Abstract
Defining complete sets of gene family members from diverse species provides the foundation for comparative studies. Using a bioinformatic approach, we have defined the entire nuclear receptor complement within the first available complete sequence of a non-human vertebrate (the teleost fish Fugu rubripes). In contrast to the human set (48 total nuclear receptors), we found 68 nuclear receptors in the Fugu genome. All 68 Fugu receptors had a clear human homolog, thus defining no new nuclear receptor subgroups. A reciprocal analysis showed that each human receptor had one or more Fugu orthologs, excepting CAR (NR1I3) and LXRβ (NR1H2). These 68 receptors add striking diversity to the known nuclear receptor superfamily and provide important comparators to human nuclear receptors. We have compared several pharmacologically relevant human nuclear receptors (FXR, LXRα/β, CAR, PXR, VDR and PPARα/γ/δ) to their Fugu orthologs. This comparison included expression analysis across five Fugu tissue types. All of the Fugu receptors that were analyzed by PCR in this study were expressed, indicating that the majority of the additional Fugu receptors are likely to be functional.
INTRODUCTION
Nuclear receptors (NRs) play key roles in integrating the complexities of homeostasis and development (1). In general, NRs function as transcriptional regulators, working in concert with co-activators and co-repressors to activate and suppress target gene expression (2,3). The common themes of ligand binding and cofactor interaction are reflected in shared structural features within the superfamily. A DNA-binding domain (DBD) targets the receptor to specific DNA sequences, termed hormone response elements. The C-terminal portion of the receptor contains a ligand-binding domain (LBD) which interacts directly with the hormone. Ligand-induced conformation changes within the LBD dictate the activity of the receptor within a particular cellular context.
NRs have been aggressively pursued as drug targets due to their central roles in metabolism and because of their potential for modulation by exogenous compounds. Examples of current NR drug targets include the steroid and thyroid hormone receptors (targets for multiple hormone replacement therapies and for immunomodulatory and cancer drugs), the peroxisome proliferator-activated receptors (PPARs) (targets for dyslipidemia and diabetes drugs) (4) and the liver X receptors (LXRs) and farnesoid X-activated receptor (FXR) (potential targets for treatment of atheroschlerosis and dyslipidemia, respectively) (5,6). Moreover, NRs such as pregnane X receptor (PXR) and constitutive androstane receptor (CAR) have been shown to be important in drug metabolism (7). A major goal of NR target discovery is to expand the knowledge of NR structure/function in order to validate additional NR family members as tractable drug targets.
Genome sequencing has recently disclosed that there are 48 functional NRs in the complete human NR set (8–10). Approximately 30 of the human NRs are still orphan with regard to function and represent an important source of untapped drug discovery targets (11,12). Aside from having important implications in evolutionary studies and in studies of the biology of distinct organisms, sequencing of the non-human genomes can be an important source of information on the function of human target class members. For example, comparison of the NR LBD amino acid residues between species can help identify conserved structural motifs and may pinpoint key functional residues. Identification of primitive receptors may also help in our understanding of the origins of ligand regulation in the superfamily, ultimately giving information regarding NR tractability. Non-human genome sequencing also identifies new models in which to generate NR functional data. The majority of NR functional analysis data have been derived from studies in genetic models such as mice, Caenorhabditis elegans and Drosophila; additional species provide new comparators for NR studies. Finally, in the case of frequently used pharmacological models such as mouse and rat, genome sequencing will help evaluate the relevance and caveats to working with these models. For example, significant differences have been observed in the LBD sequences and activation profiles of the xenobiotic receptor PXR (NR1I2) from humans versus rodents (13). This knowledge helped to explain the highly species-specific cytochrome P450 3A response of each of these species to xenobiotics (14).
The NR superfamily of genes appears to be restricted to metazoans and, to date, complete NR sets from three metazoan genomes have been defined (humans, Drosophila melanogaster and C.elegans). The C.elegans NR set (>270 members) displays a high degree of duplication and divergence relative to both the Drosophila (21 members) and human sets (48 members) (8,15). Comparison of these gene families has provided and continues to provide valuable information regarding the origin and function of specific NR family members. To extend this comparative genomic analysis, we have analyzed the complete NR set from a fish. The genome of the teleost, ray-finned fish, Fugu rubripes, represents the first complete non-human vertebrate genome to become available. In this manuscript, we have defined the complete set of Fugu NRs and have compared them to the human NR set and found that the Fugu NR set exhibits remarkable diversity and complexity. We have focused on the Fugu receptors of particular interest to current NR drug discovery research.
MATERIALS AND METHODS
Computational identification of Fugu nuclear receptor sequences
The proteome, transcriptome and genomic sequences of F.rubripes were obtained from the Fugu ftp site (http://genome.jgi-psf.org/fugu3/fugu3.info.html) and formatted for NCBI BLAST analysis. Protein homology searches were performed using nucleotide sequences of individual members of the human NR family compared against the Fugu proteome database using BLASTX (16). The top four hits for each query were analyzed. In the event that a Fugu protein was similar to multiple members of a NR family, percent identity was used to assign putative ortholog status. The DBDs of the 48 human nuclear receptors were aligned using T-Coffee (17) and the resulting multiple sequence alignments were used to generate hidden Markov models (18,19). HMMer was then used to search the Fugu database for hits to DBD HMM. Seventy-four hits were obtained above an e value cut-off of 0.001. In another HMMer search, a hidden Markov model of the LBDs of nuclear receptors identified 73 putative nuclear receptors from the Fugu database. Most of the hits corroborated the BLAST results. The hits from the BLAST and HMMer searches were combined resulting in 85 putative nuclear receptors. To rule out any redundant peptides, the sequence set was further refined using nrdb95.pl (20). This resulted in a final set of 74 putative nuclear receptors. This set contains full-length LBDs.
