Abstract
The yeast protein Rrf1p encoded by the FIL1 nuclear gene bears significant sequence similarity to Escherichia coli ribosome recycling factor (RRF). Here, we call FIL1 Ribosome Recycling Factor of yeast, RRF1. Its gene product, Rrf1p, was localized in mitochondria. Deletion of RRF1 leads to a respiratory incompetent phenotype and to instability of the mitochondrial genome (conversion to rho–/rho0 cytoplasmic petites). Yeast with intact mitochondria and with deleted genomic RRF1 that harbors a plasmid carrying RRF1 was prepared from spores of heterozygous diploid yeast. Such yeast with a mutated allele of RRF1, rrf1-L209P, grew on a non-fermentable carbon source at 30 but not at 36°C, where mitochondrial but not total protein synthesis was 90% inhibited. We propose that Rrf1p is essential for mitochondrial protein synthesis and acts as a RRF in mitochondria.
INTRODUCTION
Mitochondrial gene expression involves a complex translation system. In Saccharomyces cerevisiae more than 120 genes have been identified that code for components required for mitochondrial protein synthesis (1–5). The components of the mitochondrial translation machinery are distinct from those of the cytoplasmic system, except for three aminoacyl tRNA synthetases (6–8). The mitochondrial translation system is known to resemble the prokaryotic translation system (9). However, a number of unusual features of the mitochondrial translation system are not found in the prokaryotic translation system. For example, the mitochondrial genetic code differs in various aspects from the universal one (10), and the structural features of the mitochondrial ribosome are unusual (1,2,11). A stringent block of mitochondrial protein synthesis destabilizes the mitochondrial genome (12). The mitochondrial translation factors have been cloned either through their homology with bacterial translation factors [EF-Tu (13), human ribosome recycling factor (RRF) and RF1 (14)] or through the complementation of respiratory incompetent mutants [IF2, EF-G (15) and RF1 (16)]. However, no EF-Ts have been detected in yeast mitochondria (17). Since no functional in vitro mitochondrial translation system with natural mRNA exists (18), the mechanisms of mitochondrial protein synthesis are poorly understood.
The RRF (formally called ribosome releasing factor) is an essential component of the translation machinery in prokaryotes, where it was first characterized in 1970 in our laboratory (19,20). It catalyzes the disassembly of post-termination complexes consisting of ribosome, mRNA and tRNA, allowing the recycling of the components for another round of translation. In the absence of RRF, the ribosome stays on the mRNA and starts downstream unscheduled translation (21,22). RRF is also involved in the fidelity of translation (23). Recent work indicates that RRF binds to A/P sites of the ribosome (24,25) and is translocated on the ribosome to release tRNA (26) bound at the P/E site (27). RRF has been identified in all prokaryotic organisms examined so far except for Archaeons (for reviews see 28–31). The spinach RRF homolog has been shown to be localized in the chloroplast (32), where a stoichiometric amount of RRF homolog is associated with the 50S ribosomal subunit (33).
Kanai et al. isolated a yeast mutant that failed to derepress expression of the S.cerevisiae isocitrate lyase gene (ICL1) in acetate medium and then identified the gene, FIL1, that complemented this mutation. This gene was named FIL1 (factor for isocitrate lyase derepression) (34). In this paper, we call the gene FIL1 RRF1 because, as pointed out by Kanai et al., it is clearly a yeast homolog of the Escherichia coli gene coding for RRF (frr). According to yeast conventions, the protein encoded by RRF1 is called Rrf1p.
In this communication, we show that Rrf1p is a mitochondrial protein that is essential for mitochondrial protein synthesis and for the maintenance of the respiratory function of mitochondria. A haploid yeast dependent on plasmid-borne RRF1 for growth on non-fermentable carbon source was created. Using this strain, we changed a leucine residue at position 209 of Rrf1p to proline and obtained a temperature-sensitive yeast strain. Mitochondrial protein synthesis of this strain was severely reduced at 37°C while total cytoplasmic protein synthesis was not affected under the same conditions.
MATERIALS AND METHODS
Strains, plasmids and genetic manipulations
The yeast strains used are listed in Table 1. They are derivatives of WY344 and DS413 and the haploid of DS413. The rho0 strains were prepared by ethidium bromide treatment (35). Genetic techniques used are described by Sprague (36). The E.coli were DH5α, BL21(DE3)pLysS (Novagen) and NMS22 mutS (Pharmacia Biotech).
Table 1. Strains and plasmids.
| Strain | Genotype | Source |
|---|---|---|
| H1402 | MATα ino1 ura3-52 leu2-3,112 HIS4-lacZ | (65) |
| Strain RRF | Wild-type haploid stain with intact RRF1 [for example, WY344 (RRF1)] | |
| Strain Δrrf1 | Haploid cells lacking RRF1 [for example, WY347 (Δrrf1/ rho–)] | |
| WY344 (RRF1) | H1402 in which the TRP1 gene was disrupted using a method described in (66). MATα ino1 ura3-52 leu2-3,112 trp1::URA3 HIS4-lacZ | This study |
| WY347 (Δrrf1/rho–) | WY344 in which the RRF1 gene was disrupted using a method described in (38). MATα ino1 ura3-52 leu2-3,112 trp1::URA3 HIS4-lacZ RRF1::TRP1 | This study |
| DS413 | MATa/α ura3-52/ura3-52 leu2-3,112/leu2-3,112 ade2-101/+ his3Δ200/his3Δ200 +/trp1Δ1 | D. Roofa |
| ET1 (RRF1/Δrrf1) | DS413 in which one allele of the RRF1 gene was disrupted using a method described in (41). MATa/α ura3-52/ura3-52 leu2-3,112/leu2-3,112 ade2-101/+ his3Δ200/his3Δ200 +/trp1 Δrrf1/RRF1::kanr | This study |
| ET2 (Δrrf1/ rho+) |
Haploid meiotic progeny of ET1. MATa ura3-52 leu2-3,112 ade2-101 his3Δ200trp1 Δrrf1::kanr |
This study |
| Plasmid |
Markers |
Source |
| pRS415-ADHp | ARS_CEN6 LEU2 ADH1 promoter (low copy number, centromeric) | D. Roofa |
| pRRF1W1 | pRS415-ADHp carrying RRF1 (wild type) | This study |
| prrf1-K67R | pRS415-ADHp carrying rrf1-K67R | This study |
| prrf1-L206E | pRS415-ADHp carrying rrf1-L206E | This study |
| prrf1- L209P | pRS415-ADHp carrying rrf1-L209P | This study |
| prrf1- I228P | pRS415-ADHp carrying rrf1-I228P | This study |
| pRS426 | 2 µm origin URA3 (high copy number) plasmid | (39) |
| pRRF1W2 | pRS426 ADH1 promoter carrying RRF1 (wild type) | This study |
| pET28a | Escherichia coli expression plasmid | Novagen |
| pET28a-tRRF1 | pET28a carrying a PCR amplified fragment coding for the 194 C-terminal fragment Rrf1p | This study |
aUniversity of Pennsylvania, Philadelphia.
Media
YEP medium (1% yeast extract, 2% Bacto-Peptone) or synthetic minimal medium containing the required nutrients (0.7% yeast nitrogen base with ammonium sulfate) was supplemented with glucose (2%), glycerol (3%) or ethanol (2%) as noted (37).
Construction of Δrrf1 rho– (WY347) and Δrrf1 rho+ (ET2) strains
The RRF1 ORF was replaced with TRP1 (tryptophan synthase gene) or kanMX (kanamycin resistance gene), resulting in Δrrf1 rho–, as described in Baudin et al. (38). The TRP1 gene was amplified by PCR from the plasmid PRS314 (39). Stable transformants with the amplified fragment were prepared as in Ausubel et al. (40). Replacement of RRF1 by TRP1 was confirmed by PCR. Δrrf1 rho+ was prepared as in Wach et al. (41) by inserting kanMX into RRF1 and sporulating. The kanMX gene was amplified by PCR with pFA6a-kanMX6 (41–43). Diploid yeast strain DS413 was transformed with this fragment as in Ausubel et al. (40) and a geneticin (Gibco BRL)-resistant clone, strain ET1, Δrrf1::kanMX/RRF1 diploids, were selected.
Expression of recombinant truncated Rrf1p (His6-partial Rrf1p) in E.coli and production of antibody against Rrf1p
Histidine-tagged truncated Rrf1p (194 C-terminal residues of Rrf1p) was expressed in BL21(DE3)pLysS E.coli harboring pET28a-tRRF1 [pET28a (Novagen) carrying tRRF1 (truncated RRF1)]. His6-partial Rrf1p was used to obtain a rabbit polyclonal antibody (Cocalico Biologicals Inc.) against Rrf1p.
Low copy and multi-copy expression of Rrf1p
Yeast total DNA (strain WY344) for PCR was isolated as in Ausubel et al. (40). The amplified fragment was ligated into the BamHI site of the LEU2/CEN shuttle vector pRS415-ADHp (a gift from D. Roof), creating pRRF1W1. For multi-copy expression, pRRF1W1 was sub-cloned into the XhoI and SacI sites of the 2 µm URA3 shuttle vector pRS426 (a gift from D. Roof), generating pRRF1W2.
Complementation of Δrrf1 cells
The plasmid pRRF1W2 was introduced into Δrrf1::kanMX/RRF1 diploids (strain ET1), which were then sporulated (40) and subjected to random spore analysis. We selected ade2 cells which turn red on low adenine-containing medium if respiratory competent (44,45). The Δrrf1 haploid, RRF1, replaced with kanMX in spores containing pRRF1W2, was selected for resistance to geneticin and the ability to grow without uracil (pRRF1W2 carries the selectable marker URA3).
Site-directed mutagenesis of Rrf1p
Mutations were introduced using the unique site elimination (USE) procedure (Mutagenesis Kit; Pharmacia Biotech) developed by Deng and Nickoloff (46). ADHp-RRF1 (pRRF1W1) was mutagenized using both a target mutagenic primer introducing the desired mutation in RRF1 and a USE selection primer introducing a mutation in the unique XhoI site of the plasmid polylinker. The elimination of the XhoI site subsequently served as the basis for selection of mutated plasmids. The target mutagenic primers used to introduce the different mutations were as follows: CTTTAGAAATTCAAAGACAGAAAATG for the mutation Rrf1p-K67R, GCTGAGAGGGATGAGGAAAAACTG for the mutation Rrf1p-L206P, GGGATTTGGAAAAACCGCATAAGGATTACG for the mutation Rrf1p-L209P and GTTGAAAA AAGCCCTGTAAAATGA for the mutation Rrf1p-I228P. Underlined characters indicate the nucleotide positions changed by the site-directed mutagenesis. The phenotypes of the mutant alleles were tested by the ‘plasmid shuffle’ procedure (47).
Immunofluorescence and DAPI staining
Yeast cells were fixed and prepared for immunofluorescence microscopy as described (48). Rrf1p was detected using a rabbit antibody against Rrf1p and FITC-conjugated goat anti-rabbit secondary antibody (Sigma). DNA was stained with 4,6-diamino-2-phenylindole (DAPI) (Sigma) as described (48).
Preparation of cellular extracts, isolation of mitochondria and detection of Rrf1p
Crude yeast extracts were prepared according to Kolodziej and Young (49). Mitochondria were isolated as in Daum et al. (3). The protein Rrf1p was detected by western blot, using immunoaffinity purified rabbit antibody against His6-partial Rrf1p (dilution 1/250) and goat anti-rabbit alkaline phosphatase-conjugated antibody (dilution 1/7500; Sigma). Porin protein was detected using a rabbit antibody against S.cerevisiae porin (a gift of D. Pain) and FITC-conjugated goat anti-rabbit antibodies (Sigma).
Labeling of mitochondrial translation products in vivo
Mitochondrial translation products were labeled in vivo as described (50). Cells were incubated at 30°C and cycloheximide was added to a final concentration of 150 µg/ml. After further incubation for 2 min, the incubation temperature was shifted to 37°C and labeling with [35S]methionine (40 µCi, 1175 Ci/mmol) (NEN) was performed for 80 min and stopped by adding unlabeled methionine (10 mM). Total cell proteins were extracted by alkaline lysis (51). Proteins were separated by SDS–PAGE (12% acrylamide) and visualized by autoradiography and Coomassie staining.
[35S]methionine incorporation into whole cells
Cells (grown at 29°C in glucose medium) were diluted to OD540 = 0.05 and [35S]methionine 20 µM (39.5 µCi in 5 ml) was added and incubated at 29°C. When the OD540 reached 0.45, the culture was split for incubation at 37 and 29°C. The incorporation of [35S]methionine into the hot TCA-insoluble fraction was counted.
Homology modeling
The structural model was built using the software provided by Tripos and Associates after amino acid sequence alignment with Thermotoga maritima RRF. There is ∼20% sequence identity and ∼52% sequence similarity. Individual side chain replacements were made to the 1DD5 structure to reflect the yeast RRF sequence, as previously described (52). A loop engraftment was used to modify the template in the region of the T.maritima RRF between residues 102 and 107 (FPSPTT). The corresponding yeast sequence is LPPPTT. In searching the structural database, a small loop was found in 15-lipoxygenase (database code 1LOX) with the identical sequence LPPPTT and a suitable geometry for insertion into the structural template. After loop grafting, explicit hydrogens were added and the final structure was subjected to four cycles of dynamics (200 steps)/minimization (50 steps) followed by minimization to energy convergence. Minimization was done by the conjugate gradient method using the Kollman all-atom force field and Gasteiger–Huckle charges. The final structure was checked for appropriate bond lengths, chirality and dihedral angles using the ProTable module of the Tripos software.
RESULTS
Irreversible damage of the mitochondrial genome by RRF1 deletion
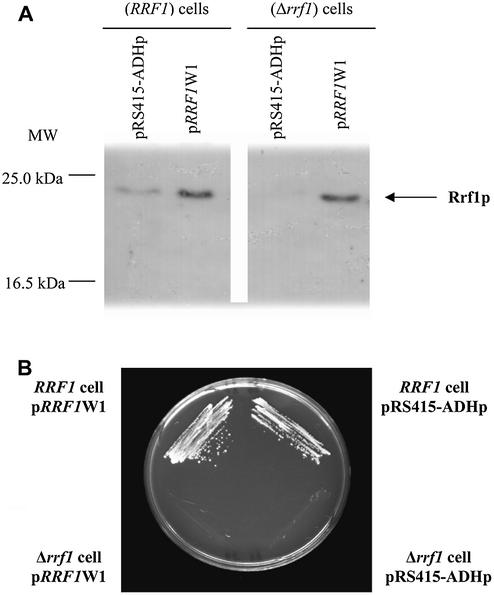
The Δrrf1 haploid strain, in which RRF1 was disrupted by replacing the complete RRF1 ORF with the TRP1 gene, grew on rich glucose medium but slower than the wild-type (data not shown). The Δrrf1 strain, however, did not grow on rich glycerol medium. These two observations suggest that the cells deleted for RRF1 are respiratory incompetents. Although data were not shown, Kanai et al. stated that a similar strain did not grow on acetate (34). The expression of Rrf1p in wild-type (RRF1 cells) and mutant S.cerevisiae (Δrrf1 cells) was examined in crude cell extracts by western blot using anti-Rrf1p antibodies as shown in Figure 1A. A protein with an apparent molecular mass of 23 kDa was detected in the wild-type strain RRF1 and in a strain, Δrrf1, transformed with a centromeric low copy number plasmid carrying the DNA corresponding to RRF1 (pRRF1W1). This band was absent in Δrrf1 transformed with the control plasmid (pRS415-ADHp). The apparent molecular mass of Rrf1p (23 kDa) was smaller than the size deduced from the RRF1 nucleotide sequence (26.4 kDa). This difference in size suggests that the Rrf1p N-terminal signal sequence acts as a removable mitochondrial targeting sequence (34). The yeast RRF gene carried by pRRF1W1 (low copy number plasmid), despite its expression, was unable to restore the capacity to grow on ethanol in the strain Δrrf1 (Fig. 1B, lower left corner).
Figure 1.
Haploid yeast with a RRF1 deletion does not grow on glycerol, a phenotype that cannot be overcome by cytoplasmic Rrf1p. (A) Immunodetection of Rrf1p in Δrrf1 haploid cells (WY347) transformed with a centromeric copy of the RRF1 gene. Total protein extracts from four different strains were analyzed, shown from left to right: wild-type RRF1 haploid cells (WY344, designated RRF1 cells) harboring a centromeric control plasmid carrying no RRF1 (pRS415-ADHp); WY344 harboring a centromeric plasmid carrying the RRF1 gene (designated pRRF1W1); Δrrf1 haploid cells (WY347, designated Δrrf1 cells) harboring pRS415-ADHp or harboring pRRF1W1. Protein extracts (50 µg) were analyzed in 12% gels by western blotting using an antibody against His6-partial Rrf1p. (B) Lack of growth of Δrrf1 haploid cells harboring a plasmid carrying RRF1 on a non-fermentable carbon source. Four different strains were streaked on an agar plate containing ethanol selective medium. Wild-type RRF1 haploid cells (WY344, designated RRF1) harboring a centromeric control plasmid carrying no RRF1 (pRS415-ADHp) (top right). WY344, harboring a centromeric plasmid carrying the RRF1 gene (pRRF1W1) (top left). Δrrf1 haploid cells (WY347, designated Δrrf1) harboring pRS415-ADHp (bottom right). WY347 harboring pRRF1W1 (bottom left). The photograph was taken after 3 days growth at 30°C.
We then mated Δrrf1 haploids with haploids that contained wild-type RRF1 but no mitochondrial DNA (rho0 strain). The diploids generated were unable to grow on non-fermentable carbon sources (data not shown). Furthermore, the mitochondrial protein porin and the mitochondrial DNA of Δrrf1 haploids were stained with DAPI and fluorescent labeled anti-porin. The results showed that mitochondria-like particles were present in Δrrf1 haploids, but the mitochondrial DNA appeared more diffuse and less abundant than in the control strain (data not shown). These results suggest that, in Δrrf1 cells, the mitochondrial DNA present may be irreversibly damaged and additional RRF1 is unable to restore the mitochondrial function necessary for growth on a non-fermentable carbon source.
RRF1 rescues the respiratory deficiency of Δrrf1 mutants that contain intact mitochondrial DNA
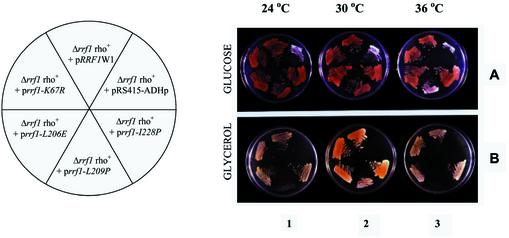
Since the simple addition of a plasmid carrying RRF1 (pRRF1W1) to the strain Δrrf1 did not complement the null disruption as indicated above, we concluded that the lack of RRF1 led to a loss of respiration-competent mitochondria. Rrf1p expression also could not restore respiration by the mitochondria. To circumvent this problem, we generated a haploid Δrrf1 strain maintained by RRF1 on a plasmid from a heterozygous diploid strain using the sporulation technique. All Δrrf1 haploids harboring pRRF1W2 obtained were able to utilize a non-fermentable carbon source (data not shown). Therefore, as predicted, pRRF1W2 was able to complement the strain Δrrf1 (haploid) if prepared in this fashion. The high copy number plasmid (pRRF1W2) was then exchanged with a low copy number centromere-based plasmid (pRRF1W1). Figure 2A shows that these haploid cells with (upper middle section) and without (upper right section) extra-chromosomal RRF1 grew on the plate containing glucose. As shown in Figure 2B (upper middle sections of plates 1–3), the Δrrf1 haploids carrying pRRF1W1 (Δrrf1 rho+ pRRF1W1) were able to grow on a non-fermentable carbon source. The control strain without wild-type RRF1 (Δrrf1 rho+, pRS415-ADHP) did not grow on the glycerol media (upper right sections of plates 1–3). This demonstrates that complementation is possible only when the plasmid with wild-type RRF1 is introduced before mitochondrial function is compromised.
Figure 2.
Complementation of Δrrf1 rho+ haploid cells with a plasmid carrying RRF1. Evidence that rrf1-L209P gives temperature-sensitive growth. Δrrf1 rho+ haploid cells harboring various plasmids carrying mutated RRF1 were created as described in Materials and Methods. These various strains are indicated in the circle with the pie cut (left). (A) Cells harboring each of the plasmids as shown in the left panel were streaked onto synthetic glucose (3%) medium lacking leucine and grown at 24, 30 and 36°C. (B) The same strains were grown overnight in synthetic glucose medium minus leucine, streaked onto YPG plates (containing 3% glycerol) and incubated at 24, 30 and 36°C. Δrrf1 rho+ haploid cells (ET2) are ade2-101. Such adenine-deficient cells are known to accumulate red pigments in the presence of low adenine when they are respiratory competent. Note that Δrrf1 rho+ + prrf-L209P could not grow on glycerol at 36°C (lower middle streak, plate 3) while the strain harboring pRS415-ADHp (no RRF1) did not grow in glycerol medium at all temperatures tested (plates 1–3, upper right streak). This indicates that Δrrf1 rho+ + pRS415-ADHp was converted to rho– because of the lack of Rrf1p.
Intracellular localization of Rrf1p
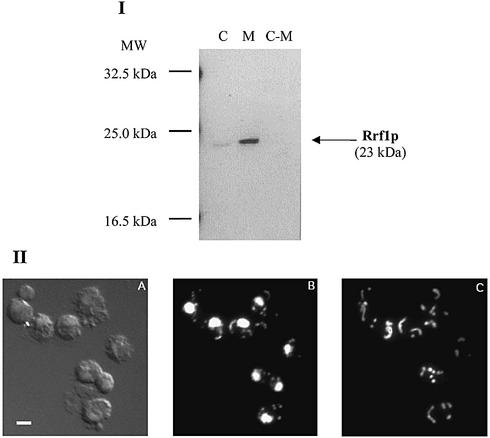
To determine the subcellular location of Rrf1p, mitochondria and post-mitochondrial supernatant fractions were isolated from wild-type diploid cells (DS413). The presence of Rrf1p in each fraction was examined by immunoblotting with affinity purified Rrf1p polyclonal antibodies. Western blot analysis revealed Rrf1p in mitochondria, but not in the post-mitochondrial supernatant (Fig. 3I). In a separate experiment, anti-Put2p serum (a gift from D. Pain) detected Put2p, a protein known to localize to mitochondria, only in the mitochondrial fraction obtained by the same method (data not shown). This established the validity of our biochemical fractionation method for the isolation of mitochondria used in Figure 3I. The size of Rrf1p detected in the mitochondria was the same as that of Rrf1p detected in the crude extract, 23 kDa. This suggests that cleavage of the mitochondrial signal peptide takes place in the mitochondria and that the major percentage of Rrf1p is in the mitochondria.
Figure 3.
(I) Rrf1p is localized in the fraction containing mitochondria. Western blot analysis of crude yeast extracts (designated column C), isolated mitochondria (column M) and post-mitochondrial supernatant fraction (C-M) obtained from wild-type strain DS413. Proteins (40 µg) of each preparation were analyzed by western blotting using antibody against His6-partial Rrf1p. (II) Cytological localization of Rrf1p to mitochondria. A representative microscopic field of Δrrf1 rho+ haploid cells (strain ET2) harboring a high copy number plasmid carrying RRF1 (pRRF1W2) is shown. (A) Differential interference contrast view. (B) DAPI staining showing the locations of nuclear and mitochondrial DNA. (C) Indirect immunofluorescence using antibodies to His6-partial Rrf1p and FITC- conjugated secondary antibody. Bar represents 1 µm.
To further confirm mitochondrial localization of Rrf1p, Rrf1p was detected in situ by immunofluorescence microscopy of wild-type cells and cells overexpressing Rrf1p. Cells were treated with polyclonal anti-Rrf1p antibodies and with DAPI. As shown in Figure 3II, in the cells overexpressing Rrf1p, Rrf1p co-localized with mitochondrial DNA stained with DAPI. It should be emphasized that Rrf1p was detected exclusively in mitochondria. There was no fluorescent signal associated with either the nuclei or cytoplasm. The signals associated with anti-Rrf1p in wild-type cells expressing native levels of Rrf1p were very faint, but the results indicate a similar mitochondrial distribution as in cells containing overexpressed Rrf1p (data not shown).
Isolation of a temperature-sensitive RRF1 allele
To establish the role of Rrf1p in mitochondrial protein synthesis, it was desirable to obtain a temperature-sensitive allele of RRF1. We reasoned that, because of the sequence similarity between Rrf1p and E.coli RRF, site-specific mutagenesis of RRF1 at residues corresponding to temperature-sensitive alleles of E.coli frr may also produce temperature-sensitive yeast strains. Twelve temperature- sensitive mutations of E.coli frr gene have been characterized (21). The position and the amino acid change for each of these mutations are known. Out of the 12 mutations, four were selected using two criteria: (i) the amino acid residue at the mutated position was conserved among most RRF homolog sequences and (ii) the corresponding mutation in RRF1 requires the modification of less than 3 nt. The four selected mutations were frr4, frr7, frr15 and frr17 in E.coli (21) and correspond to L163P, V160E, K21R and L182P, respectively.
We constructed the alleles rrf1-K67R, rrf1-L206E, rrf1-L209P and rrf1-I228P, each of which was confirmed by sequencing to be otherwise identical to RRF1. Low copy number centromere plasmids each bearing either rrf1-K67R (prrf1-K67R), rrf1-L206E (prrf1-L206E), rrf1-L209P (prrf1-L209P) or rrf1-I228P (prrf1-I228P) were prepared. Each of these plasmids was introduced into haploid Δrrf1::kanMX (ade2–) (rho+) cells (strain ET2) harboring pRRF1W2 by the ‘plasmid shuffle’ scheme. Haploid Δrrf1 cells transformed with wild-type RRF1 (pRRF1W1), as described in the preceding section, provided the respiratory positive control for analysis of the RRF1 mutants. Haploid Δrrf1 cells transformed with the empty plasmid (pRS415-ADHp) was a respiratory negative control.
At 24, 30 and 36°C, cells harboring either prrf1-K67R, prrf1-L206P, prrf1-L209P or prrf1-I228P grew as well as the cells containing pRRF1W1 on media containing a fermentable carbon source (glucose) and low levels of adenine. As shown in Figure 2A, they turn pink, as expected for respiratory competent ade2 strains growing on low adenine-containing medium (45). However, cells deleted for RRF1 (upper right corner of Fig. 2A, rrf1 rho+ + pRS415-ADHp) and cells harboring prrf1-L209P grown at 36°C (lower middle section of plate 3 of Fig. 2A) were white. This suggested that expression of rrf1-L209P led to a respiratory deficiency at 36°C but not at 30°C. Supporting this conclusion, cells containing prrf1-L209P did not grow on glycerol plates at 36°C (Fig. 2B, plate 3, lower middle section) but grew at lower temperatures (plates 1 and 2). The control strain, rrf1 rho+ + pRS415-ADHp, did not grow on glycerol at any of the temperatures tested, confirming the requirement of RRF1 for growth on glycerol (Fig. 2B, upper right section of plates 1–3).
In contrast, plasmids carrying mutated copies of RRF1 [prrf1-K67R (upper left section of the plates, Fig. 2B), prrf1-L206E (lower left section) and prrf1-I228P (lower right section of the plates)] complemented the null disruption of RRF1 at 24, 30 and 36°C. These all grew on glycerol-containing media (Fig. 2B) and turned pink on glucose plates at each temperature, indicating their respiratory competence.
A strain carrying prrf1-L209P stops mitochondrial but not cytoplasmic major protein synthesis at the non-permissive temperature
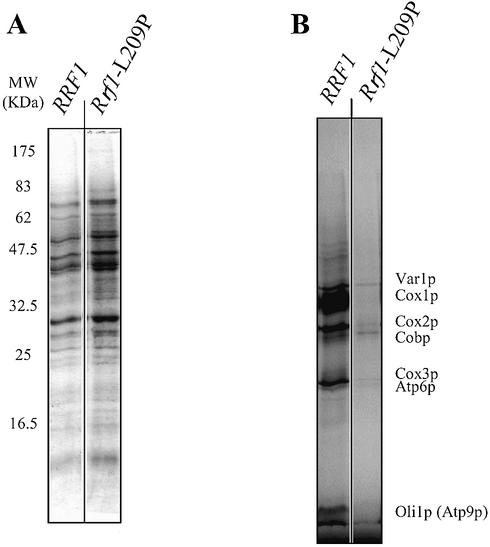
The effect of rrf1-L209P mutation on mitochondrial translation activity was examined by in vivo labeling of mitochondrial protein with [35S]methionine after inhibiting cytoplasmic protein synthesis with cycloheximide (53). After [35S]methionine incorporation at 37°C, the whole cell extract was subjected to gel electrophoresis. Because cycloheximide inhibits cytosolic translation, only proteins synthesized in mitochondria are labeled (Fig. 4B). At 37°C, the total proteins shown by Coomassie blue staining (Fig. 4A) of the extracts of cells carrying wild-type RRF1 are very similar to those of cells carrying rrf1-L209P. On the other hand, a striking difference was observed between cells with wild-type and mutant Rrf1p; the mutant showed a very low level of [35S]methionine incorporation into seven mitochondrial proteins, as identified in Figure 4B. Autoradiography shows that mitochondrial translation activity at 37°C is at least 90% less in cells expressing rrf1-L209P compared to wild-type cells (Fig. 4B). At the semi-permissive temperature (30°C) similar but less inhibition of synthesis of three of these proteins was observed (data not shown).
Figure 4.
Cells with mutant Rrf1p (L209P) synthesize much less mitochondrial protein than those with wild-type Rrf1p. Haploid Δrrf/rho+ cells expressing wild-type Rrf1p (ET2 + pRRF1W1) and thermosensitive Rrf1p (ET2 + prrf1-L209P) were grown overnight at 30°C in minimal glucose medium lacking methionine until mid-exponential phase. Cells were then labeled with [35S]methionine for 80 min in the presence of cycloheximide at 37°C. Whole cell extracts (∼0.5 OD600 units) were separated on a 12% polyacrylamide gel. The gel was Coomassie stained, dried and exposed to X-ray film. (A) and (B) are Coomassie staining and autoradiography, respectively. The mitochondrial proteins revealed by autoradiography were identified as Cox1p, Cox2p, Cox3p, Cobp (cytochrome b), Atp6p and Oli1p (Atp9p) (subunits 6 and 9 of the F0F1-ATPase, respectively) and Var1p (variant ribosomal protein). The positions of the molecular weight markers are indicated on the left (in kDa).
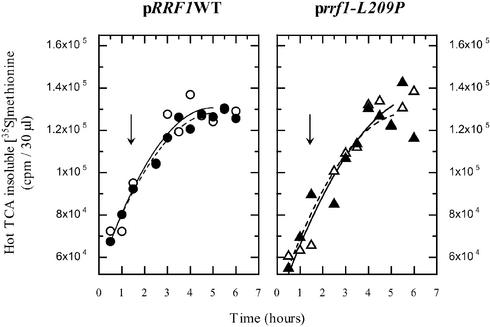
In contrast, cytoplasmic translation activity is not appreciably affected in the rrf1-L209P mutant, as shown by the measure of total [35S]methionine incorporation at both permissive and non-permissive temperatures. At different time points, aliquots of cells were taken and the corresponding incorporated radioactivity was measured (Fig. 5). This is consistent with Figure 2A, where cells with rrf1-L209P still grew on glucose at 37°C, although at a somewhat reduced rate.
Figure 5.
Incorporation of [35S]methionine into total proteins by haploid cells expressing thermosensitive Rrf1p at a permissive (29°C) and a non-permissive temperature (37°C) are similar. Translation activities of haploid Δrrf1 rho+ cells expressing wild-type Rrf1p (ET2 + pRRF1W1, left panel) and thermosensitive Rrf1p (ET2 + prrf1-L209P, right panel) were examined. Overnight cultures in glucose were diluted to 0.05 OD540 and [35S]methionine was added. Cells were incubated at 29°C until the OD540 reached 0.45. Then (indicated by arrow), one sample was exposed to 37°C (closed circles or triangles and dot line) while the other was kept at 29°C (open circles or triangles and solid line). The incorporation of radioactive methionine into the hot TCA-insoluble fraction after the temperature shift is shown.
DISCUSSION
Kanai et al. (34) identified a gene named FIL1 (factor for isocitrate lyase depression). This gene, tentatively renamed here RRF1, has sequence similarity to E.coli RRF and the putative N-terminal mitochondrial targeting sequence. As shown in the present paper and stated by Kanai et al., the null RRF1 mutants are viable but cannot grow on a non-fermentable carbon source. Kanai et al. further showed that cytochrome c oxidase was severely reduced in the null RRF1 mutants.
Isocitrate lyase is a key enzyme in the glyoxylate cycle that plays a vital role in producing succinate for mitochondria. When yeast are placed on a non-fermentable carbon source, a message from functional mitochondria is transmitted to the nucleus to initiate the synthesis of the enzyme(s) necessary for succinate production for mitochondria. Since functional mitochondria are required to communicate non-availability of fermentable sugars to the nucleus (54), it is understandable that FIL1 turned out to code for yeast RRF because, as shown in this paper, RRF1 is essential for the function of mitochondria.
Interestingly, of the many genes necessary for mitochondrial maintenance, only mitochondrial RRF was identified as causing the phenotype, derepression of isocitrate lyase. Theoretically, any gene specifically required for the maintenance of mitochondria should exhibit the same phenotype and can be ‘FIL1’. Since Kanai et al. did not indicate whether or not they identified another mitochondrial maintenance gene as ‘FIL1’, it would be of interest to examine whether other genes involved in mitochondrial protein synthesis could become ‘FIL1’. This may reveal a possible second role of RRF other than as a protein synthesis factor in mitochondria.
In this paper, we propose that Rrf1p, the yeast homolog of bacterial RRF, functions in mitochondrial protein synthesis in a manner similar to bacterial RRF. Mitochondrial localization of Rrf1p is supported by four observations. (i) Immuno chemical studies revealed Rrf1p in isolated mitochondria but not in post-mitochondrial supernatants. (ii) Rrf1p could only be detected in situ in mitochondria by immunofluorescence. (iii) The molecular mass of Rrf1p detected in yeast crude extract and in yeast isolated mitochondria is 23 kDa, a size similar to that predicted from the cDNA sequence, assuming that Rrf1p undergoes maturation in mitochondria. (iv) The N-terminal sequence of the protein deduced from the cDNA sequence is identified as a mitochondrial targeting signal (55).
The following observations also support the above proposal regarding the function of Rrf1p. (i) Deletion of RRF1 yielded a phenotype (rho–) very similar to those exhibited by deleting other mitochondrial proteins needed for mitochondrial protein synthesis [e.g. threonine and tryptophan tRNA synthetases, mitochondrial EF-G and mitochondrial ribosomal protein (12)]. Our results indicate that RRF1 encodes a component essential for yeast mitochondrial DNA maintenance as observed by Myers et al. (12). (ii) The Δrrf1 strain expressing Rrf1p-L209P exhibited a respiratory deficient phenotype at the non-permissive temperature (37°C) because this strain did not grow at 37°C. However, it grew at the lower culture temperature (Fig. 2B). (iii) Mitochondrial protein synthesis was markedly reduced at the non-permissive temperature in the Rrf1p-L209P strain but not in the wild-type strain. No significant inhibition of cytoplasmic protein synthesis was observed with the mutant strain (Figs 4 and 5).
The structural biology of Rrf1p also supports the above proposal. First, Rrf1p has sequence homology with E.coli RRF. Both proteins are of similar size, sharing 23% identical residues (34). The crystal structure of RRF was first solved by us (T.maritima) (56) and confirmed by others with RRF from other organisms (57–59). We have also obtained the solution structure of Aquifex aeolicus RRF (60) using a novel NMR technique. Based on the crystal structure of T.maritima RRF (accession no. 1DD5), a 3-dimensional structure of Rrf1p was built using an homology modeling approach. The modeled Rrf1p structure as shown in Figure 6, just like other RRFs, is remarkably similar to that of tRNA (56). It is important to recall that mammalian mitochondrial tRNA has no or a small D loop (61). The modeled yeast RRF structure does contain the portion corresponding to the D loop, shown in green.
Figure 6.
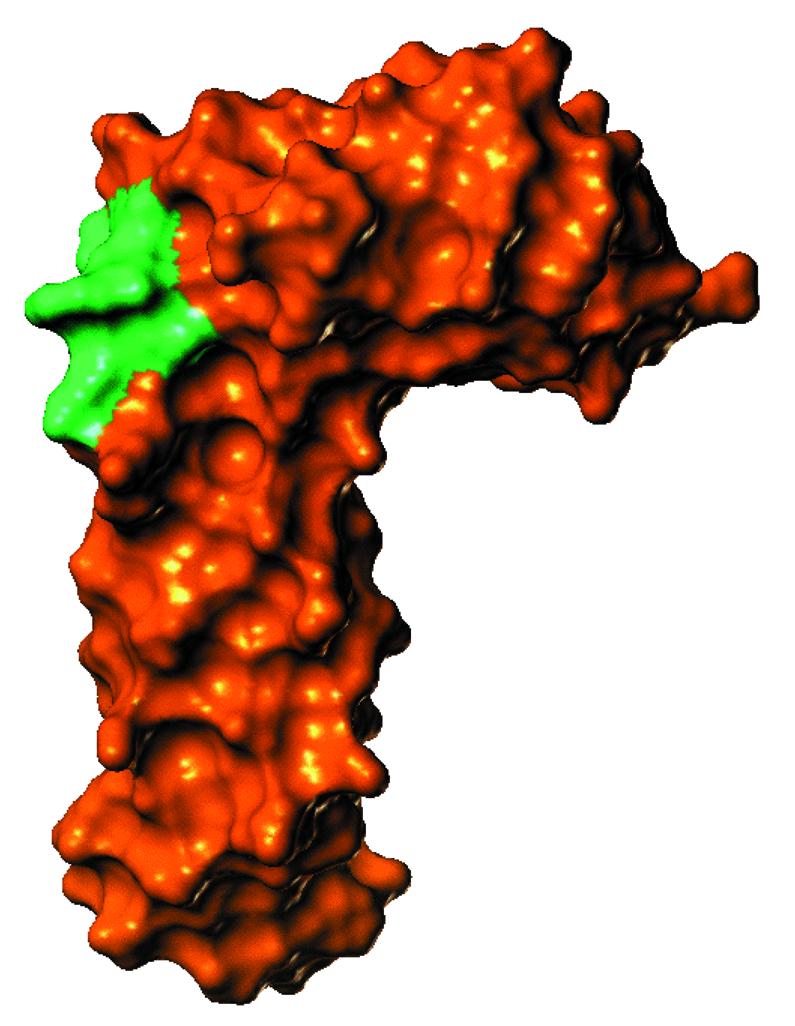
Predicted surface of Rrf1p. A Connolly surface was calculated as described in Materials and Methods for the Rrf1p protein using a theoretical sphere of 2.4 Å. The green color indicates the portion corresponding to the D loop of tRNA.
Second, the mutation rrf1-L209P resulted in temperature-sensitive yeast on a non-fermentable carbon source. This is due to temperature-sensitive protein synthesis in mitochondria (Fig. 4). The corresponding mutation in E.coli RRF, L163P, also resulted in a temperature-sensitive E.coli (21). This finding supports the notion that Rrf1p has a structure very similar to that of E.coli RRF, since changing Leu209 to Pro influences the structure of both proteins in a similar way. In addition, although the function and cytological localization of the human RRF homolog remain obscure, it has been cloned using its sequence similarity to E.coli RRF and has been suggested to be a mitochondrial enzyme (14).
Our proposal can be widened to state that all eukaryotic RRF homologs localize in organelles containing ribosomes and function as a ribosome recycling factor. In our previous paper, we showed that mature RRFHCP (the RRF homolog in chloroplasts) was found in the chloroplasts of spinach (32). The chloroplast RRF homolog is very similar to Rrf1p in the following aspects. (i) The molecular mass is 26.5 kDa. (ii) A larger precursor with an N-terminal signal sequence guides the protein to chloroplasts. (iii) RRFHCP was exclusively localized to chloroplasts and does not exist in the cytoplasm. (iv) The fact that RRFHCP appears to compete with E.coli RRF in E.coli strongly suggests both functional and structural homology between these proteins.
As stated above, Rrf1p appears to function in mitochondria as RRF does in E.coli. However, we should hasten to point out that there may be important functional and structural differences between Rrf1p and bacterial RRF. For example, the sequence identity between bacterial RRF and the eukaryotic homolog is rather low. In addition, as shown in Figure 2B, rrf1-K67R, rrf1-L206E and rrf1-I228P did not yield a temperature-sensitive phenotype despite the fact that these mutations resulted in a temperature-sensitive bacterial RRF. Clearly, subtle but distinct differences in the structure of Rrf1p and bacterial RRF exist. In addition, the protein synthesis machinery of mitochondria is not exactly the same as that in bacteria (2,5). This may be related to the evolution and diversification of mitochondrial genomes (62). Different ribosomal proteins are present in mitochondrial ribosomes compared to bacterial ribosomes (2,11,63,64). These considerations lead us to believe that enough structural and functional differences may exist between eukaryotic mitochondrial and prokaryotic RRF to make it possible to devise antibacterial agents targeted against bacterial RRF. Finally, our present results confirm the notion that the disassembly of post-termination complexes in the cytoplasm of eukaryotes must take place under the control of factors other than RRF homologs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs D. Roof, D. Gordon, D. Pain and A. Dancis of the University of Pennsylvania for plasmids, yeast strains, antibodies and their precious advice. We thank Dr R. Wek of Indiana University School of Medicine for kind advice and supply of mutant yeast strains during the initial phase of this study. We are also grateful to Dr James Kocsis of Thomas Jefferson University and to Dr Michael Kiel of the University of Pennsylvania for their linguistic as well as scientific help. This work was supported by NIH grant GH60429 (to A.K.) and by the Nippon Paint Research Fund (to H.K.).
REFERENCES
- 1.Suzuki T., Terasaki,M., Takemoto-Hori,C., Hanada,T., Ueda,T., Wada,A. and Watanabe,K. (2001) Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J. Biol. Chem., 11, 33181–33195. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien T.W., Liu,J., Sylvester,J.E., Mougey,E.B., Fischel-Ghodsian,N., Thiede,B., Wittmann-Liebold,B. and Graack,H.R. (2000) Mammalian mitochondrial ribosomal proteins (4). Amino acid sequencing, characterization and identification of corresponding gene sequences. J. Biol. Chem., 275, 18153–18159. [DOI] [PubMed] [Google Scholar]
- 3.Daum G., Bohni,P.C. and Schatz,G. (1982) Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem., 257, 13028–13033. [PubMed] [Google Scholar]
- 4.Landrieu I., Vandenbol,M., Hartlein,M. and Portetelle,D. (1997) Mitochondrial asparaginyl-tRNA synthetase is encoded by the yeast nuclear gene YCR24c. Eur. J. Biochem., 243, 268–273. [DOI] [PubMed] [Google Scholar]
- 5.Fox T.D. (1996) Genetics of mitochondrial translation. In Hershey,J.W.B., Mathews,M.B. and Sonnenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 6.Natsoulis G., Hilger,F. and Fink,G.R. (1986) The HTS1 gene encodes both the cytoplasmic and mitochondrial histidine tRNA synthetases of S. cerevisiae. Cell, 46, 235–243. [DOI] [PubMed] [Google Scholar]
- 7.Chatton B., Walter,P., Ebel,J.P., Lacroute,F. and Fasiolo,F. (1988) The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J. Biol. Chem., 263, 52–57. [PubMed] [Google Scholar]
- 8.Turner R.J., Lovato,M. and Schimmel,P. (2000) One of the two genes encoding glycyl-tRNA synthetase in Saccharomyces cerevisiae provides mitochondrial and cytoplasmic functions. J. Biol. Chem., 275, 27681–27688. [DOI] [PubMed] [Google Scholar]
- 9.Pel H.J. and Grivell,L.A. (1994) Protein synthesis in mitochondria. Mol. Biol. Rep., 19, 183–194. [DOI] [PubMed] [Google Scholar]
- 10.Osawa S., Jukes,T.H., Watanabe,K. and Muto,A. (1992) Recent evidence for evolution of the genetic code. Microbiol. Rev., 56, 229–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graack H.R. and Wittmann-Liebold,B. (1998) Mitochondrial ribosomal proteins (MRPs) of yeast. Biochem. J., 329, 433–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers A.M., Pape,L.K. and Tzagoloff,A. (1985) Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J., 4, 2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagata S., Tsunetsugu-Yokota,Y., Naito,A. and Kaziro,Y. (1983) Molecular cloning and sequence determination of the nuclear gene coding for mitochondrial elongation factor Tu of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 80, 6192–6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y.L. and Spremulli,L.L. (1998) Identification and cloning of human mitochondrial translational release factor 1 and the ribosome recycling factor. Biochim. Biophys. Acta Gene Struct. Expr., 1443, 245–250. [DOI] [PubMed] [Google Scholar]
- 15.Vambutas A., Ackerman,S.H. and Tzagoloff,A. (1991) Mitochondrial translational-initiation and elongation factors in Saccharomyces cerevisiae. Eur. J. Biochem., 201, 643–652. [DOI] [PubMed] [Google Scholar]
- 16.Pel H.J., Maat,C., Rep,M. and Grivell,L.A. (1992) The yeast nuclear gene MRF1 encodes a mitochondrial peptide chain release factor and cures several mitochondrial RNA splicing defects. Nucleic Acids Res., 20, 6339–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenthal L.P. and Bodley,J.W. (1987) Purification and characterization of Saccharomyces cerevisiae mitochondrial elongation factor Tu. J. Biol. Chem., 262, 10955–10959. [PubMed] [Google Scholar]
- 18.Dekker P.J., Papadopoulou,B. and Grivell,L.A. (1993) In-vitro translation of mitochondrial mRNAs by yeast mitochondrial ribosomes is hampered by the lack of start-codon recognition. Curr. Genet., 23, 22–27. [DOI] [PubMed] [Google Scholar]
- 19.Hirashima A. and Kaji,A. (1970) Factor dependent breakdown of polysomes. Biochem. Biophys. Res. Commun., 41, 877–883. [DOI] [PubMed] [Google Scholar]
- 20.Hirashima A. and Kaji,A. (1972) Purification and properties of ribosome-releasing factor. Biochemistry, 11, 4037–4044. [DOI] [PubMed] [Google Scholar]
- 21.Janosi L., Mottagui-Tabar,S., Isaksson,L.A., Sekine,Y., Ohtsubo,E., Zhang,S., Goon,S., Nelken,S., Shuda,M. and Kaji,A. (1998) Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J., 17, 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inokuchi Y., Hirashima,A., Sekine,Y., Janosi,L. and Kaji,A. (2000) Role of ribosome recycling factor (RRF) in translational coupling. EMBO J., 19, 3788–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janosi L., Ricker,R. and Kaji,A. (1996) Dual functions of ribosome recycling factor in protein biosynthesis: disassembling the termination complex and preventing translational errors. Biochimie, 78, 959–969. [DOI] [PubMed] [Google Scholar]
- 24.Lancaster L., Kiel,M.C., Kaji,A. and Noller,H.F. (2002) Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell, 111, 129. [DOI] [PubMed] [Google Scholar]
- 25.Hirokawa G., Kiel,M.C., Muto,A., Kawai,G., Igarashi,K., Kaji,H. and Kaji,A. (2002) Binding of ribosome recycling factor to ribosomes—comparison with tRNA. J. Biol. Chem., 277, 35847–35852. [DOI] [PubMed] [Google Scholar]
- 26.Hirokawa G., Kiel,M.C., Muto,A., Selmer,M., Raj,V.S., Liljas,A., Igarashi,K., Kaji,H. and Kaji,A. (2002) Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J., 21, 2272–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noller H.F., Yusupov,M.M., Yusupova,G.Z., Baucom,A. and Cate,J.H. (2002) Translocation of tRNA during protein synthesis. FEBS Lett., 514, 11–16. [DOI] [PubMed] [Google Scholar]
- 28.Kaji A., Kiel,M.C., Hirokawa,G., Muto,A., Inokuchi,Y. and Kaji,H. (2001) The fourth step of protein synthesis: disassembly of the post-termination complex is catalyzed by elongation factor G and ribosome recycling factor, RRF, a near perfect mimic of tRNA. Cold Spring Harb. Symp. Quant. Biol., 66, 515–529. [DOI] [PubMed] [Google Scholar]
- 29.Kaji A., Teyssier,E. and Hirokawa,G. (1998) Disassembly of the post-termination complex and reduction of translational error by ribosome recycling factor (RRF)—a possible new target for antibacterial agents. Biochem. Biophys. Res. Commun., 250, 1–4. [DOI] [PubMed] [Google Scholar]
- 30.Janosi L., Hara,H., Zhang,S. and Kaji,A. (1996) Ribosome recycling by ribosome recycling factor (RRF)—an important but overlooked step of protein biosynthesis. Adv. Biophys., 32, 121–201. [DOI] [PubMed] [Google Scholar]
- 31.Kaji A. and Hirokawa,G. (2000) Ribosome recycling factor: an essential factor for protein synthesis. In Garret,R.A., Douthwaite,S.R., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press, Washington, DC, pp. 527–539. [Google Scholar]
- 32.Rolland N., Janosi,L., Block,M.A., Shuda,A., Teyssier,E., Miege,C., Cheniclet,C., Carde,J., Kaji,A. and Joyard,J. (1999) Plant ribosome recycling factor homologue is a chloroplastic protein and is bactericidal in Escherichia coli carrying temperature-sensitive ribosome recycling factor. Proc. Natl Acad. Sci. USA, 96, 5464–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi K. and Subramanian,A.R. (2000) The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J. Biol. Chem., 275, 28466–28482. [DOI] [PubMed] [Google Scholar]
- 34.Kanai T., Takeshita,S., Atomi,H., Umemura,K., Ueda,M. and Tanaka,A. (1998) A regulatory factor, Fil1p, involved in derepression of the isocitrate lyase gene in Saccharomyces cerevisiae—a possible mitochondrial protein necessary for protein synthesis in mitochondria. Eur. J. Biochem., 256, 212–220. [DOI] [PubMed] [Google Scholar]
- 35.Fox T.D., Folley,L.S., Mulero,J.J., McMullin,T.W., Thorsness,P.E., Hedin,L.O. and Costanzo,M.C. (1991) Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol., 194, 149–165. [DOI] [PubMed] [Google Scholar]
- 36.Sprague G.F., Jr (1991) Assay of yeast mating reaction. Methods Enzymol., 194, 77–93. [DOI] [PubMed] [Google Scholar]
- 37.Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- 38.Baudin A., Ozier-Kalogeropoulos,O., Denouel,A., Lacroute,F. and Cullin,C. (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1990) Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York, NY. [Google Scholar]
- 41.Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 42.Wach A. (1996) PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast, 12, 259–265. [DOI] [PubMed] [Google Scholar]
- 43.Wach A., Brachat,A., Alberti-Segui,C., Rebischung,C. and Philippsen,P. (1997) Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast, 13, 1065–1075. [DOI] [PubMed] [Google Scholar]
- 44.Smirnov M.N., Smirnov,V.N., Budowsky,E.I., Inge-Vechtomov,S.G. and Serebrjakov,N.G. (1967) Red pigment of adenine-deficient yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun., 27, 299–304. [DOI] [PubMed] [Google Scholar]
- 45.Fisher C.R. (1969) Enzymology of the pigmented adenine-requiring mutants of Saccharomyces and Schizosaccharomyces. Biochem. Biophys. Res. Commun., 34, 306–310. [DOI] [PubMed] [Google Scholar]
- 46.Deng W.P. and Nickoloff,J.A. (1992) Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem., 200, 81–88. [DOI] [PubMed] [Google Scholar]
- 47.Boeke J.D., LaCroute,F. and Fink,G.R. (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet., 197, 345–346. [DOI] [PubMed] [Google Scholar]
- 48.Burke D. (2000) Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 49.Kolodziej P.A. and Young,R.A. (1991) Epitope tagging and protein surveillance. Methods Enzymol., 194, 508–519. [DOI] [PubMed] [Google Scholar]
- 50.van Dyck L., Neupert,W. and Langer,T. (1998) The ATP-dependent PIM1 protease is required for the expression of intron-containing genes in mitochondria. Genes Dev., 12, 1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yaffe M.P. and Schatz,G. (1984) Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl Acad. Sci. USA, 81, 4819–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jameson B.A. (1989) Modelling in peptide design. Nature, 341, 465–466. [DOI] [PubMed] [Google Scholar]
- 53.Clark-Walker G.D. and Linnane,A.W. (1966) In vivo differentiation of yeast cytoplasmic and mitochondrial protein synthesis with antibiotics. Biochem. Biophys. Res. Commun., 25, 8–13. [DOI] [PubMed] [Google Scholar]
- 54.Epstein C.B., Waddle,J.A., Hale,W.IV., Dave,V., Thornton,J., Macatee,T.L., Garner,H.R. and Butow,R.A. (2001) Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell, 12, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakai K. and Kanehisa,M. (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics, 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Selmer M., Al-Karadaghi,S., Hirokawa,G., Kaji,A. and Liljas,A. (1999) Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science, 286, 2349–2352. [DOI] [PubMed] [Google Scholar]
- 57.Kim K.K., Min,K. and Suh,S.W. (2000) Crystal structure of the ribosome recycling factor from Escherichia coli. EMBO J., 19, 2362–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakano H., Yoshida,T., Uchiyama,S., Kawachi,M., Matsuo,H., Kato,T., Ohshima,A., Yamaichi,Y., Honda,T., Kato,H. et al. (2003) Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium. J. Biol. Chem., 278, 3427–3436. [DOI] [PubMed] [Google Scholar]
- 59.Toyoda T., Tin,O.F., Ito,K., Fujiwara,T., Kumasaka,T., Yamamoto,M., Garber,M.B. and Nakamura,Y. (2000) Crystal structure combined with genetic analysis of the Thermus thermophilus ribosome recycling factor shows that a flexible hinge may act as a functional switch. RNA, 6, 1432–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida T., Uchiyama,S., Nakano,H., Kashimori,H., Kijima,H., Ohshima,T., Saihara,Y., Ishino,T., Shimahara,H., Yoshida,T. et al. (2001) Solution structure of the ribosome recycling factor from Aquifex aeolicus. Biochemistry, 40, 2387–2396. [DOI] [PubMed] [Google Scholar]
- 61.Helm M., Brule,H., Friede,D., Giege,R., Putz,D. and Florentz,C. (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA, 6, 1356–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gray M., Burger G. and Lang B.F. (1999) Mitochondrial evolution. Science, 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- 63.O’Brien T.W., Fiesler,S.E., Denslow,N.D., Thiede,B., Wittmann-Liebold,B., Mougey,E.B., Sylvester,J.E. and Graack,H.-R. (1999) Mammalian mitochondrial ribosomal proteins (2). Amino acid sequencing, characterization and identification of corresponding gene sequences. J. Biol. Chem., 274, 36043–36051. [DOI] [PubMed] [Google Scholar]
- 64.Bui D.M., Jarosch,E. and Schweyen,R.J. (1997) The yeast ORF YDL202w codes for the mitochondrial ribosomal protein YmL11. Curr. Genet., 31, 396–400. [DOI] [PubMed] [Google Scholar]
- 65.Hannig E.M., Williams,N.P., Wek,R.C. and Hinnebusch,A.G. (1990) The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics, 126, 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alani E., