Abstract
Hybridization properties of oligodeoxyxylonucleotides (OXNs) built from pyrimidine monomers with an inverted 3′-OH group of the furanose have been studied using the gel mobility shift, UV melting and circular dichroism (CD) spectroscopy methods. Pyrimidine OXNs form triple helices with complementary purine RNA in which one OXN is parallel and another is antiparallel with respect to the RNA target. Surprisingly, no duplex formation between the pyrimidine OXNs and purine RNAs is detected. The modified triplexes are stable at pH 7. Their thermal stability depends on the number of C(G-C) triplets and, for G-rich RNA sequences, it is comparable with the stability of native DNA–RNA duplexes. The CD spectra of triplexes formed by OXNs with purine RNA targets are similar to spectra of A-type helices. A pyrimidine OXN having a clamp structure efficiently inhibits reverse transcription of murine pim-1 mRNA in vitro mediated by the Mo-MuLV reverse transcriptase.
INTRODUCTION
One approach to nucleic acid recognition is the formation of triple helices. Interaction of triplex-forming oligonucleotides with regions of double-stranded DNA possessing regulatory activity often results in gene expression inhibition (1–3). Single-stranded RNA molecules are also attractive targets for triplex formation. In this case two oligonucleotides, the first one complementary in the Watson–Crick sense and the other complementary in the Hoogsteen sense, have to be used. Application of clamp or circle oligonucleotides containing two domains that are complementary to a target RNA sequence in both senses has also been described (4–6). Single-stranded pyrimidine RNAs have been demonstrated to form stable triplexes (7,8). However, polypurine RNA sequences cannot usually be used as a target for triplex-forming oligodeoxyribonucleotides (9–12). Therefore, a search for oligonucleotide modifications permitting targeting of polypurine RNA for triplex formation is of interest.
Earlier we showed that oligonucleotides built from 1-(β-d-2′-deoxy-threo-pentafuranosyl)thymine (2′-deoxyxylothymidine) and 1-(β-d-2′-deoxy-threo-pentafuranosyl)cytosine (2′-deoxyxylocytidine) residues (Fig. 1a), so-called oligodeoxyxylonucleotides (OXNs), formed triplexes with single-stranded purine DNA targets which were stable at pH 7.0 (13,14). Triplex formation by OXNs was confirmed by gel retardation experiments, DMS footprinting and thermal denaturation studies. Two important and unexpected results were obtained from this investigation. Firstly, pyrimidine OXNs formed triplexes with purine DNA targets demonstrating a high stability at pH 7.0, in contrast to native pyrimidine oligodeoxyribonucleotides, whose triplexes with the same DNA targets were only stable at acidic pH. Indeed, protonation of N3 atoms of cytosines is necessary to generate hydrogen bonds between guanosine residues of the Watson– Crick duplex and the third strand (15). Therefore, C+(GC) triplets are usually formed at pH values near the pKa of cytosine (pKa = 4.5). Secondly, neither antiparallel nor parallel duplexes were detected between the DNA targets and OXNs using the methods mentioned above (14).
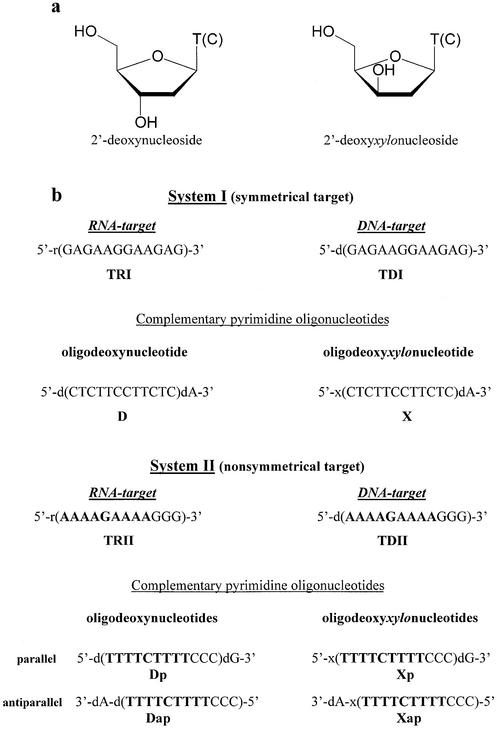
Figure 1.
Structure of oligonucleotides used in the study. (a) Structure of 2′-deoxynucleosides and 2′-deoxyxylonucleosides. (b) Oligonucleotide sequences.
The main goal of the present work is to find out if OXNs could form triplexes with single-stranded purine RNAs and to study their structure.
MATERIALS AND METHODS
Oligonucleotide synthesis and purification
All oligonucleotides were synthesized on an ASM-102U DNA synthesizer (Biosset, Novosibirsk, Russia). The standard phosphoramidite procedure was utilized to prepare oligodeoxyribonucleotides. Deprotection of t-butyldimethylsilyl groups blocking the 2′-hydroxyl of oligoribonucleotides was carried out using 1 M tetra-n-butylammoniumfluoride solution in tetrahydrofuran for 24 h at room temperature, followed by gel-filtration on a Sephadex G-25 column. Fully protected 1-(β-d-2′-deoxy-threo-pentafuranosyl)thymine-3′-O-phosphoramidite and 1-(β-d-2′-deoxy-threo-pentafuranosyl)cytosine-3′-O-phosphoramidite were prepared as described (16,17) and incorporated in OXNs using standard phosphoramidite chemistry. Condensation time was increased to 10 min to improve the modified oligonucleotide yield. All OXNs were synthesized using a commercial 500 Å CPG support with one immobilized nucleoside, for which reason they possessed an additional 2′-deoxyribonucleoside at their 3′-ends.
After deprotection, the oligonucleotides were purified in 20% PAGE, 7 M urea in TBE buffer.