Based on sequence alignment, several pairs of Fugu identifiers were combined to make complete LBDs: liver receptor homolog 1 (LRH-1) (fr006605 + fr083170), LRH-1 (fr057871 + fr070767), retinoic acid receptor (RAR) (fr079596+ fr088133), retinoid X receptor (RXR) (fr087764 + fr087766) and estrogen receptor (ER) (fr062438 + fr62437). To confirm that these sequences were derived from the same DNA transcript, we designed primers from each sequence and used them to amplify contiguous DNA from cDNA reverse transcribed from a pool of Fugu liver, ovary and brain RNA (MCR Geneservice) using Superscript II RNase H– (Invitrogen). Amplified bands were confirmed by sequence analysis.
For the comprehensive comparison of the Fugu and human NR sets shown in Table 1, each Fugu LBD sequence was identified beginning with the Gly-Met linker sequence that is well conserved in the NR superfamily. Each Fugu LBD sequence was then aligned with each human LBD sequence individually using the Needleman–Wunsch algorithm as implemented in the program MVP (21). The percent identity of each pair of nuclear receptors was calculated as i/n, where i is the number of identities in the sequence alignment and n = (ni + nj)/2, the average number of amino acids in the two sequences being aligned.
Table 1. Comparison of PPAR sequences.
| Position in human hPPARγ | PPARα | PPARγ | PPARδ | ||||
|---|---|---|---|---|---|---|---|
| |
Human |
fr074373 |
fr087148 |
Human |
fr054267 |
Human |
fr07671 |
| Helix3-289 | Ser | Ser | Ser | Ser | Ser | Thr | Thr |
| Helix5-323 | Tyr | Tyr | Tyr | His | Ile | His | His |
| Helix10-449 | His | His | His | His | His | His | Asn |
| HelixAF2-473 | Tyr | Tyr | Tyr | Tyr | Met | Tyr | Tyr |
Italics specify amino acids that differ from the consensus. Bold specifies amino acids predicted to prevent fatty acid ligand binding.
Expression analysis of Fugu NR mRNAs
Fugu brain, gill, gut, heart and ovary RNAs were ordered from MCR Geneservice. Dr Greg Elgar (UK-HGMP-RC) generously provided Fugu liver tissue. Liver RNA was prepared using the Trizol reagent (Invitrogen). RNAs were reversed transcribed using Superscript II RNase H– (Invitrogen). Thirty cycle PCR reactions were then run with specific primers designed from the genomic sequence, as follows: fr078207 (PXR) 224 bp fragment from exon 5: forward, 5′-tctttgctgtgcgcggcagc-3′, reverse, 5′-agagcctcacggacttcaagt-3′; fr073157 [vitamin D receptor (VDR)] 161 bp fragment from exon 5: forward, 5′-gacaccaagttgaatctcag-3′, reverse, 5′-cggctttgccaagatgattc-3′; fr064850 (VDR) 181 bp fragment from exon 8: forward, 5′-cctacatccggctgcaccat-3′, reverse, 5′-cggcagcgaggtctcc-3′; fr070795 (LXR) 176 bp fragment from exon 4: forward, 5′-gtggccgcagagtcaggact-3′, reverse, 5′-ctgaagacttcgaccatcga-3′; fr072134 (FXR) 233 bp fragment from exon 8: forward, 5′-cgaccttatgttaaggatc-3′, reverse, 5′-gtgagatctgggatgtgcag-3′; fr082389 (FXR) 160 bp fragment from exon 9: forward, 5′-cctgcaggcggcacactg-3′, reverse, 5′-ggaccctccaccacaactac-3′. Reactions were run on a 1% agarose gel and the image was acquired using ChemiDoc with QuantityOne software (Bio-Rad). A control without reverse transcriptase was run to verify that the Fugu mRNA samples were not contaminated with genomic DNA (data not shown).
RESULTS AND DISCUSSION
General features
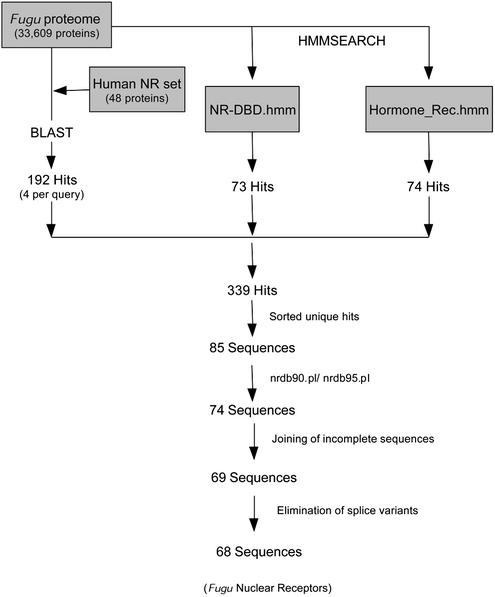
The F.rubripes genome contains a relatively small 365 Mb sequence. Recent analysis of the complete Fugu genome (>95% sequence coverage) (22) revealed a non-redundant set of 31 059 predicted gene loci within this sequence, similar to the number of predicted gene loci within the human genome (23,24). In order to identify the complete set of nuclear receptor genes within the Fugu genome, we developed a bioinformatic strategy (Fig. 1) involving sequence comparison between human nuclear receptors and the 33 609 predicted peptides in the Fugu proteome. In a first pass analysis, BLAST was used to scan for orthologs of the 48 human nuclear receptors in the Fugu proteome. Once putative orthologs were identified, sequence alignments were constructed to find conserved domains as well as novel protein family members. Using this approach we identified several new family members in both the COUP transcription factor (NR2F) family as well as in the RAR-related receptor (NR1F) family. We followed up the approach using a BLAST analysis with a HMMer-based analysis. A distinguishing feature about nuclear receptors is the presence of a DBD characterized by C4-type zinc fingers contained in the N-terminal half of the proteins. Hidden Markov models were generated using specifically the DBDs and this resulted in the identification of 10 new receptors not seen in the BLAST analyses. Similarly, using a second HMM that represented the LBD, we were able to identify six more receptors not seen by the BLAST analyses.
