disease linked to gain-of-function mutations has been described, and in vitro studies on CAM AT1 have implicated only Asn111 (third TM) in receptor activation (19–21), although it may interact with Asn295 (seventh TM) (22). As predicted by theoretical models, the agonist properties of CGP42112A, a classical AT2 agonist, depend on whether the AT1 receptor is in its WT or constitutively active form (19, 22, 23). We used this compound as a highly discriminating ligand for CAM detection and monitored receptor activation with æquorin, a sensitive luminescent sensor of intracellular calcium mobilization. This approach was very efficient and enabled us to identify several new CAM AT1A receptors.
Materials and Methods
Materials.
Æquorin and Gαq plasmids were gifts from R. Rizzuto (Univ. of Ferrara, Ferrara, Italy) (24) and B. Conklin (Univ. of California, San Francisco). The plasmids used for inducible expression (25), pTRE and pTetOff, were from CLONTECH. The pTRE HindIII site was removed and the polylinker modified to give EcoRI-EcoRV-HindIII-NotI-PstI-PmlI-XbaI, making it possible to insert the HindIII-XbaI fragment of the previously described synthetic AT1A cDNA (26). Losartan was kindly provided by Merck Sharp Dohme-Chibret. CGP42112A was purchased from Neosystem (Strasbourg, France), cœlenterazin from Molecular Probes, and other chemicals from Sigma.
Mathematical Predictions for an Optimized Random Mutant Library.
Mathematical analysis was used (i) to determine the optimal mutation rate and (ii) to predict the number of clones that must be tested for exhaustive screening.
(i) We aimed to maximize the number of clones with one amino acid substitution and to minimize the number of clones with no mutation or more than 5 mutations. The sequence was long (359 aa), and the probability of an amino acid mutation at a given position was low (≈0.01). Therefore, clone distribution could be predicted by using Poisson statistics, showing that the number of unwanted clones was minimal for 2.6 amino acid mutations per clone. The PCR conditions we used gave a mean rate of 3.1 DNA mutations per clone (see below). Further analysis indicated that 80% of the DNA mutations resulted in amino acid substitution (caused by genetic code redundancy and PCR bias), giving a frequency of 2.5 amino acid mutations per clone, close to the optimum.
(ii) The maximum number of amino acid substitutions for a protein with 359 amino acids is 6,821 (359 × 19). But PCR can produce no more than one mutation per codon, thereby limiting the number of amino acid substitutions that can really be achieved to ≈5 substitutions per AT1A residue (because of genetic code properties, they are, however, representative of the different classes of amino acids). Systematic analysis of the codon composition of the synthetic AT1A cDNA showed that only 2,137 amino acid substitutions could be obtained by PCR. The mutation rate of the library was 2.5 amino acid mutation per clone, so we screened ≈5,000 clones to ensure that the entire library of mutations was represented several times.
Construction of a Library of AT1A Mutants.
We used a modified PCR protocol (27): 30 cycles (1 min at 96°C; 1 min at 55°C; 1 min at 72°C) in 50 μl of PCR buffer (50 mM KCl/10 mM Tris-HCl, pH 8.3) containing 1 ng of pTRE-AT1A, 50 pmol of primers (flanking sequences), 7 mM MgCl2, 0.2 mM dRTP, 1 mM dYTP, and 5 units of Taq polymerase (Boehringer Mannheim). The PCR product was purified (high purification kit, Boehringer) and inserted into the HindIII-XbaI-digested pTRE. Individual clones were isolated after transformation in XL-1 blue strain. Twenty-three randomly selected clones were sequenced, showing that the cloning efficiency was 100% and the mean number of DNA mutations per clone was 3.1.
Cell Culture and Transfection.
CHO-TetOff (CLONTECH) and HEK-293 (American Type Culture Collection, CRL 1573) cells were cultured in Ham's F-12 medium or Dulbecco's modified eagle medium (Life Technologies, Rockville, MD), supplemented with 10 mM glutamine (Life Technologies) and 7.5% FCS (Boehringer Mannheim). A stable CHO-ÆQ cell line was established as described (28) by transfecting the CHO-TetOff cell line with the æquorin plasmid (24) and selecting with puromycin.
Transient transfection was performed by using lipofection reagents: Dosper (Boehringer Mannheim) for HEK-293 and FuGene6 (Boehringer Mannheim) for CHO. The day before transfection, cells were plated in white opaque 96-well plates (Culturplate, Packard) at a density of 4 × 104 cells/well or in 24-well plates at a density of 2 × 105 cells/well. Transfections were performed in serum-free medium by using 0.1 μg (96-well plates) or 0.5 μg (24-well plates) of DNA and 5 μl of lipofection reagent per 1 μg of DNA. After 6 h (Dosper) or 24 h (FuGene6) of incubation, the medium was changed, and assays were then performed. Doxycycline was added at a concentration of 0.04, 0.2, or 1 μg/ml to repress receptor expression and was maintained until the end of the experiment.
Screening of the AT1A Mutants Library.