Gel retardation assay
The oligonucleotides were labeled with T4 polynucleotide kinase (Promega) using [γ-32P]ATP (Amersham, Russia). The labeled pyrimidine strand (∼1–2 pmol) was mixed with the same non-labeled strand in an amount necessary to form the corresponding duplex or triplex and complementary purine oligonucleotides (90 pmol). To carry out experiments in which the strand ratio was 1:1, 90 pmol of the ‘cold’ pyrimidine oligonucleotide was used; when the ratio was 1:2 or 1:3 the amount of this oligonucleotide was doubled or tripled, respectively. All mixtures were dissolved in 10 µl of TA buffer (10 mM Tris–acetate, pH 7.0, 100 mM Na acetate, 10 mM Mg acetate), heated to 80°C for 5 min, slowly cooled to 5°C, incubated at 5°C overnight and then supplemented with 1 µl of 70% glycerol containing xylene cyanol and bromophenol blue. The products were resolved in 20 × 20 cm slabs of non-denaturing 20% acrylamide gel (19:1 acrylamide/bis-acrylamide) in TA buffer at 24°C for 18 h (9 V/cm). Oligonucleotide positions were visualized by the autoradiography method.
Thermal denaturation studies
The targeted purine oligonucleotides were mixed with complementary pyrimidine oligonucleotides in 50 mM Tris–HCl buffer, 100 mM NaCl, 10 mM MgCl2 (pH 7.0) or 50 mM sodium cacodylate buffer, 100 mM NaCl, 10 mM MgCl2 (pH 7.0 or 5.0). The concentration of each oligonucleotide strand was 2 µM. These solutions were heated to 80°C and slowly cooled to 10°C. The melting studies were carried out in Teflon-stoppered 1 cm pathlength quartz cells in an Hitachi 100-20 spectrophotometer equipped with a thermoprogrammer. Absorption at 260 nm was monitored while the temperature was increased at a rate of 0.5°C/min. Melting temperatures (Tm) were determined by computer fit of the first derivative of absorbance with respect to 1/T. The uncertainty in the Tm values is estimated to be ±0.5°C, based on triplicate experiments.
Circular dichroism (CD)
CD spectra were recorded on a modified Mark V dichrograph (Jobin & Yvon) within the 210–340 nm wavelength range at a scanning rate of 50 nm/min. Samples were placed in a quartz cell (200 µl, 2 mm pathlength) thermostated at 20°C. To avoid oxygen absorption, dry nitrogen was flushed through the cell block. In typical experiments, a 10 µM concentration of oligonucleotide complexes in 50 mM sodium cacodylate buffer, 100 mM NaCl, 10 mM MgCl2 (pH 7.0 or 5.0) was used. The molar extinction coefficient difference (ΔE) was calculated by using the equation ΔE = ΔA/lC (M–1cm–1), where ΔA is the absorbency difference, l is the optical pathlength and C is the oligonucleotide concentration.
Reverse transcription
pim-1 mRNA was produced by SP6 RNA polymerase (New England Biolabs) transcription from 1 µg of HindIII-linearised pSP64 plasmid containing the EcoRI–HindIII pim-1 cDNA fragment as indicated by the shipper in a final volume of 20 µl for 2 h at 37°C. Then a further incubation with 1 µl of RQ1 RNase-free DNase (Promega) was done for 15 min at 37°C to hydrolyze the DNA matrices. The proteins were then extracted by addition of 3 µl of 2 M Na acetate buffer, pH 4, 100 µl of water-saturated phenol and 15 µl of chloroform:isoamyl alcohol (49:1). The aqueous fraction was discarded and precipitated with 24 µl of isopropanol for 30 min at –20°C. After centrifugation for 15 min at 13 000 r.p.m., the pellet was washed with 100 µl of 70% ethanol, dried and dissolved in 20 µl of water containing 0.5 U RNasin (Promega). The synthesized pim-1 RNA was analysed on 1% agarose gel electrophoresis in TAE buffer and its concentration was estimated by comparison with a RNA standard. Reverse transcription was performed in a 10 µl volume containing 5 mM MgCl2, 1× reaction buffer (Perkin Helmer), 1 µM each dNTP, 1 U RNasin, 2.5 U Mo-MuLV reverse transcriptase, 0.5 µl [α-32P]dCTP (3000 Ci/mM), 0.5 µg pim-1 mRNA, 0.2 µM oligonucleotide primer and 1 µM OXN to be tested. After 1 h incubation at 37°C, the reaction was stopped at –20°C and the cDNA produced was analysed by 8% PAGE, 6.5 M urea in TBE buffer or 1% agarose gel electrophoresis in TAE buffer, followed by autoradiography with a phosphorimager (Storm 840; Molecular Dynamics).
RESULTS AND DISCUSSION
It is generally accepted that triplets based on direct Hoogsteen hydrogen bonding require the parallel orientation of the third strand with respect to the purine-rich strand of the duplex, whereas the opposite orientation is true for reverse Hoogsteen based triplets (15). The triple helix, containing T and C in the third strand, is based on the triplets C+(GC) and T(AT), where the parentheses contain the Watson–Crick base pairs. The third strand represented by native pyrimidine ribo- and 2′-deoxyribooligonucleotides has a parallel orientation that imposes direct (cis)-Hoogsteen pairing of the triplet to the Watson–Crick base pair. The type of triplet pairing in the triple helix formed by OXNs is difficult to predict because of inverted 3′-hydroxyl groups in the xylose residues. As for the strand orientation in OXN-containing triplexes, our previous results showed that in the triple helix formed by OXNs with a purine DNA target one modified oligonucleotide has a parallel and another one an antiparallel orientation with respect to the purine strand (14).
In order to investigate interactions of OXNs with polypurine RNA targets, two oligonucleotide systems have been synthesized (Fig. 1b). The target from system I (TRI) has a symmetrical sequence, so the same complementary polypyrimidine sequence (D or X) can interact with the target in the antiparallel as well as in the parallel orientation, thus resulting in both duplex and triplex formation. The second target (TRII), with a non-symmetrical sequence, represents a fragment of the polypurine tract of the HIV RNA LTR sequence, and its interaction with antiparallel (Xap) and parallel (Xp) oligonucleotides was investigated. Earlier, hybridization of these OXNs with DNA counterparts of both RNA targets (TDI and TDII) was studied and formation of stable triplexes X-TDI-X and Xp-TDII-Xap was detected (14).
Gel mobility shift assays
In order to investigate whether OXNs are able to interact with RNA targets, gel mobility shift assays were performed. Native (ODN) and modified (OXN) polypyrimidine oligonucleotides were 5′-32P-labeled and mixed with the same non-labeled oligonucleotides, RNA targets and polypyrimidine strands of the other polarity, if necessary. The quantity of a labeled oligonucleotide in the mixture was not more than 2% of the total amount of the same non-labeled oligonucleotide.
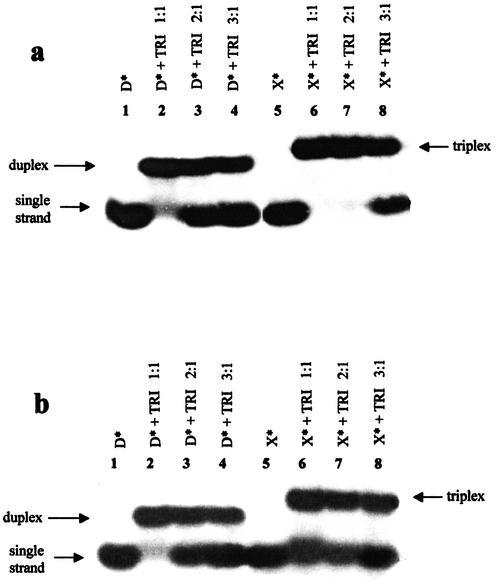
Since triple helices containing T and C in the third strand are usually stable at acidic pH, hybridization of oligonucleotides D and X with the target TRI was firstly studied at pH 5 (Fig. 2a). As expected from the literature (9–11), the only complex formed by the native symmetrical oligonucleotide D with its target TRI corresponded to a duplex (Fig. 2a, lanes 2–4). The OXN/RNA complex migrated slower than the duplex TRI-D. The OXN X only formed a complex with TRI when the pyrimidine–purine strand ratio was 1:1 or 2:1 (Fig. 2a, lanes 6 and 7). Only when X was in 3-fold excess did the single-stranded oligonucleotide appear (Fig. 2a, lane 8). This result may signify that at pH 5 the ratio of X and TRI in the complex was 2:1, and it was, therefore, a triple helix. Further, the same experiment was carried out at pH 7, and the only difference observed, if compared with the gel obtained at acidic pH, was that X was not completely involved in complex formation (Fig. 2b, lanes 6–8). The radioactive intensity ratio for the bands corresponding to single-stranded X and this oligonucleotide involved in a complex stayed, however, constant when X and TRI were in the ratio 1:1 or 2:1 (Fig. 2b, lanes 6 and 7). When the ratio X:TRI was 3:1, the radioactive intensity of the single-stranded OXN band increased (Fig. 2b, lane 8). This result allowed us to suppose that X also formed a triplex with TRI at pH 7, but its stability is lower than at pH 5.
Figure 2.
Analysis of complexes formed by oligonucleotides X and D with TRI by gel mobility shift assay in a buffer containing 50 mM Tris–acetate, 100 mM Na acetate, 10 mM Mg acetate at 24°C, at pH 5.0 (a) and 7.0 (b). The complex composition is indicated at the top. A 5′-32P-labeled oligonucleotide is marked by an asterisk (*).
To confirm this supposition, interactions of the 5′-32P-labeled OXNs Xap and Xp with the RNA target TRII were investigated at pH 7. Neither antiparallel nor parallel OXN alone formed any complex with the target (Fig. 3, lanes 9 and 13). No complex formation was observed when two OXNs were mixed without the target (Fig. 3, lanes 8 and 12). However, the simultaneous presence of antiparallel and parallel OXNs and the RNA target resulted in complex formation (Fig. 3, lanes 10 and 14). These data confirmed that pyrimidine OXNs are able to form triplexes with purine RNA targets and showed that both parallel and antiparallel OXNs are necessary for triplex generation. It should be noted that formation of a triple helix without a detectable double-helical intermediate was earlier found for some polypyrimidine oligonucleotides generating very stable triplexes with a purine target, for example N3′→P5′ phosphoramidate oligodeoxythymidylates (18).
Figure 3.
Analysis of complexes formed by native and modified oligonucleotides with TRII by gel mobility shift assay in a buffer containing 50 mM Tris–acetate, 100 mM Na acetate, 10 mM Mg acetate, pH 7.0 at 24°C. The complex composition is indicated at the top. A 5′-32P-labeled oligonucleotide is marked by an asterisk (*).