Figure 1.
Bioinformatics strategy. All 48 human NR cDNAs were compared against the Fugu proteome database using BLASTX. In a complementary approach, hidden Markov models for the DBD and the LBD were used in a HMMer search. Combining the BLAST results and HMM results, 85 putative Fugu NRs were identified. These hits were reduced to 74 putative orthologs after removing redundant peptide sequences with nrdb95.pl (see Materials and Methods). Partial sequences that represented different sections of the same receptor mRNA sequence were identified by cloning and sequence analysis. The joining of these partial sequences reduced the number of receptor sequences to 69. Finally, one pair of sequences was determined to represent splice variants, reducing the final number of NR sequences to 68.
Overall, we identified 68 total nuclear receptors in Fugu, 20 more than are in the human NR set. Broad sequence comparison of the Fugu and human NR sets (Fig. 2) showed that all of the 68 Fugu receptors displayed significant similarity to a human NR. The lack of novel subgroups within the expanded Fugu nuclear receptor set (relative to the human NR set) contrasts strikingly with a previous analogous analysis of the C.elegans NR set (8). The C.elegans NR family underwent a dramatic proliferation and diversification producing multiple novel subfamilies relative to other non-nematode genomes analyzed to date (25). Among the high number of C.elegans receptors that did not cluster within the traditional six subfamilies seen in humans, a comparatively low degree of sequence homology with human receptors is observed (15). Moreover, many of the novel C.elegans NR DBDs contained unique P-box regions, key determinants of DNA binding specificity. Thus, C.elegans NRs would be predicted to bind a larger repertoire of response elements compared to vertebrate NRs. One hypothesis that has been put forward is that the diversified C.elegans NRs expanded to fill a much broader niche in transcriptional regulation. The closer relationship between the Fugu NRs and the human NRs with respect to sequence supports the concept that proliferation of the Fugu nuclear receptor set did not occur in response to selection pressure for broader function of nuclear receptors as may have occurred in C.elegans, but is more likely to represent the result of larger scale genome-wide events. Our findings were consistent with the general conclusion that most teleosts contain an excess of duplicate genes in comparison with tetrapods (9,26).
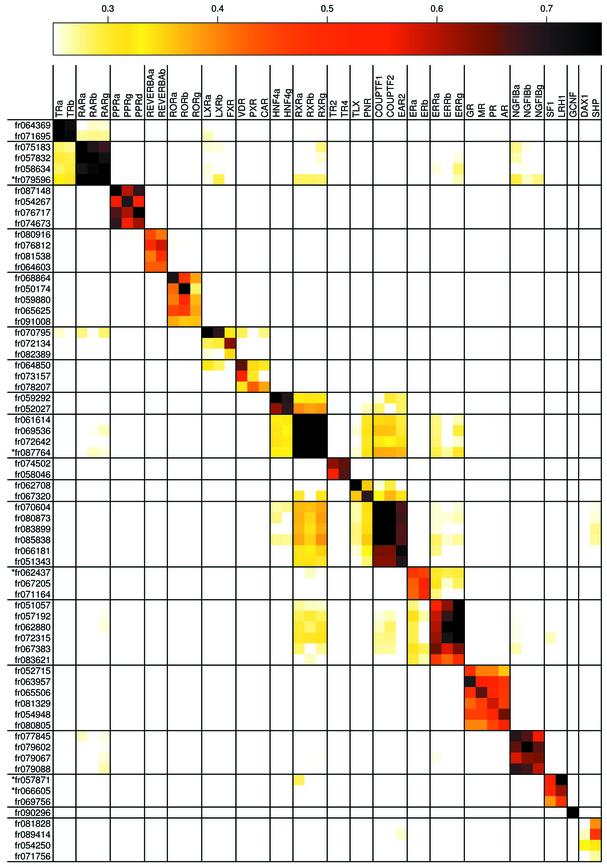
Figure 2.
(Opposite) Sequence comparison. The percent identity of each Fugu LBD sequence was compared to the complete set of human NR LBDs. Percent identities are indicated by color, with white indicating <25% identity, black indicating ≥75% identity and intermediate percent identities indicated as in the legend above the table. The human NRs are listed in order of their subfamily numbers and the Fugu sequences are ordered according to their closest human ortholog. Sequences that resulted from combining two separate Fugu sequence identifiers are marked with an asterisk.