Isolated bacterial colonies were cultured in 1 ml of LB in deep 96-well plates (CLP, San Diego) for 20 h at 37°C under oxygenation (Airpore, Qiagen, Hilden, Germany) and agitation. The alkaline lysis procedure was adapted to 96-well plates and was used to prepare DNA (final concentration of ≈15 ng/μl). Each clone was transiently transfected in CHO-ÆQ cells by using four different wells (8 μl of DNA/well) and was tested in duplicate for æquorin activity in two sets of conditions: (i) two wells were successively stimulated with 0.01, 1, and 25 nM AngII; and (ii) two wells with 200 and 5,000 nM CGP42112A and 100 μM ATP (positive control involving endogenous purinergic receptors).
Clones were automatically classified according to phenotype by using Microsoft excel software (see Results). In less than 1% of transfections, there was no ATP stimulation because of rare low-quality DNA preparations toxic to the cells. The cDNAs of clones giving a strong signal were sequenced, and the mutations were identified. If clones harbored multiple mutations, single mutants were constructed by using the many restriction sites in the synthetic AT1A cDNA (26).
Æquorin Assay.
Intracellular calcium mobilization assays were performed in 96-well plates with transiently transfected CHO-ÆQ cells, using the bioluminescent calcium-sensitive protein æquorin (24). Forty-eight hours after transfection, cells were incubated for two hours at 37°C in 50 μl/well medium supplemented with 0.5 μM cœlenterazin, were washed twice, and then were incubated for 30 min at room temperature in 50 μl of buffer (0.3 mM CaCl2/1 mM MgCl2/125 mM NaCl/5 mM KCl/5.5 mM glucose/20 mM Hepes, pH 7.3). Cells were stimulated by adding 50 μl of agonist per well (increasing concentrations in one well for library screening; one concentration per well for EC50 determinations) and determining luminescence 30 s later in a TopCount counter (Packard).
Binding Assays.
Protocols for Kd and Ki determination (29) were adapted to transiently transfected HEK-293 cells in 96-well plates. Briefly, 48 h after transfection, cells were incubated for 45 min at room temperature in 50 μl of binding buffer supplemented with [125I]AngII and ligands. Cells were washed three times, and 250 μl/well Microscint 20 (Packard) was added to determine bound [125I]AngII in the TopCount.
IP Accumulation Assay.
IP levels were determined in HEK-293 cells transiently transfected with a 1/1/1 mix of TetOff/receptor/Gαq plasmids in 24-well plates. Twenty-four hours after transfection, cells were incubated for twenty-four hours in 500 μl of medium supplemented with 2 μCi/ml myo-[3H]inositol, then were washed and incubated for 40 min at 37°C with or without Losartan [in 24 mM NaCl, 0.96 mM KCl, 0.24 mM MgSO4, 3.12 mM NaH2PO4, 0.3 mM CaCl2, 1 mM LiCl, and 0.1% BSA (pH 7.6)]. Cell lysis and IP separation on a Dowex AG1-X8 column (Bio-Rad) were performed as described (29). In parallel, the binding of 4 nM [125I]AngII was measured for the same transfection to estimate receptor expression by extrapolation from the Kd determined in an independent experiment.
Results
Construction of an Optimized Library of AT1A Receptor Mutants.
Studies of CAM AT1A provide insight into the molecular mechanisms of AT1A receptor activation. In this study, we aimed to identify such mutants systematically by testing a library of randomly generated mutants of the entire AT1A cDNA. Two parameters had to be determined for the screening to be efficient: (i) the optimal mutation rate, and (ii) the number of clones to be tested for exhaustive coverage of mutations generated. We had to find a suitable compromise between low and high mutation rate to limit the number of undesired clones: those with no mutations, behaving like the WT, and highly mutated clones, which were more likely to be inactive. Mathematical analysis was performed to answer the two questions (see Materials and Methods): (i) the optimal library is obtained with a mean of 2.6 amino acid substitutions per clone; and (ii) ≈5,000 clones must be tested for the screening to be exhaustive. Several modified PCR protocols based on a previous study (27) were used to produce several AT1A mutant libraries differing in mutation rates (data not shown). The library closest to the optimum had 2.5 amino acid substitutions per clone and was chosen for screening. The distribution of the clones was predicted by Poisson statistics to be as follows: 9% nonsense, 9% WT, 22% one amino acid substitution, 26% two substitutions, 21% three substitutions, and 13% more than three substitutions. The mutated cDNAs were inserted into the doxycycline-inducible mammalian expression vector pTRE (25).
Screening of the AT1A Mutant Library.