Thermal stability of complexes formed by oligodeoxyxylonucleotides with RNA targets
The thermal stabilities of complexes generated by OXNs with RNA targets TRI and TRII were measured by UV absorption spectroscopy and compared with those of earlier studied triplexes of OXNs with DNA targets TDI and TDII and also the corresponding native complexes (Table 1). To calculate Tm values, thermal denaturation curves were applied. All triplexes assembled from OXNs and DNA or RNA targets had monophasic melting profiles. Comparing the data obtained for complexes containing modified and native oligonucleotides, some important conclusions may be drawn: (i) the thermal stabilities of the complexes formed by OXNs with their DNA targets at pH 7 are comparable with those of the native DNA duplexes; (ii) the complexes of OXNs with their RNA targets at pH 7 are slightly less stable than the corresponding complexes with the DNA targets and significantly less stable than the DNA–RNA duplexes; (iii) the OXN-containing complexes are less stable at pH 7 than at pH 5, which corresponds with the electrophoretic data (Fig. 2); (iv) complexes formed by OXNs with RNA and DNA targets from the same system at pH 5 have about the same thermal stability, which is considerably above the stability of the non-modified triplex D-TDI-D. The thermal dissociation curve of the native triplex D-TDI-D at pH 5 had one transition, demonstrating similar stabilities of the triplex and corresponding duplex (not shown). It is well known that triplexes containing C and T oligonucleotides as a third strand are stable under acidic conditions, when the N3 atoms of the cytosines may be protonated and C+(GC) triplets are formed (15). For this reason, native Pyr-Pur-Pyr triplexes are most stable at pH values near the cytosine pKa value, i.e. near pH 5, and their thermal stability decreases strongly with increasing pH. The stability of OXN-containing triplexes also decreased significantly when the buffer pH was increased from 5 to 7 under the same saline conditions. Nevertheless, triplexes X-TRI-X and X-TDI-X exist at pH 7 while the native triplex D-TDI-D does not (14). This indicates that even at pH 7 there is at least partial 2′-deoxyxylocytidine protonation sufficient for hydrogen bonding in OXN-containing complexes. It is also important that complete 2′-deoxyxylocytidine protonation at pH 5 results in significant complex stabilization. Such a dramatic augmentation of the thermal stability with decreasing pH is typical of pyrimidine triplexes, and shows once more that complexes of OXNs with their RNA targets are really triple helices.
Table 1. Thermal stability of oligonucleotide complexes (50 mM sodium cacodylate, 100 mM NaCl, 10 mM MgCl2).
| Oligonucleotide system | Complex composition | Tm (°C) (±0.5°C) | |
|---|---|---|---|
| pH 7.0 | pH 5.0 | ||
| System I | TDI-D | 48.2 | 49.0a |
| X-TDI-X | 47.1 | 71.0 | |
| TRI-D | 58.7 | ||
| X-TRI-X | 40.7 | 71.1 | |
| System II | TDII-Dap | 44.6 | |
| Xp-TDII-Xap | 38.4 | 63.8 | |
| TRII-Dap | 49.2 | ||
| Xp-TRII-Xap | 36.0 | 62 | |
aThe oligonucleotide ratio at pH 5 was D:TDI = 2:1.
Modified triplexes from system I, X-TRI-X and X-TDI-X, were more stable than triplexes Xp-TRII-Xap and Xp-TDII-Xap. This result may reflect the difference in nucleotide composition of these complexes. Indeed, the targets from system I are more G-rich than the targets from system II (six G residues in target I and four G in target II). So one can conclude that the thermal stability of triplexes containing OXNs increases with the number of C+(GC) triplets even at neutral pH. The situation is completely the opposite in the case of triple helices formed by native oligonucleotides, because the N3 atoms of 2′-deoxyribo- and ribonucleotides are not protonated at pH 7 and thus cannot form Hoogsteen base pairs (19).
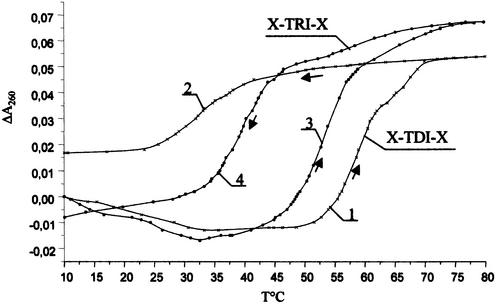
It is also worthwhile noting that triplexes formed by OXNs were found to be stable in the presence of K+, which is considered to be a triplex destabilizing ion (20). The triplex X-TDI-X was slightly less stable in K+-containing than in Na+-containing buffer (Table 2). The difference in Tm was more pronounced for the RNA-containing triplex X-TRI-X. The rate of complex formation is an important parameter to evaluate the hybridization properties of modified oligonucleotides, so thermal denaturation–renaturation processes were studied for triplexes X-TRI-X and X-TDI-X (Fig. 4). The temperature was increased and then decreased at a constant rate of 0.5°C/min. One can see that the melting curves are non-reversible for both triplexes and that the hysteresis is greater for the triplex X-TDI-X. Surprisingly, the triplex X-TRI-X, having a lower stability, had a higher formation rate. This might be attributed to the better conformational similarity of OXN and RNA when compared with DNA. To clarify the structure of the complexes formed by OXNs with both DNA and RNA targets, the CD method was used.
Table 2. The influence of monovalent ions on the thermal stability of the triplexes X-TDI-X and X-TRI-X in a buffer containing 50 mM Tris–HCl, pH 7.0, 10 mM MgCl2 and 100 mM NaCl or KCl.
| Complex | Tm (°C) (±0.5°C) in the presence of ions | |
|---|---|---|
| Na+ | K+ | |
| X-TDI-X | 59.2 | 56.3 |
| X-TRI-X | 54.5 | 47.7 |
Figure 4.
Thermal denaturation–renaturation study of the triplexes X-TDI-X and X-TRI-X in a buffer containing 50 mM Tris–HCl, 100 mM NaCl, 10 mM MgCl2, pH 7.0. Curves 1 and 3 correspond to denaturation; curves 2 and 4 correspond to renaturation of triplexes X-TDI-X and X-TRI-X, respectively.
Circular dichroism study
The CD method has earlier been applied to analyze the structure of oligodeoxyxylothymidylates as well as their complexes with oligodeoxyadenylates (16,17,21). Their spectra showed a negative band at 280 nm that allowed a left-handed helix structure resembling Z-type DNA to be attributed to these compounds (16,17).