Presumably NR pseudogenes are present within the Fugu genome. Our bioinformatic scheme was designed to identify only receptors containing a full-length LBD (amino acid start prior to a predicted helix 1 region) and thus would not have identified pseudogenes. In four cases, only partial LBD sequences were present on a single scaffold, while remaining sequence information from the same receptor subtype was found on a different scaffold (see Materials and Methods). These fragments could have represented independent pseudogene fragments or a contiguous sequence spanning two sequencing scaffolds. We showed that the latter was the case for all four of these receptors by amplifying a single cDNA fragment using 5′ and 3′ primers from the different scaffold sequences, thus ruling these out as pseudogene sequences. Future efforts to identify all NR sequences in the Fugu genome, including pseudogenes, should disclose more information regarding the nature of the origin of the NR gene family in teleosts.
Within each NR subgroup, it would be possible to assign specific orthologs based solely on percent sequence identity [for example, matching a human estrogen-related receptor α (ERRα) with Fugu ERRα], but such an assignment is likely to change as the systematic nomenclature for Fugu NRs is debated. Also, the final determination of nomenclature will depend on a broader set of data, including expression and other functional data. To avoid ambiguity, this manuscript identifies each Fugu receptor by its SINFRUP number indicated as fr (22) (http://genome.jgi-psf.org/fugu3/fugu3.info.html) combined with the name of the human NR with the highest sequence similarity. The variety of non-human NR orthologs provides the basis for comparative analysis of specific receptors from the human nuclear receptor complement. To begin the more detailed comparison of human and Fugu NRs, we have examined in more detail multiple subgroups of current pharmacological interest.
NR1I subfamily (PXR, CAR, VDR)
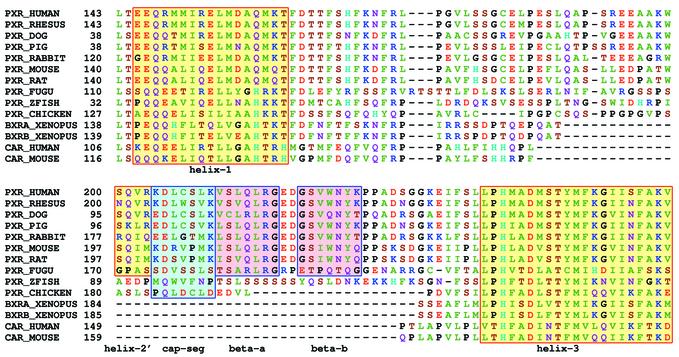
The human NR1I subfamily includes the xenobiotic receptors PXR (NR1I2) and CAR (NR1I3) as well as the VDR. The PXRs are activated by a broad range of xenobiotics, steroids and bile acids, while the CARs are less promiscuous, have higher basal activities than the PXRs and are more subject to inverse activation. These functional differences effectively classify PXR and CAR as pharmacologically distinct receptors (27). The X-ray structure of PXR revealed an unusually large and nearly spherical binding pocket that could accommodate a wide range of lipophilic ligands (28). In contrast, modeling work suggested that the CAR ligand-binding pocket would be smaller and more selective in terms of ligand binding (27). The hypothesized pocket expansion in PXR was thought to be caused primarily by a 50–60 amino acid insert that lies between helix 1 and helix 3 in the PXR sequence. This ‘helix 1–3 insert’ adopts a conformation with two strands of β-sheet that effectively displace helix 6, one of the α-helices that normally line the ligand-binding pocket, thereby expanding the pocket. The helix 1–3 insert is absent in CAR, suggesting that its helix 6 should remain undisplaced, acting as a wall to constrict the ligand-binding pocket as in most other NRs. Support for this hypothesis came from functional studies of zebrafish (Danio rerio) and chicken NR1I family members. These NRs each have comparable 45–55% sequence identity with both human PXR and human CAR, but each contains a helix 1–3 insert (27). Functionally, the chicken and zebrafish NR1I receptors were activated by a wide range of xenobiotics and steroids. These functional data classify these NR1I receptors as PXRs rather than CARs, in agreement with the structural classification based on the presence of the helix 1–3 insert. Importantly, these studies allow us to use sequence alone to characterize newly found NR1I sequences as either PXR-like or CAR-like receptors.
With the completed Fugu sequence, we can now ask whether the Fugu has separate CAR and PXR genes or whether a single progenitor exists, presumably the representative of the separate, diverged mammalian CAR and PXR genes. When the Fugu NR set was searched for potential PXR sequences, a single CAR/PXR gene was identified (fr078207). Sequence analysis showed that it displayed the hallmark structural characteristics of the PXR family members, including the helix 1–3 insert (Fig. 3). In fact, the Fugu helix 1–3 insert displayed a higher degree of sequence similarity to the human PXR helix 1–3 insert than did the zebrafish PXR. From these data, we predict that the Fugu receptor is the functional analog of PXR. This suggests that CAR diverged from PXR at a later point in vertebrate evolution or was lost in some or all of the teleost lines.
Figure 3.
Comparison of amino acid sequence of NR1I family members. α-Helix and β-strand residues are identified with yellow and red background highlighting, respectively. The capping segment of the PXR helix 1–3 insert is highlighted in blue. The secondary structure of human PXR was determined by examination of the crystal structure (28). For the remaining sequences, the secondary structure was predicted by the modeling analysis of Moore et al. (27).
In mice and humans, PXR and CAR expression is highest in liver and intestine, consistent with their role in xenobiotic protection. We examined expression of the Fugu PXR in various tissue cDNA libraries using non-quantitative PCR. In contrast to the mammalian receptors, Fugu PXR was expressed in a broad range of tissues, including brain, gill, gut, heart, liver and ovary (Fig. 4).
Figure 4.