We set up a functional test for detecting constitutive activity by using the N111A mutant as a positive control. This test had to meet two strict specifications: (i) it had to be easy to perform with thousands of mutants, and (ii) it had to be sensitive, with a high signal/noise ratio. Classical measurement of agonist-independent IP production was not suitable for screening and was poorly sensitive, as demonstrated with the N111A mutant, with only 50–200% increase over control (data not shown; ref. 19). A luciferase reporter gene sensitive to receptor activation (28) was more suitable for large-scale screening but was not sensitive enough to detect constitutive activity reliably (data not shown). We therefore developed a new strategy, using the sensitivity of CAM receptors to partial agonists as an indicator of their activity (4, 16, 17). For the AT1A receptor mutant N111A, changes in affinity and efficacy have been experimentally demonstrated with CGP42112A, a classical AT2 ligand (19, 22, 23). A test based on the properties of CGP42112A fully satisfied the required specifications: (i) by using æquorin, a luminescent protein sensitive to intracellular calcium mobilization, it was easy to perform on thousands of clones; and (ii) in this assay, the EC50 for CGP42112A was ≈500 nM for the WT and ≈10 nM for the N111A mutant, resulting in luminescence levels 100-fold higher for the N111A mutant than for the WT, after stimulation with 200 nM CGP42112A.
By using scaled-down and standardized techniques, 4,800 clones were transfected transiently in CHO cells stably expressing æquorin and were tested for intracellular calcium mobilization on stimulation with various concentrations of AngII or CGP42112A. Clones were screened individually because preliminary experiments indicated that the WT receptor had an inhibitory effect on CAM receptors if pooled in the same transfection (data not shown). About 6% of the tested clones were not responsive to AngII, and 10% were misresponsive (responsive to 25 nM AngII, but not to lower doses). In contrast, 24 of the 4,800 clones tested were highly responsive to 200 nM CGP42112A (more than 30% of AngII-induced maximal levels), and 58 clones were slightly responsive (less than 30% AngII-induced levels). The 24 clones that reacted strongly were characterized further. Their phenotype was confirmed by determining the EC50 of CGP42112A in the æquorin assay, and their sequence was determined to identify the amino acid mutations responsible (Table 1). The EC50 of CGP42112A for the WT receptor was 375 nM whereas the mutants had greatly shifted EC50 values, between 5 and 30 nM.
Table 1.
Isolated AT1A mutants highly sensitive to CGP42112A
| Harbored amino acid substitutions | |||
|---|---|---|---|
| Major | Others |
EC50, nM | Basal IP, % |
| WT | 375 | 99 ± 2 | |
| F77S(2)† | 25.7 | 102 ± 1 | |
| F77S(2) | G203A-D273N | n.d. | n.d. |
| F77S(2) | (I38V)-W253R-(L316P) | n.d. | n.d. |
| F77Y(2) | W253R | 10.9 | 111 ± 1* |
| L78F(2) | L191P-L212H-F293L | 12.3 | 101 ± 4 |
| S107F(3) | 7.20 | 102 ± 1 | |
| F110C(3) | I201T-(F304I) | 11.3 | 90 ± 17 |
| N111S(3) | N231S-F293L | 7.18 | 170 ± 9* |
| L112H(3) | V29A-L143P | 15.0 | 141 ± 3* |
| L112H(3) | F44S-T175A-F182Y-K325* | 4.94 | 120 ± 1* |
| L112F(3) | V164A | 7.20 | 105 ± 2 |
| L118H(3) | (K318E) | 11.7 | 116 ± 1* |
| E173G(e2) | 21.1 | 92 ± 3 | |
| P162Q-A181V | 7.15 | 99 ± 4 | |
| I193K(5) | M142R-S186P | 11.2 | 104 ± 1 |
| L195P(5) | (G306R) | 7.94 | 118 ± 3* |
| L195R(5) | T178S-S329G | 14.8 | 100 ± 4 |
| T198I(5) | (K310N) | 10.1 | 99 ± 6 |
| I245T(6)† | 20.7 | 111 ± 1* | |
| I245T(6) | M142T-H256F | 4.36 | 149 ± 8* |
| I245T(6) | (F44L) | n.d. | n.d. |
| L305Q(7) | 7.39 | 143 ± 4* | |
| L305Q(7) | (S326T) | n.d. | n.d. |
| L305Q(7) | D9E | n.d. | n.d. |
| L305Q(7) | A221V | n.d. | n.d. |
| L305Q(7) | Q15R-V52D-S189P-Q229R | n.d. | n.d. |
Characteristics of the isolated clones (†, except for the F77S and I245T mutants, which were constructed subsequently): list of amino acid substitutions (one letter amino acid code; Z, stop codon), either resulting in much (“Major”) or slightly (underlined) higher sensitivity to CGP42112A, in no change (amino acid substitutions in brackets) or untested (others); EC50 of CGP42112A in the æquorin assay (representative of at least two independent determinations); basal IP production (see Fig. 2). The positions of the mutated amino acids (for major substitutions) in the putative secondary structure of AT1A receptor are indicated in brackets, either TM (2, 3, 5, 6, or 7) or second extracellular loop (e2).
Significantly different from WT.
Identification of the Amino Acid Substitutions Responsible for CGP42112A Sensitivity.