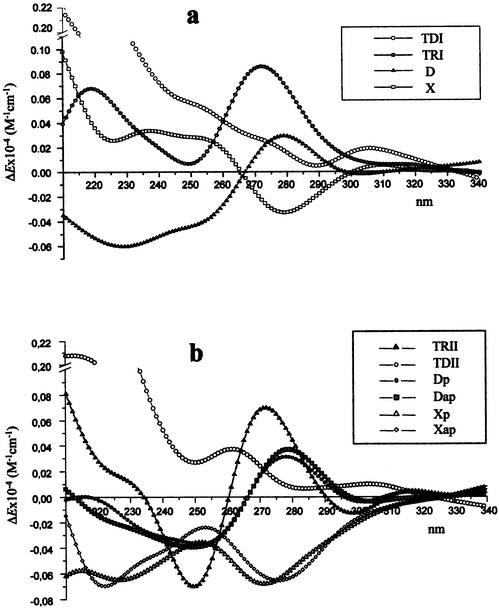
CD spectra of C and T containing OXNs were found to depend on the oligonucleotide composition, but all of them had a negative band at 270–280 nm (Fig. 5a and b). At the same time all complexes formed by the OXNs with DNA and RNA targets gave CD spectra with positive maxima in this wavelength region (Fig. 6).
Figure 5.
CD spectra of single-stranded oligonucleotides TDI, TRI, D and X (a); TRII, TDII, Dp, Dap, Xp and Xap (b) in a buffer containing 50 mM sodium cacodylate, 100 mM NaCl, 10 mM MgCl2, pH 7.0 at 20°C.
Figure 6.
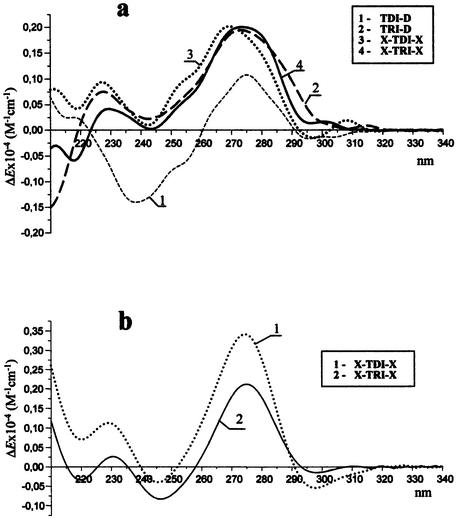
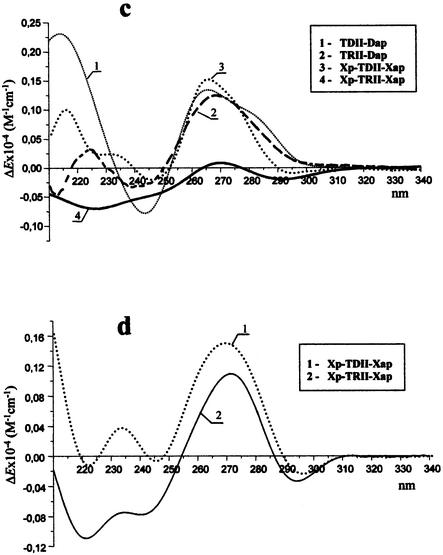
CD spectra of complexes from system I at pH 7.0 (a) and 5.0 (b) and from system II at pH 7.0 (c) and 5.0 (d) in a buffer containing 50 mM sodium cacodylate, 100 mM NaCl, 10 mM MgCl2 at 20°C.
As shown in Figure 6a, at pH 7 the duplex TDI-D (curve 1) gave a CD spectrum with a positive band at 276 nm, a negative band with about the same intensity at 240 nm and a long wavelength crossover at 260 nm. It corresponds to the typical CD spectrum of the antiparallel B-type DNA (22). The duplex formed by D with its RNA target (TRI-D) had a CD spectrum characterized by a positive maximum at 273 nm and a minimum at 244 nm (Fig. 6a, curve 2), which resembles the CD spectrum of the A-type double helix typical of DNA–RNA duplexes with a purine-rich RNA strand (23). Surprisingly, both triplexes generated by X with DNA and RNA targets (Fig. 6a, curves 3 and 4) gave about the same type of CD spectrum in the region of 220–300 nm, which is nearly identical to that of the RNA–DNA duplex TRI-D. It means that the helix geometry of these complexes resembles the geometry of the A form. At pH 5 where the modified triplexes X-TRI-X and X-TDI-X are more stable, they have about the same type of CD spectrum as at pH 7 (Fig. 6b). This result additionally shows that complexes formed by OXNs with RNA and DNA targets have the similar type of helix structure and this is the same at acidic and neutral pH.
The triplex Xp-TDII-Xap at pH 7 had a CD spectrum characterized by a positive band with a maximum at 263 nm and a negative band at 245 nm (Fig. 6c, curve 3). The CD spectrum of the complex Xp-TRII-Xap at pH 7 showed a maximum at 270 nm but its amplitude was too small (Fig. 6c, curve 4). So, identification of the spectrum type (A or B) was not as evident as in the case of system I, probably because of their lower stability. Indeed, all spectra were recorded at room temperature where triplexes with Tm = 36–38°C may be partially destroyed and their CD signature may represent the sum of the spectra of a triplex and single-stranded oligonucleotides. However, at pH 5 where triplexes Xp-TDII-Xap and Xp-TRII-Xap are much more stable, their CD spectra looked similar (Fig. 6d) and resembled the CD spectra of OXN-containing triplexes from system I (Fig. 6b). All triplexes formed by OXNs display a positive band at 270 (system II) or 275 nm (system I) and two minima at 220 and 245 nm. The intensities of the bands at 270–275 nm are significantly greater than the intensity of the band at 245 nm, which is similar to what is obtained for the A form. The negative band at 220 nm is also more typical of the A-like helix geometry (usually RNA demonstrates a CD spectrum with a minimum at 212 nm) (22) than of the B-like one (generally showing a positive band at this wavelength) (Fig. 6a and c, curves 1) (22).
The similarity of the CD spectra of triplexes formed by OXNs with both DNA and RNA targets points to their structural similarity, which may be a result of the rigid structure of OXNs that compels both RNA and DNA complementary strands to accept a conformation favorable for the OXN.
Based on the data obtained, a hypothesis concerning the sugar conformation in OXNs may be suggested. As was mentioned above, formation of a triple helix without a detectable double-helical intermediate was earlier found for N3′→P5′ phosphoramidate oligodeoxythymidylates, generating very stable triplexes with oligodeoxyadenylates (18). In these triplexes phosphoramidate oligodeoxythymidylates are shown to have a C3′-endo sugar conformation (24). This conformation is typical of ribooligonucleotides and consequently of the A form of helices, while the B form is characterized by the C2′-endo (or C3′-exo) conformation. The similarity of CD spectra of triplexes in which OXNs are involved with the A-type like CD spectra as well as their enhanced stability allow us to presume that in these triplexes the 2′-deoxyxylose possesses a C3′-endo or related conformation, as happens with 2′-deoxyribose in phosphoramidate oligodeoxynucleotides. This possible structural similarity of sugar puckerings in deoxyxylo- and ribooligonucleotides might explain the involvement of a purine RNA target in the triplex with pyrimidine OXNs. Indeed, purine RNAs are usually excluded from triplex formation with DNA but they form stable triplexes with pyrimidine RNAs (10,25,26). It is worth noting that a C3′-endo conformation was earlier shown for deoxyxylonucleosides, whereas the sugar puckering in single-stranded OXNs was likely C2′-endo (27). However, as was mentioned above, all single-stranded OXNs studied to date show CD spectra with a negative band at 270–280 nm (Fig. 5a and b), which is rather typical of a left-handed helix (16,17,27). At the same time the CD spectra of OXN complexes with complementary DNA and RNA are typical of a right-handed helix, and this change in the helix type may evidently be accompanied by a configurational change at the C3′ atom. Indeed, a C2′-endo type sugar puckering of the furanose ring is typical for single-stranded oligodeoxyxylocytidines, while the C3′-endo type conformer population exceeds 95% in homoduplexes (or tetraplexes) formed by these OXNs at acidic pH (27). These data strongly point to the possibility of a C3′-endo conformation of the deoxyxylose ring in complexes formed by OXNs with complementary DNA and RNA. If OXNs do have RNA-like sugar puckering they can generate a xylo-ribo-xylo triplex, as three RNA strands do. Additional experiments are now in progress in order to confirm this hypothesis and to make clear the helix structure of OXN-containing triplexes.
Inhibition of reverse transcription
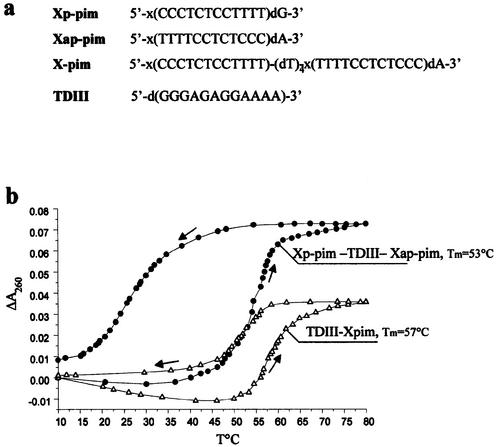
Due to the capacity of OXNs to form triple helices stable at neutral pH with purine RNA tracts, they may be considered useful tools to inhibit gene expression at the levels of pre-mRNA splicing, translation or reverse transcription. It should also be mentioned that owing to their unusual sugar structure, OXNs are stable to nucleolytic digestion (27,28). Here we have studied the ability of OXNs to stop reverse transcription. A 12mer polypurine tract (5′-GGGAGAGGAAAA-3′) at positions 1599–1611 of the mRNA of the murine oncogene pim-1 was chosen as a target for triplex formation. An oligonucleotide d(ACCCAGGCAGAGTTTGAG) complementary to positions 1743–1760 of this mRNA was used as a primer for reverse transcription. Three pyrimidine OXNs were synthesized to form a triplex with pim-1 mRNA, i.e. parallel (Xp-pim), antiparallel (Xap-pim) and clamp (X-pim) oligonucleotides, composed of parallel and antiparallel sequences linked by four deoxythymidine residues (Fig. 7a). The clamp OXN was synthesized because it was shown that triple helix formation with such compounds has an entropic advantage over triplex formation by two separate pyrimidine oligonucleotides (28). Indeed, the joining together in the proper orientation of two out of the three strands of the triple helix gives a considerable advantage in terms of free energy.
Figure 7.
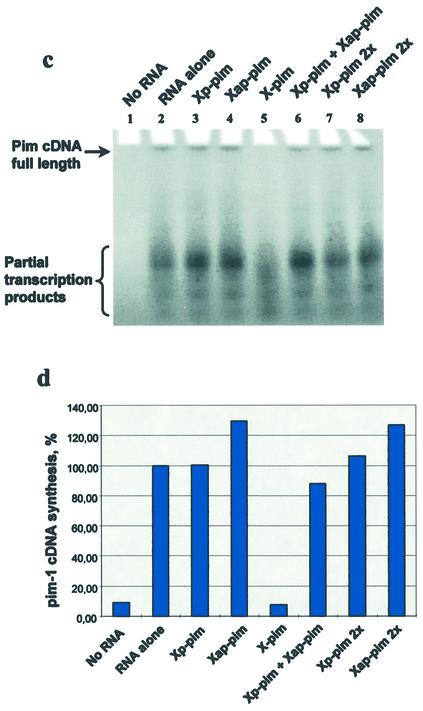
Study of reverse transcription inhibition by OXNs. (a) Oligo nucleotide sequences. (b) Thermal denaturation–renaturation study of triplexes formed by the DNA target TDIII with two separate OXNs (Xp-pim and Xap-pim) and the clamp X-pim in a buffer containing 50 mM Tris–acetate, 100 mM Na acetate, 10 mM Mg acetate, pH 7.0. (c) PAGE analysis of the pim-1 mRNA reverse transcription products in the absence (lane 2) and presence of OXNs (lanes 3–8). (d) Calculated efficiency of pim-1 cDNA synthesis in the absence and presence of OXNs.