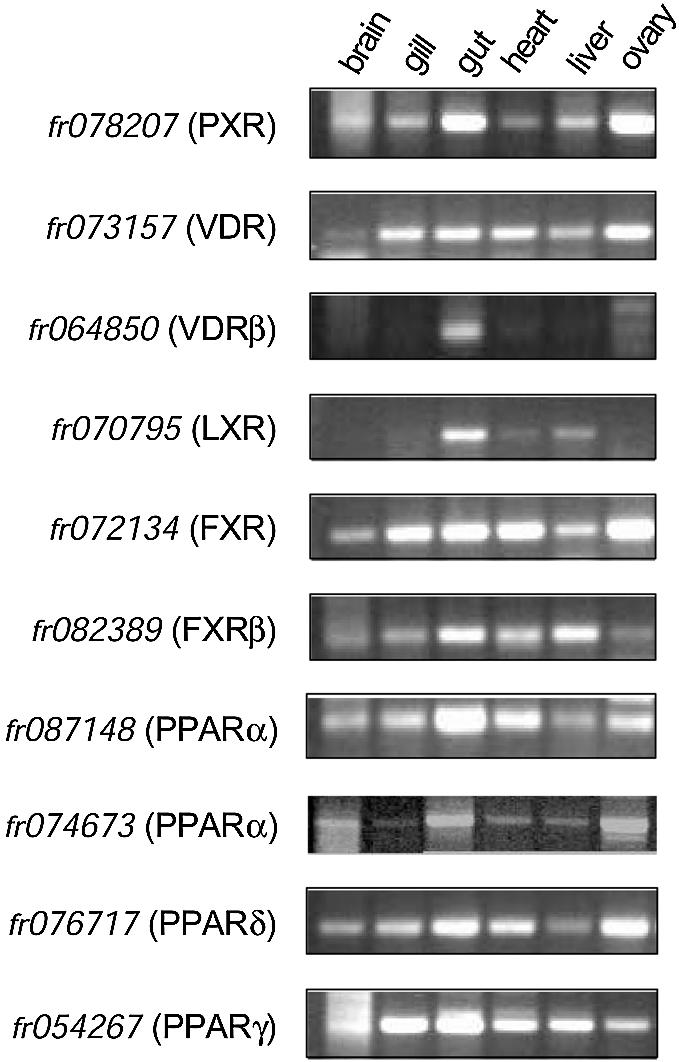
Expression analysis. Fugu brain, gill, gut, heart and ovary RNAs were prepared from frozen tissues and subjected to PCR analysis. Specific primers were chosen based on exon sequences representing fr078207 (PXR), fr073157 (VDR), fr064850 (VDR), fr070795 (LXR), fr072134 (FXR) and fr082389 (FXRβ), respectively. After 30 cycles of PCR, products were resolved on 1% agarose gels and visualized by ethidium bromide staining. In each case, PCR product size matched the predicted size based on sequence from the Fugu genome database (http://genome.jgi-psf.org/fugu3/fugu3.info.html).
Two related NR1I receptors, the benzoate X receptors (BXRα and BXRβ), have been identified in Xenopus laevis (29,30). The Xenopus BXRs are specialized for the benzoate class of ligands and are pharmacologically distinct from both PXR and CAR (27,31). When the human NR set was analyzed, no BXR ortholog was found (8). Until the availability of the complete Fugu sequence, it has not been possible to assess whether the BXRs are common among non-mammalian species. In our analysis, a BXR ortholog in Fugu was also not found, suggesting that this receptor could be unique to amphibians or specific to Xenopus. In the absence of the fully sequenced Xenopus genome, we cannot yet conclude whether amphibians have separate CAR and PXR genes in addition to the BXRs.
The VDRs are also NR1I family members, and in fact have recently been found to bind bile acids overlapping with the set that is also bound by PXR/CAR (32). The human VDR LBD contains a helix 1–3 insert different from that of PXR. The Fugu genome contained two VDR sequences, each displaying comparable sequence similarity with the human VDR. The existence of two VDRs has been noted previously in another species of fish (flounder) (33). As in the flounder, the two Fugu VDRs are more closely related to each other than to the PXRs or any other receptors.
The two VDRs in Fugu had distinct expression patterns. The receptor that was most closely related to the flounder VDRα, fr064850 (VDR), was expressed in all of the tissues tested, including brain, gill, gut, heart, liver and ovary (Fig. 4). In contrast, fr073157 (VDR) displayed a much more restricted pattern of expression (Fig. 4). Of the tissues we tested, VDRβ was expressed only in the gut. Interestingly, the sole mammalian VDR is expressed in a broad range of tissues, including brain, gut, skeletal muscle and heart, liver, pancreas and immune tissues, comparable to the broad expression pattern of fr064850 (VDR). It is possible that fr073157 (VDR) has taken on a more specialized role in Fugu in the gut.
NR1H subfamily (LXR, FXR)
The human NR1H subfamily contains the oxysterol receptors (LXRα and LXRβ) and the bile acid receptor (FXR). LXRs regulate genes involved in cholesterol and lipid metabolism while FXR regulates genes involved in bile acid metabolism (5). Within the Fugu NR set, an interesting contrast exists; the Fugu genome contains only one LXR, but two FXR sequences.
In mammals, LXRα and LXRβ have contrasting expression patterns. In mouse, LXRα is expressed most predominantly in liver, macrophages, brown and white adipose tissue and intestine, while LXRβ is expressed in virtually all tissues. In our analysis of the Fugu NR set, we have found that the Fugu genome contains only a single LXR gene (fr070795). Sequence analysis showed that the Fugu LXR was most closely related to the vertebrate LXRα gene. Fugu LXR displayed 75% sequence identity to the human LXRα LBD and 65% to the human LXRβ LBD.