Of the 24 clones, 3 had only one amino acid substitution (S107F, E173G, or L305Q). Thus, the phenotype of each of these three clones was clearly caused by the mutation. For the other 21 clones, the analysis was more complex, as they had mutations in several positions (10 clones with two mutations, 7 with three, 3 with four, and 1 with five). Four of these 21 clones harbored the L305Q mutation, so their phenotype was attributable to this mutation. For the other 17 clones, the mutations actually responsible for CGP42112A sensitivity were unknown. Therefore, cDNAs with single mutations were constructed with the various candidate amino acid substitutions, and the EC50 of CGP42112A was determined and compared with that of the initial clone. By using this strategy, the phenotype of 16 of the 17 clones was attributed to one of the following 13 amino acid substitutions: F77S, F77Y, L78F, F110C, N111S, L112F, L112H, L118H, I193K, L195R, L195P, T198I, and I245T. Thus, 23 of 24 clones had one major mutation sufficient to explain their phenotype (Table 1 and Fig. 1). One of these mutations was the already described N111S substitution (22).
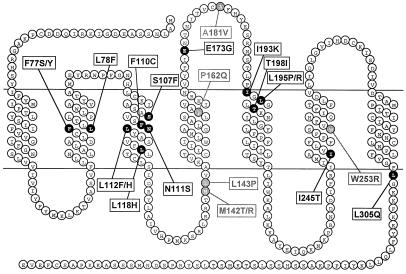
Figure 1.
Amino acid substitutions resulting in an increase in sensitivity to CGP42112A. shown are isolated amino acid substitutions resulting in much (black) or slightly (gray) higher sensitivity to CGP42112A.
The 24th clone was different in that it harbored two mutations (P162Q and A181V) contributing equally and additively to the phenotype: each of these two mutations individually caused only a slight change in CGP42112A sensitivity. Similar minor mutations were also found in analysis of the other 23 clones (Table 1). In these clones, already harboring a major mutation, some minor mutations were found to make a minor contribution to CGP42112A sensitivity (2- to 5-fold shifts in dose-response curves). Finally, 16 of the 62 mutations present in the 24 strongly reacting clones were found to have major effects on CGP42112A sensitivity, 6 had minor effects, and 8 had no effect. There may be other active mutations among the 32 remaining untested amino acid substitutions. The mutated amino acids were located essentially in TM 2, 3, 5, 6, and 7 (Fig. 1).
Constitutive Activity of the Isolated AT1A Mutants as Measured by Agonist-Independent IP Production.
Parallel to the isolation of substitutions responsible for increased CGP42112A sensitivity, constitutive activity was assessed by determining agonist-independent IP production in transiently transfected HEK-293 cells. Except for the F77S and I245T mutations and the three clones with single mutations, S107F, E173G, and L305Q, experiments were performed by using native clones with multiple mutations. The mutated or WT AT1A cDNAs were transfected together with the TetOff plasmid to allow doxycycline-inducible expression (25), and the Gαq cDNA to increase assay sensitivity (30). The difference between IP levels measured after doxycycline pretreatment (no receptor expression) and in the absence of pretreatment (receptor expression) was a reliable indicator of basal IP levels. The results were normalized to the expression levels of the receptor to allow accurate comparison, except for three clones with low levels of expression (E173G, P162Q-A181V, and L191P-L212H-F293L-L78F).
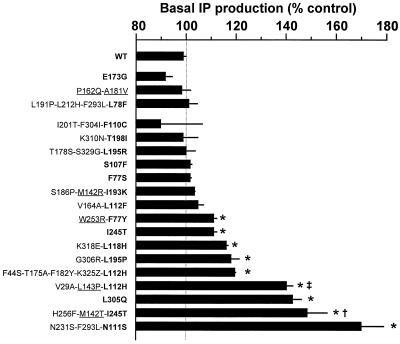
Basal IP levels measured after WT AT1A transfection were not different from those of mock-transfected cells (data not shown) and were not sensitive to doxycycline pretreatment, consistent with very low levels of spontaneous AT1A activity (19, 22). For half the clones, those harboring N111S, I245T, L305Q, L112H, L195P, L118H, or F77Y substitutions, agonist-independent IP production was significantly different from WT (P < 0.05) and was up to 70% higher than control (Fig. 2). Three of the other clones were poorly expressed, and no conclusion could be drawn for these. Thus, about half of the substitutions responsible for high sensitivity to CGP42112A resulted in constitutive activity if assessed for agonist-independent IP production, demonstrating that the screening test (response to a dose of the partial agonist CGP42112A below the EC50 for WT receptor) was efficient for detecting CAM receptors.
Figure 2.
The agonist-independent IP production of clones highly responsive to CGP42112A. Bars indicate the levels obtained without doxycycline pretreatment (receptor expression) as a percentage of the levels obtained after pretreatment with 1 ng/ml doxycycline (no receptor expression), normalized to 200,000 sites/cell (except for the first three clones, which were poorly expressed). The amino acid substitutions (one letter amino acid code; Z, stop codon) found to be responsible for much or slightly higher sensitivity to CGP42112A are in bold or underlined, respectively. Results are means ± SEM of three independent experiments. *, different from WT (P < 0.05); †, different from I245T mutant (P < 0.05); ‡, different from F44S-T175A-F182Y-K325Z-L112H mutant (P < 0.05).