The thermal stability and triplex formation rate were evaluated by melting experiments using a DNA analog d(GGGAGAGGAAAA) (TDIII) of the mRNA purine tract (Fig. 7). Use of the clamp OXN for triplex formation instead of two separate OXNs had at least two serious advantages: (i) the thermal stability of the bimolecular triplex (TDIII-X-pim) was higher; and (ii) hysteresis was smaller for the bimolecular triplex. These results are in good agreement with the literature data demonstrating a higher stability of bimolecular triplexes composed of native oligonucleotides (29,30). These data allow us to suppose that the thermal stability of a triplex between X-pim and the mRNA purine tract will be higher than the stability of a triplex formed by this RNA and two separate OXNs, Xp-pim and Xap-pim. Moreover, taking into account the fact that the rate of triplex formation was higher for OXN-containing triplexes with a RNA target than with a DNA one (Fig. 4), one could consider that the rate of X-pim hybridization with the targeted mRNA sequence would be rather quick.
Reverse transcription was carried out using pim-1 mRNA as the template, murine Moloney retrovirus (Mo-MuLV) reverse transcriptase and the 18mer primer indicated above (Fig. 7). It should be noted that due to a complicated and stable secondary structure of the pim-1 mRNA, synthesis of the full-length cDNA was not efficient and products of partial transcription were obtained as the main products. In the absence of OXNs pim-1 cDNA synthesis occurred efficiently (Fig. 7c, lane 2). Neither parallel Xp-pim nor antiparallel Xap-pim oligonucleotides inhibited reverse transcription (Fig. 7c and d). The mixture of both OXNs, which could form a triplex with the mRNA, had only a slight effect on reverse transcription (Fig. 7c, lane 6, and d), and this effect was significantly augmented with the clamp oligonucleotide X-pim, which efficiently inhibited synthesis of the full-length pim-1 cDNA and one of the partial transcription products (Fig. 7c, lane 5, and d). Therefore, reverse transcription inhibition only occurs when parallel and antiparallel OXNs are added to the mRNA, and the inhibition efficiency is greatly increased when the parallel and antiparallel OXN fragments are joined. The latter effect may arise from the higher hybridization rate and stability of triplexes formed by clamps compared with two separate oligonucleotides (Fig. 7a) (28,29). To be sure that the OXN effect arises from the oligonucleotide interaction with the mRNA and not with the protein, reverse transcription was carried out using another primer complementary to positions 1532–1560 of the mRNA. Since this position was nearer to the 5′-end of the mRNA than the polypurine sequence targeted by X-pim, the latter could influence reverse transcription from this primer only if the OXN interacted with the protein. It was found that X-pim had no inhibitory effect when the primer 1532–1560 was used (data not shown); hence, the reverse transcription inhibition caused by X-pim was a result of its interaction with the mRNA. This result is one more indication of the capacity of OXNs to generate a triple helix structure with RNA targets. Moreover, it demonstrates that a clamp OXN can bind an extended mRNA with a complicated spatial structure and form a stable triplex under conditions close to the physiological ones.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr E. Gromova for helpful discussions on the CD experiments. We acknowledge the Centre National de la Recherche Scientifique and Association pour la Recherche sur le Cancer (grant no. 4310). This work was also supported by the Russian Foundation for Basic Research (grant RFBR-CNRS no. 00-04-22003 and RFBR no. 02-04-48797).
REFERENCES
- 1.Knauert M.P. and Glazer,P.M. (2001) Triplex forming oligonucleotides: sequence-specific tools for gene targeting. Hum. Mol. Genet., 10, 2243–2251. [DOI] [PubMed] [Google Scholar]
- 2.Vasquez K.M. and Wilson,J.H. (1998) Triplex-directed modification of genes and gene activity. Trends Biochem. Sci., 23, 4–9. [DOI] [PubMed] [Google Scholar]
- 3.Praseuth D., Guieysse,A.L. and Helene,C. (1999) Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta, 1489, 181–206. [DOI] [PubMed] [Google Scholar]
- 4.Wang S. and Kool,E.T. (1994) Recognition of single-stranded nucleic acids by triplex formation: the binding of pyrimidine-rich sequences. J. Am. Chem. Soc., 116, 8857–8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vokmann S., Jendis,J., Frauendorf,A. and Moelling,K. (1995) Inhibition of HIV-1 reverse transcription by triple-helix forming oligonucleotides with viral RNA. Nucleic Acids Res., 23, 1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maksimenko A.V., Volkov,E.M., Bertrand,J.-R., Porumb,H., Malvy,C., Shabarova,Z.A. and Gottikh,M.B. (2000) Targeting of single-stranded DNA and RNA containing adjacent pyrimidine and purine tracts by triple helix formation with circular and clamp oligonucleotides. Eur. J. Biochem., 267, 3592–3603. [DOI] [PubMed] [Google Scholar]
- 7.Vo T., Wang,S. and Kool,E.T. (1995) Targeting pyrimidine single strands by triplex formation: structural optimization of binding. Nucleic Acids Res., 23, 2937–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morvan F., Imbach,J.-L. and Rayner,B. (1997) Comparative stability of eight different triple helices formed by differently modified DNA or RNA pyrimidine strands and a DNA hairpin. Antisense Nucleic Acid Drug Dev., 7, 327–334. [DOI] [PubMed] [Google Scholar]
- 9.Semerad C.L. and Maher,L.J. (1994) Exclusion of RNA strands from a purine motif triple helix. Nucleic Acids Res., 22, 5321–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escude C., Francois,J.C., Sun,J.S., Ott,G., Sprinzl,M., Garestier,T. and Helene,C. (1993) Stability of triple helices containing RNA and DNA strands: experimental and molecular modeling studies. Nucleic Acids Res., 21, 5547–5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han H. and Dervan,P.B. (1993) Sequence-specific recognition of double helical RNA and RNA-DNA by triple helix formation. Proc. Natl Acad. Sci. USA, 90, 3806–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noronha A. and Damha,M.J. (1998) Triple helices containing arabinonucleotides in the third (Hoogsteen) strand: effects of the inverted stereochemistry at the 2′-position of the sugar moiety. Nucleic Acids Res., 26, 2665–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottikh M.B., Alekseev,Y.I., Perminov,A.V., Pinskaya,M.D. and Shabarova,Z.A. (1999) Oligodeoxyxylonucleotides form stable triplexes with single-stranded DNA. Nucleosides Nucleotides, 18, 1625–1627. [Google Scholar]
- 14.Ivanov S.A., Alekseev,Y.I. and Gottikh,M.B. (2002) Capacity of pyrimidine oligodeoxyxylonucleotides for triplex formation at neutral pH. Mol. Biol. (Mosk.), 36, 160–170. [PubMed] [Google Scholar]
- 15.Porumb H. (1998) Triple-helix structure. In Malvy,C., Harel-Bellan,A. and Pritchard,L.L. (eds), Triple-Helix-Forming Oligonucleotides. Kluwer Academic, Boston, MA, pp. 17–32.
- 16.Rosemeyer H. and Seela,F. (1991) (2′-Deoxy-β-d-xylofuranosyl)thymine building blocks for solid-phase synthesis and properties of oligo(2′-deoxyxylonucleotides). Helv. Chim. Acta, 74, 748–760. [Google Scholar]
- 17.Rosemeyer H., Krecmerova,M. and Seela,F. (1991) (2′-Deoxy-β-d-xylofuranosyl)adenine building blocks for solid-phase synthesis and properties of oligo(2′-deoxyxylonucleotides). Helv. Chim. Acta, 74, 2054–2067. [Google Scholar]
- 18.Zhou-Sun B.-W., Sun,J.-S., Gryaznov,S.M., Liquier,J., Garestier,T., Helene,C. and Taillandier,E. (1997) A physico-chemical study of triple helix formation by an oligodeoxythymidylate with N3′→P5′ phosphoramidate linkages. Nucleic Acids Res., 25, 1782–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keppler M.D. and Fox,K.R. (1997) Relative stability of triplexes containing different numbers of T·AT and C+·GC triplets. Nucleic Acids Res., 25, 4644–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanvey J.C., Williams,E.M. and Besterman,J.M. (1991) DNA triple-helix formation at physiologic pH and temperature. Antisense Res. Dev., 1, 307–317. [DOI] [PubMed] [Google Scholar]
- 21.Schöppe A., Hinz,H.-J., Rosemeyer,H. and Seela,F. (1996) Xylose-DNA: comparison of the thermodynamic stability of oligo(2′-deoxyxylonucleotide) and oligo(2′-deoxyribonucleotide) duplexes. Eur. J. Biochem., 239, 33–41. [DOI] [PubMed] [Google Scholar]
- 22.Gray D.M., Hung,S.-H. and Johnson,K.H. (1995) Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Methods Enzymol., 246, 19–35. [DOI] [PubMed] [Google Scholar]
- 23.Gyi J.I., Lane,A.N., Corn,G.L. and Brown,T. (1998) Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes. Biochemistry, 37, 73–80. [DOI] [PubMed] [Google Scholar]
- 24.Matray T., Gamsey,S., Pongracz,K. and Gryaznov,S. (2000) A remarkable stabilization of complexes formed by 2,6-diaminopurine oligonucleotide N3′→P5′ phophoramidates. Nucleosides Nucleotides Nucleic Acids, 19, 1553–1567. [DOI] [PubMed] [Google Scholar]
- 25.Vuyisich M. and Beal,P.A. (2000) Regulation of the RNA-dependent protein kinase by triple helix formation. Nucleic Acids Res., 28, 2369–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beban M. and Miller,P.S. (2000) Pyrimidine motif triplexes containing polypurine RNA or DNA with oligo 2′-O-methyl or DNA triplex forming oligonucleotides. Biochim. Biophys. Acta, 1492, 155–162. [DOI] [PubMed] [Google Scholar]
- 27.Seela F., Worner,K. and Rosemeyer,H. (1994) 1-(2′-Deoxy-β-d-xylofuranosyl)cytosine: base pairing of oligonucleotides with a configurationally altered sugar-phosphate backbone. Helv. Chim. Acta, 77, 883–896. [Google Scholar]
- 28.Oretskaya T.S., Ibragim,H.K.H., Volkov,E.M., Romanova,E.A., Tashlitsky,V.N. and Shabarova,Z.A. (1994) Synthesis of oligonucleotides containing 1-(β-d-3′-deoxy-threo-pentofuranosyl)pyrimidines and their resistance to the action of snake venom phosphodiesterase. Bioorgan. Khim., 20, 967–974. [PubMed] [Google Scholar]
- 29.Kandimalla E.R., Manning,A. and Agrawal,S. (1996) Single stand targeted triplex formation: physicochemical and biochemical properties of foldback triplexes. J. Biomol. Struct. Dyn., 14, 79–90. [DOI] [PubMed] [Google Scholar]
- 30.Giovannangeli C., Montenay-Garestier,T., Rouge,M., Chassignol,M., Thuong,N.T. and Helene,C. (1991) Single stranded DNA as target for triple helix formation. J. Am. Chem. Soc., 113, 7775–7777. [Google Scholar]