The expression pattern of Fugu LXR was assessed across a panel of tissues (Fig. 4). Despite a higher sequence similarity to vertebrate LXRα, the pattern of expression of the Fugu LXR was more reminiscent of vertebrate LXRβ. Fugu LXR was expressed in all of the tissues tested, including brain, gill, gut, heart, liver and ovary. These data may indicate that the progenitor of the mammalian LXRs may have carried out a physiological role more analogous to mammalian LXRβ than LXRα. A second LXR gene with a more restricted tissue expression pattern may have arisen to function in specific tissues with key roles in cholesterol/lipid metabolism, such as liver, macrophages and adipose tissue. Interestingly, LXRα has been shown to be dramatically up-regulated in macrophages through regulation of the LXRα gene by LXRβ (34,35), consistent with LXRα playing a role in the amplification of LXR response in certain tissues.
In contrast to the LXRs, Fugu had two FXR genes (fr072134 and fr082389) compared to the single mammalian FXR. In humans, two FXR gene sequences are present in the genome, but one sequence represents an FXR pseudogene (8). A second FXR sequence was recently reported to exist in non-primate vertebrates (36) and we have identified the mammalian FXRβ sequence in the mouse genome sequence. Fugu may represent a good model in which to contrast the function of FXRα and FXRβ to gain insight into the physiological role of FXRβ. In Fugu, we found that expression of the FXR (fr072134) most closely related to mammalian FXR was restricted to gut and liver (Fig. 4). This expression pattern correlates well with the expression of mammalian FXR. In contrast, Fugu FXRβ (fr082389) was expressed in a broad range of tissues (Fig. 4). Ligand activation studies will be needed to determine whether the Fugu FXRβ and the mammalian FXRβ are functionally analogous.
PPARs
The human NR1C family consists of three members, PPARα, PPARδ and PPARγ (NR1C1, NR1C2 and NR1C3). The PPARs are receptors for endogenous and dietary fatty acids and mediate their effects on various cellular processes (37–39). Although the three PPAR subtypes are closely related and bind similar hormone response elements, each subserves a distinct role in fatty acid metabolism. PPARα agonists result in lowered triglycerides and increased high density lipoprotein cholesterol levels, while PPARγ agonists are insulin sensitizers, partly due to their role in promoting adipocyte differentiation. Relatively little is known about the PPARδ subtype, though it too appears to play a role in lipid homeostasis. The expression patterns of PPARα and PPARγ reflect their different physiological functions. PPARα is expressed mainly in liver, kidney and heart, while PPARγ is expressed mainly in adipose, colon and monocyte/ macrophage cells and PPARδ is expressed broadly.
Fugu contained a single ortholog of the human PPARγ and PPARδ subtypes and two orthologs of human PPARα (fr087148 and fr074673). The expression of the four Fugu PPAR genes was examined by PCR (Fig. 4). All four Fugu PPAR genes were broadly expressed (Fig. 4), contrasting with the human pattern, where only PPARδ is broadly expressed.
Mammalian PPARs contain four highly conserved residues that make hydrogen bonds to the acid head-group of their fatty acid ligands (40). Table 1 lists the amino acids at these key positions (numbering relative to the human PPARγ sequence) in the human receptors and their Fugu counterparts. Both Fugu PPARα orthologs (fr074373 and fr087148) have identical residues to human PPARα at these four key positions. The Fugu PPARδ (fr07671) has identical residues at three of the four positions relative to human PPARδ. At position 449 Fugu PPARδ (fr07671) contains an Asn rather than the His residue seen in human PPARδ, but this amino acid change would likely still allow binding of acidic ligands. Interestingly, the Fugu PPARγ (fr054267) has two amino acid differences within these key residues that would likely prevent binding of an acidic ligand. These changes are not unique to Fugu, but are seen in the PPARγ from salmon (41) as well as two other bony fish (flounder and the flatfish Pleuronectes platessa). Overall, these results indicate that fatty acids are not likely to activate fish PPARγ.
Final notes
In summary, we have found striking differences between the NR sets of the Fugu and human genomes. This analysis has focused on full-length NR coding sequences predicted by in silico methods. It is possible that some of these genes, though predicted to code for full-length proteins, may turn out to be pseudogenes due to an aberrant promoter (no expression) or aberrant post-transcriptional processing (incorrect splicing). The former possibility is not likely to be true for the majority of the Fugu NR genes since all of the 10 genes whose expression was assessed by PCR were expressed.
Overall, comparison of the complete human and Fugu NR sets highlights the remarkable diversity within the NR superfamily among vertebrates. It is possible that a small number of genes may yet lie in an unfinished portion of the Fugu genome. This is unlikely due to the extent of the coverage of the Fugu genome coupled with the fact that repetitive regions with low gene content are typically the last sequences to be derived in genome sequencing projects.
The human and Fugu NR sets were very analogous in terms of NR subfamilies, but not in terms of the total number of receptors within those subfamilies. Analysis of the 68 fish NRs help link NRs to function, to identify key sequence motifs, to identify conserved and likely physiologically relevant mRNA variants, to understand differences between humans and pharmacological model species and to provide clues as to the potential tractability of particular receptor subgroups. Thus, the teleost genome with its expanded set of NRs provides an additional and interesting model to study both the evolution and function of the conserved vertebrate NRs. Comparisons such as these at the nucleic acid level reflect only one level of complexity of NR sets. The definition and comparison of the Fugu NR proteome (the total set of NRs resulting from differential post-transcriptional processing) will be a subject of future studies.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Dr Ann Sluder (Cambria Biotech) for her insightful comments and suggestions during the preparation of this manuscript. The authors would also like to thank Dr Greg Elgar (UK-HGMP-RC) for the generous gift of liver samples from Fugu rubripes.
REFERENCES
- 1.Willson T.M. and Moore,J.T. (2002) Minireview: genomics versus orphan nuclear receptors—a half-time report. Mol. Endocrinol., 16, 1135–1144. [DOI] [PubMed] [Google Scholar]
- 2.Robyr D., Wolffe,A.P. and Wahli,W. (2000) Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol. Endocrinol., 14, 329–347. [DOI] [PubMed] [Google Scholar]
- 3.McKenna N.J. and O’Malley,B.W. (2002) Minireview: nuclear receptor coactivators—an update. Endocrinology, 143, 2461–2465. [DOI] [PubMed] [Google Scholar]
- 4.Willson T.M., Jones,S.A., Moore,J.T. and Kliewer,S.A. (2001) Chemical genomics: functional analysis of orphan nuclear receptors in the regulation of bile acid metabolism. Med. Res. Rev., 21, 513–522. [DOI] [PubMed] [Google Scholar]
- 5.Lu T.T., Repa,J.J. and Mangelsdorf,D.J. (2001) Orphan nuclear receptors as eLiXiRs and FiXeRs of sterol metabolism. J. Biol. Chem., 276, 37735–37738. [DOI] [PubMed] [Google Scholar]
- 6.Edwards P.A., Kast,H.R. and Anisfeld,A.M. (2002) BAREing it all: the adoption of LXR and FXR and their roles in lipid homeostasis. J. Lipid Res., 43, 2–12. [PubMed] [Google Scholar]
- 7.Willson T.M. and Kliewer,S.A. (2002) PXR, CAR and drug metabolism. Nature Rev. Drug Discov., 1, 259–266. [DOI] [PubMed] [Google Scholar]
- 8.Maglich J.M., Sluder,A., Guan,X., Shi,Y., McKee,D.D., Carrick,K., Kamdar,K., Willson,T.M. and Moore,J.T. (2001) Comparison of complete nuclear receptor sets from the human, Caenorhabditis elegans and Drosophila genomes. Genome Biol., 2, 29.1–29.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson-Rechavi M., Carpentier,A.S., Duffraisse,M. and Laudet,V. (2001) How many nuclear hormone receptors are there in the human genome? Trends Genet., 17, 554–556. [DOI] [PubMed] [Google Scholar]
- 10.Enmark E. and Gustafsson,J.A. (2001) Comparing nuclear receptors in worms, flies and humans. Trends Pharmacol. Sci., 22, 611–615. [DOI] [PubMed] [Google Scholar]
- 11.Kliewer S.A., Lehmann,J.M. and Willson,T.M. (1999) Orphan nuclear receptors: shifting endocrinology into reverse. Science, 284, 757–760. [DOI] [PubMed] [Google Scholar]
- 12.Blumberg B. and Evans,R.M. (1998) Orphan nuclear receptors—new ligands and new possibilities. Genes Dev., 12, 3149–3155. [DOI] [PubMed] [Google Scholar]
- 13.Kliewer S.A., Moore,J.T., Wade,L., Staudinger,J.L., Watson,M.A., Jones,S.A., McKee,D.D., Oliver,B.B., Willson,T.M., Zetterstrom,R.H., Perlmann,T. and Lehmann,J.M. (1998) An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell, 92, 73–82. [DOI] [PubMed] [Google Scholar]
- 14.Jones S.A., Moore,L.B., Shenk,J.L., Wisely,G.B., Hamilton,G.A., McKee,D.D., Tomkinson,N.C., LeCluyse,E.L., Lambert,M.H., Willson,T.M., Kliewer,S.A. and Moore,J.T. (2000) The pregnane X receptor: a promiscuous xenobiotic receptor that has diverged during evolution. Mol. Endocrinol., 14, 27–39. [DOI] [PubMed] [Google Scholar]
- 15.Sluder A.E. and Maina,C.V. (2001) Nuclear receptors in nematodes: themes and variations. Trends Genet., 17, 206–213. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Notredame C., Higgins,D.G. and Heringa,J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol., 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 18.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 20.Holm L. and Sander,C. (1998) Removing near-neighbour redundancy from large protein sequence collections. Bioinformatics, 14, 423–429. [DOI] [PubMed] [Google Scholar]
- 21.Lambert M.H. (1997) Docking conformationally flexible molecules into protein binding sites. In Charifson,P.S. (ed.), Practical Application of Computer-aided Drug Design. Marcel Dekker, New York, NY, pp. 243–303.
- 22.Aparicio S., Chapman,J., Stupka,E., Putnam,N., Chia,J.M., Dehal,P., Christoffels,A., Rash,S., Hoon,S., Smit,A. Gelpke,M.D., Roach,J., Oh,T., Ho,I.Y., Wong,M., Detter,C., Verhoef,F., Predki,P., Tay,A., Lucas,S., Richardson,P., Smith,S.F., Clark,M.S., Edwards,Y.J., Doggett,N. et al. (2002) Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science, 297, 1301–1310. [DOI] [PubMed] [Google Scholar]
- 23.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W., Funke,R., Gage,D., Harris,K., Heaford,A., Howland,J., Kann,L., Lehoczky,J., LeVine,R., McEwan,P., McKernan,K., Meldrim,J., Mesirov,J.P., Miranda,C., Morris,W., Naylor,J. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 24.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. Gocayne,J.D., Amanatides,P., Ballew,R.M., Huson,D.H., Wortman,J.R., Zhang,Q., Kodira,C.D., Zheng,X.H., Chen,L., Skupski,M., Subramanian,G., Thomas,P.D., Zhang,J., Gabor Miklos,G.L., Nelson,C. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 25.Sluder A.E., Mathews,S.W., Hough,D., Yin,V.P. and Maina,C.V. (1999) The nuclear receptor superfamily has undergone extensive proliferation and diversification in nematodes. Genome Res., 9, 103–120. [PubMed] [Google Scholar]
- 26.Taylor J.S., Van de Peer,Y. and Meyer,A. (2001) Revisiting recent challenges to the ancient fish-specific genome duplication hypothesis. Curr. Biol., 11, R1005–R1008. [DOI] [PubMed] [Google Scholar]
- 27.Moore L.B., Maglich,J.M., McKee,D.D., Wisely,B., Willson,T.M., Kliewer,S.A., Lambert,M.H. and Moore,J.T. (2002) Pregnane X receptor (PXR), constitutive androstane receptor (CAR) and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol. Endocrinol., 16, 977–986. [DOI] [PubMed] [Google Scholar]
- 28.Watkins R.E., Wisely,G.B., Moore,L.B., Collins,J.L., Lambert,M.H., Williams,S.P., Willson,T.M., Kliewer,S.A. and Redinbo,M.R. (2001) The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science, 292, 2329–2333. [DOI] [PubMed] [Google Scholar]
- 29.Blumberg B., Kang,H., Bolado,J.,Jr, Chen,H., Craig,A.G., Moreno,T.A., Umesono,K., Perlmann,T., De Robertis,E.M. and Evans,R.M. (1998) BXR, an embryonic orphan nuclear receptor activated by a novel class of endogenous benzoate metabolites. Genes Dev., 12, 1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishikawa J., Saito,K., Sasaki,M., Tomigahara,Y. and Nishihara,T. (2000) Molecular cloning and functional characterization of a novel nuclear receptor similar to an embryonic benzoate receptor BXR. Biochem. Biophys. Res. Commun., 277, 209–215. [DOI] [PubMed] [Google Scholar]
- 31.Grun F., Venkatesan,R.N., Tabb,M.M., Zhou,C., Cao,J., Hemmati,D. and Blumberg,B. (2002) Benzoate X receptor alpha and beta are pharmacologically distinct and do not function as xenobiotic receptors. J. Biol. Chem., 26, 43691–43697. [DOI] [PubMed] [Google Scholar]
- 32.Makishima M., Lu,T.T., Xie,W., Whitfield,G.K., Domoto,H., Evans,R.M., Haussler,M.R. and Mangelsdorf,D.J. (2002) Vitamin D receptor as an intestinal bile acid sensor. Science, 296, 1313–1316. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki T., Suzuki,N., Srivastava,A.S. and Kurokawa,T. (2000) Identification of cDNAs encoding two subtypes of vitamin D receptor in flounder, Paralichthys olivaceus. Biochem. Biophys. Res. Commun., 270, 40–45. [DOI] [PubMed] [Google Scholar]
- 34.Whitney K.D., Watson,M.A., Goodwin,B., Galardi,C.M., Maglich,J.M., Wilson,J.G., Willson,T.M., Collins,J.L. and Kliewer,S.A. (2001) Liver X receptor (LXR) regulation of the LXRalpha gene in human macrophages. J. Biol. Chem., 276, 43509–43515. [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Bolten,C., Bhat,B.G., Woodring-Dietz,J., Li,S., Prayaga,S.K., Xia,C. and Lala,D.S. (2002) Induction of human liver X receptor alpha gene expression via an autoregulatory loop mechanism. Mol. Endocrinol., 16, 506–514. [DOI] [PubMed] [Google Scholar]
- 36.Otte K., Kranz,H,., Kober,I., Thompson,P., Hoefer,M., Haubold,B., Remmel,B., Voss,H., Kaiser,C., Albers,M., Cheruvallath,Z., Jackson,D., Casari,G., Koegl,M., Paabo,S., Mous,J., Kremoser,C. and Deuschle,U. (2003) Identification of farnesoid X receptor beta as a novel mammalian nuclear receptor sensing lanosterol. Mol. Cell. Biol., 23, 864–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kliewer S.A., Xu,H.E., Lambert,M.H. and Willson,T.M. (2001) Peroxisome proliferator-activated receptors: from genes to physiology. Recent Prog. Horm. Res., 56, 239–263. [DOI] [PubMed] [Google Scholar]
- 38.Willson T.M., Lambert,M.H. and Kliewer,S.A. (2001) Peroxisome proliferator-activated receptor gamma and metabolic disease. Annu. Rev. Biochem., 70, 341–367. [DOI] [PubMed] [Google Scholar]
- 39.Hihi A.K., Michalik,L. and Wahli,W. (2002) PPARs: transcriptional effectors of fatty acids and their derivatives. Cell. Mol. Life Sci., 59, 790–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willson T.M., Brown,P.J., Sternbach,D.D. and Henke,B.R. (2000) The PPARs: from orphan receptors to drug discovery. J. Med. Chem., 43, 527–550. [DOI] [PubMed] [Google Scholar]
- 41.Andersen O., Eijsink,V.G. and Thomassen,M. (2000) Multiple variants of the peroxisome proliferator-activated receptor (PPAR) gamma are expressed in the liver of Atlantic salmon (Salmo salar). Gene, 255, 411–418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.