Some of the minor substitutions (responsible for only slightly elevated sensitivity to CGP42112A) also seemed to increase basal IP production, but in an additive manner. Indeed, two clones harboring one mutation that greatly increased sensitivity to CGP42112A (I245T or L112H) together with one minor mutation (M142T or L143P, respectively) resulted in significantly higher basal IP levels than were observed with the respective major mutations alone (Fig. 2) (P < 0.05). Other mutations present in these clones may be involved in the observed effects, but, in any case, this shows that the substitutions I245T and L112H are not the only mutations responsible for the basal IP levels of the corresponding clones.
Characterization of L305Q Mutant.
The L305Q mutant was characterized further as an example of a novel CAM AT1A with a clear-cut phenotype. The Ki of various compounds for the WT and L305Q mutant were determined in [125I]AngII competitive binding experiments (Table 2). As expected, the Ki for CGP42112A was much lower for the L305Q mutant than for the WT receptor. The Ki for the nonpeptide AT1 agonist L162,313 was also significantly lower but the difference was smaller. The Ki for AngII, the peptide antagonist Sar1-Ile8-AngII, and the nonpeptide antagonist Losartan were not affected.
Table 2.
Pharmacological characterization of L305Q AT1A mutant
|
Ki, nM
|
||
|---|---|---|
| WT | L305Q | |
| AngII | 0.77 ± 0.38 | 0.67 ± 0.25 |
| Sar1-Ile8-AngII | 0.10 ± 0.04 | 0.24 ± 0.08 |
| CGP42112A | 540 ± 60 | 99 ± 42* |
| L162,313 | 51 ± 5 | 20 ± 3* |
| Losartan | 5.9 ± 1.7 | 3.7 ± 0.4 |
Results are mean ± SEM of three independent experiments.
Significantly different from WT (P < 0.01).
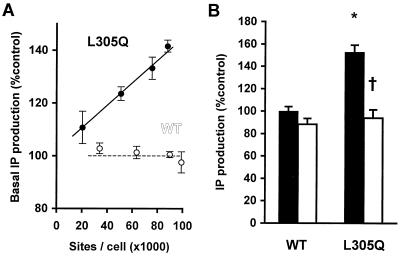
The constitutive activity of the L305Q mutant was then characterized further. By using various doxycycline doses, different levels of receptor expression were achieved for the same transfection. Agonist-independent IP levels were then plotted as a function of the number of sites per cell. For the WT receptor, agonist-independent IP production did not vary significantly with increasing expression levels whereas it was linearly related to receptor expression for the L305Q mutant (Fig. 3A). Finally, Losartan was tested for its inverse agonist activity: i.e., its ability to stabilize the inactive conformation of the receptor and consequently decrease the basal IP levels of a CAM receptor, as previously demonstrated for the N111A mutant (19). As shown in Fig. 3B, Losartan efficiently abolished the agonist-independent IP production of the L305Q mutant.
Figure 3.
Characterization of the constitutive activity of the L305Q AT1A mutant. (A) Basal IP production of the WT AT1A (white) or the L305Q mutant (black) as a function of receptor expression, after 48 h of treatment with various doses of doxycycline (0, 0.04, 0.2, and 1 ng/ml). Results (means ± SEM) are expressed as a percentage of the IP levels in mock transfected cells and are representative of three independent experiments giving similar slopes. (B) Inverse agonist effects (control, black; 1 μM Losartan, white) on the basal IP production of the WT AT1A and the L305Q mutant. Results are means ± SEM of three independent experiments. *, significantly different from WT (P < 0.01); †, significantly different from control (P < 0.01).
Discussion
In this study, we developed a specific and very sensitive bioassay, based on an original concept, for detecting constitutively active 7TMR mutants. We used this assay for the systematic screening of a random mutant library of a given 7TMR to map exhaustively the amino acids that may be involved in receptor activation.
Random mutagenesis, also referred to as “in vitro evolution,” is a powerful tool for structure/function studies (31–33). It has often been used in bacteria and yeast, but it is difficult to apply this strategy to mammalian systems. This is particularly limiting for 7TMRs: they are not easily expressed in bacteria, and, in yeast, the setting up of genetic screens is limited by the need for correct expression and heterologous coupling of the receptor. The only previous report of a systematic screening of a mutant library to find CAM receptors involved yeast α-factor receptor (34). Partial random saturation mutagenesis for four TMs of the C5a receptor has also been performed by using a yeast genetic screen, leading to the identification of potential CAM receptors (35). All other CAM 7TMRs have been identified in diseases (6–12) and site-directed mutagenesis studies (2–5, 19, 22). In the present work, we used an original strategy to make possible a systematic approach for mammalian CAM identification.
This strategy is mostly based on a new criteria for CAM detection. Classical measurement of agonist-independent second messenger production was not sensitive enough for such a large screening. Instead, we made use of the unusual pharmacological properties of CAM receptors, for which partial agonists have higher apparent affinity and efficacy than for the WT. Experimentally, this property has been reported for many CAM 7TMRs but it has only rarely been interpreted to be a characteristic of constitutive activity. Models such as the extended ternary complex model (4) and the more general cubic ternary complex model (15) have been used to explain how apparent affinity (16) and apparent efficacy (17) are modified for CAM receptors (36). In such models, the pharmacological effects are an allosteric consequence of modification of the equilibrium R↔R* between the inactive (R) and active (R*) states of the receptor. For the AT1A receptor mutant N111A, changes in apparent affinity and efficacy have been demonstrated for the pseudopeptidic AT2 ligand CGP42112A (19, 22, 23). We thought it likely that this property would be characteristic of CAM receptors, and therefore we used response to a sub-EC50 dose of CGP42112A (200 nM) for their detection. Both the sensitivity and specificity of this test were predicted to be optimal.
Combined with miniaturized techniques, this idea allowed the systematical identification of new CAM AT1A without a priori reasoning about the position of the mutated amino acids. The screening of 4,800 AT1A random mutants led to the successful identification of 16 mutations responsible for high sensitivity to CGP42112A (Fig. 1), among which 7 also significantly increased agonist-independent IP production (Fig. 2). This demonstrates the feasibility of such a pharmacological approach for systematic CAM identification, showing that high agonist sensitivity can be used as a reliable indicator of constitutive activity. One interesting observation is the relatively low number of isolated mutations. At the present time, this number can only be compared with two other 7TMRs, for which quite exhaustive analyses are available: i.e., the yeast α-factor receptor and the thyrotropin receptor. Whereas only two adjacent residues have been identified as α-factor CAM receptors (34), many activating mutations of the human thyrotropin receptor have been identified because of their role in thyroid adenomas (9, 10, 37). Despite a similar number of amino acids involved for the thyrotropin and AT1A receptor (19 and 16 amino acids), their locations are completely different, excluding an identical mechanism for their activation.
The isolated mutants should be useful for molecular modeling of AT1A activation and should help to define the role of the mutated amino acids in this mechanism. Except Trp253 and Pro162, the identified amino acids are not conserved among 7TMRs, suggesting that, if there are common mechanisms for activation of 7TMRs, they do not imply highly conserved amino acid motives. As for many CAM receptors identified to date, these residues are essentially located in transmembrane regions, confirming that TM movements play a key role in the transition between the inactive and active states of this receptor. A particularly striking cluster of mutations was observed in the third TM, around Asn111, indicating that this helix plays a key role in activation. The L305Q mutant is also very interesting in that the mutated amino acid lies at the extreme cytoplasmic end of the seventh TM and cannot interact directly with ligands. The high CGP42112A sensitivity of this mutant is thus not attributable to a direct modification of the binding site but, rather, to indirect allosteric effects of constitutive activity. Finally, the potentially additive effects of the mutations identified are also of importance, as our results clearly demonstrate that the observed basal IP levels of at least two mutants do not entirely result from a single amino acid substitution.
The identified mutants should also make it possible to test the limits of the theoretical models accounting for the pharmacological features of CAM receptors. Some intriguing discrepancies remain to be understood. An increase in apparent affinity should be observed for all agonists, but no change was observed for AngII. Shifts in CGP42112A sensitivity were larger in the æquorin functional assay (factor of ≈50-fold difference between WT and CAM) than in the binding assay (factor of ≈5-fold), suggesting that kinetic aspects may have to be taken into account. Finally, our work raises the question of how constitutive activity should be defined, both in theoretical models and in experimental results. Theoretically, “constitutive activity” phenotype is defined as a change in the number of receptors spontaneously adopting an active conformation, which cannot be measured directly. Experimentally, agonist-independent second messenger production defines a “gain-of-function” phenotype, which indirectly indicates constitutive activity. Our study strongly suggests that greater sensitivity to partial agonists (4, 16, 17) is another reliable indicator of constitutive activity. However, there are differences between this phenotypic trait and the classical gain-of-function phenotype. Some of the clones identified were positive for one phenotypic trait, high CGP42112A sensitivity, but not for the classical phenotypic trait, high basal IP levels, suggesting that the high CGP42112A sensitivity may be characteristic of a particular class of CAM AT1A receptors or, more interestingly, that the two phenotypic traits of constitutive activity have a different threshold. Indeed, basal IP levels for WT AT1A are very low, showing that the R↔R* equilibrium strongly favors the inactive R state. There would therefore need to be major mutation-induced modification of this equilibrium for significant basal IP levels to be detected. Conversely, whatever the position of the equilibrium R↔R* for the WT receptor, its modification should immediately result in a detectable increase in agonist sensitivity (4, 16). Therefore, we speculate that, for all of the mutants, the amount of receptors spontaneously adopting an active conformation is increased (the R↔R* equilibrium is modified in favor of R* state), thus involving the mutated residues in the activation process of the receptor. For all of the mutants, this modification was sufficient to result in high CGP42112A sensitivity. But for some of the mutants, it was not sufficient to result in detectable basal IP levels.
In conclusion, this work strongly suggests that agonist sensitivity is a more direct indicator of the position of R↔R* equilibrium than basal levels of second messenger, particularly if the WT receptor has very low basal activity. The AT1 receptor has a very low basal activity and appears to be difficult to activate constitutively by a single mutation, as suggested by this work (small number of frequently weak mutants) and the absence of natural mutations responsible for disease, such as the Conn adenoma (38). Appropriate associations of mutations should result in higher levels of constitutive activity. In contrast, several other 7TMRs have molecular structures more sensitive to activation, such as the β2-adrenergic receptor, for which both WT and CAM receptors give readily detectable basal second messenger production (4), and thyrotropin receptors, for which an extraordinary large number of natural mutations have been detected (37).
These considerations should be borne in mind for the interpretation of previous and future mutagenesis studies. Pharmacological changes observed on mutation may result from allosteric effects of changes in the R↔R* equilibrium and not just from binding site modifications. Studying the CAM receptors isolated in this work should help us to understand these aspects of 7TMRs. Combined with molecular modeling, such studies should enable us to link the macroscopic and microscopic features of receptor activation. In addition, studying the effects of the mutations on other aspects of AT1A function, such as internalization, proliferation pathways, and dimerization, may help us to understand their relationship to G-protein activation. The use of different CAM AT1A for transgenic models would then make it possible to link these pharmacological and functional findings to physiopathology.
Acknowledgments
We thank Dr. Rachel Malienou for methodological assistance with the æquorin assay. We also thank Dr. Susanna Cotecchia, Dr. Catherine Monnot, and Dr. Zsolt Lenkei for helpful discussions. This study was supported in part by Grant 98126 from Hœchst Marion Roussel and Grant 5254 from Association pour la Recherche contre le Cancer. C.P. was awarded a Ph.D. fellowship from Direction Générale de l'Armement and Association pour la Recherche contre le Cancer.
Abbreviations
- 7TMR
seven transmembrane receptor
- AngII
angiotensin II
- AT1A
AngII type 1A
- CAM
constitutively active mutant
- IP
inositol phosphate
- TM
transmembrane domain
- WT
wild-type
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110142297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110142297
References
- 1.Scheer A, Cotecchia S. J Recept Signal Transduct Res. 1997;17:57–73. doi: 10.3109/10799899709036594. [DOI] [PubMed] [Google Scholar]
- 2.Cotecchia S, Exum S, Caron M G, Lefkowitz R J. Proc Natl Acad Sci USA. 1990;87:2896–2900. doi: 10.1073/pnas.87.8.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kjelsberg M A, Cotecchia S, Ostrowski J, Caron M G, Lefkowitz R J. J Biol Chem. 1992;267:1430–1433. [PubMed] [Google Scholar]
- 4.Samama P, Cotecchia S, Costa T, Lefkowitz R J. J Biol Chem. 1993;268:4625–4636. [PubMed] [Google Scholar]
- 5.Perez D M, Hwa J, Gaivin R, Mathur M, Brown F, Graham R M. Mol Pharmacol. 1996;49:112–122. [PubMed] [Google Scholar]
- 6.Dryja T P, Berson E L, Rao V R, Oprian D D. Nat Genet. 1993;4:280–283. doi: 10.1038/ng0793-280. [DOI] [PubMed] [Google Scholar]
- 7.Shenker A, Laue L, Kosugi S, Merendino J J, Jr, Minegishi T, Cutler G B., Jr Nature (London) 1993;365:652–654. doi: 10.1038/365652a0. [DOI] [PubMed] [Google Scholar]
- 8.Schipani E, Kruse K, Juppner H. Science. 1995;268:98–100. doi: 10.1126/science.7701349. [DOI] [PubMed] [Google Scholar]
- 9.Parma J, Duprez L, Van Sande J, Cochaux P, Gervy C, Mockel J, Dumont J, Vassart G. Nature (London) 1993;365:649–651. doi: 10.1038/365649a0. [DOI] [PubMed] [Google Scholar]
- 10.Duprez L, Parma J, Van Sande J, Allgeier A, Leclere J, Schvartz C, Delisle M J, Decoulx M, Orgiazzi J, Dumont J, et al. Nat Genet. 1994;7:396–401. doi: 10.1038/ng0794-396. [DOI] [PubMed] [Google Scholar]
- 11.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka E G, Gutkind J S, Asch A S, Cesarman E, Gershengorn M C, Mesri E A, Gerhengorn M C. Nature (London) 1998;391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 12.Xie J, Murone M, Luoh S M, Ryan A, Gu Q, Zhang C, Bonifas J M, Lam C W, Hynes M, Goddard A, et al. Nature (London) 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 13.Milano C A, Dolber P C, Rockman H A, Bond R A, Venable M E, Allen L F, Lefkowitz R J. Proc Natl Acad Sci USA. 1994;91:10109–10113. doi: 10.1073/pnas.91.21.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schipani E, Lanske B, Hunzelman J, Luz A, Kovacs C S, Lee K, Pirro A, Kronenberg H M, Juppner H. Proc Natl Acad Sci USA. 1997;94:13689–13694. doi: 10.1073/pnas.94.25.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss J M, Morgan P H, Lutz M W, Kenakin T P. J Theor Biol. 1996;178:151–167. doi: 10.1006/jtbi.1996.0139. [DOI] [PubMed] [Google Scholar]
- 16.Weiss J M, Morgan P H, Lutz M W, Kenakin T P. J Theor Biol. 1996;178:169–182. doi: 10.1006/jtbi.1996.0139. [DOI] [PubMed] [Google Scholar]
- 17.Weiss J M, Morgan P H, Lutz M W, Kenakin T P. J Theor Biol. 1996;181:381–397. doi: 10.1006/jtbi.1996.0139. [DOI] [PubMed] [Google Scholar]
- 18.Chen G, Way J, Armour S, Watson C, Queen K, Jayawickreme C K, Chen W J, Kenakin T. Mol Pharmacol. 2000;57:125–134. [PubMed] [Google Scholar]
- 19.Groblewski T, Maigret B, Larguier R, Lombard C, Bonnafous J C, Marie J. J Biol Chem. 1997;272:1822–1826. doi: 10.1074/jbc.272.3.1822. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y H, Miura S, Husain A, Karnik S S. Biochemistry. 1998;37:15791–15798. doi: 10.1021/bi980863t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miura S, Feng Y H, Husain A, Karnik S S. J Biol Chem. 1999;274:7103–7110. doi: 10.1074/jbc.274.11.7103. [DOI] [PubMed] [Google Scholar]
- 22.Balmforth A J, Lee A J, Warburton P, Donnelly D, Ball S G. J Biol Chem. 1997;272:4245–4251. doi: 10.1074/jbc.272.7.4245. [DOI] [PubMed] [Google Scholar]
- 23.Monnot C, Bihoreau C, Conchon S, Curnow K M, Corvol P, Clauser E. J Biol Chem. 1996;271:1507–1513. doi: 10.1074/jbc.271.3.1507. [DOI] [PubMed] [Google Scholar]
- 24.Rizzuto R, Simpson A W, Brini M, Pozzan T. Nature (London) 1992;358:325–327. doi: 10.1038/358325a0. [DOI] [PubMed] [Google Scholar]
- 25.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conchon S, Monnot C, Sirieix M E, Bihoreau C, Corvol P, Clauser E. Biochem Biophys Res Commun. 1994;199:1347–1354. doi: 10.1006/bbrc.1994.1379. [DOI] [PubMed] [Google Scholar]
- 27.Cadwell R C, Joyce G F. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 28.Parnot C, Le Moullec J M, Cousin M A, Guedin D, Corvol P, Pinet F. Hypertension. 1997;30:837–844. doi: 10.1161/01.hyp.30.4.837. [DOI] [PubMed] [Google Scholar]
- 29.Conchon S, Barrault M B, Miserey S, Corvol P, Clauser E. J Biol Chem. 1997;272:25566–25572. doi: 10.1074/jbc.272.41.25566. [DOI] [PubMed] [Google Scholar]
- 30.Burstein E S, Spalding T A, Brauner-Osborne H, Brann M R. FEBS Lett. 1995;363:261–263. doi: 10.1016/0014-5793(95)00323-2. [DOI] [PubMed] [Google Scholar]
- 31.Brink M F, Nels R N, Verbeet M P, de Boer H A. J Mol Biol. 1994;237:368–377. doi: 10.1006/jmbi.1994.1240. [DOI] [PubMed] [Google Scholar]
- 32.Chen K, Arnold F H. Proc Natl Acad Sci USA. 1993;90:5618–5622. doi: 10.1073/pnas.90.12.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crameri A, Whitehorn E A, Tate E, Stemmer W P. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 34.Konopka J B, Margarit S M, Dube P. Proc Natl Acad Sci USA. 1996;93:6764–6769. doi: 10.1073/pnas.93.13.6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baranski T J, Herzmark P, Lichtarge O, Gerber B O, Trueheart J, Meng E C, Iiri T, Sheikh S P, Bourne H R. J Biol Chem. 1999;274:15757–15765. doi: 10.1074/jbc.274.22.15757. [DOI] [PubMed] [Google Scholar]
- 36.Kenakin T. Pharmacol Rev. 1996;48:413–463. [PubMed] [Google Scholar]
- 37.Duprez L, Parma J, Van Sande J, Rodien P, Sabine C, Abramowicz M, Dumont J E, Vassart G. J Pediatr Endocrinol Metab. 1999;12, Suppl. 1:295–302. [PubMed] [Google Scholar]
- 38.Davies E, Bonnardeaux A, Plouin P F, Corvol P, Clauser E. J Clin Endocrinol Metab. 1997;82:611–615. doi: 10.1210/jcem.82.2.3764. [DOI] [PubMed] [Google Scholar]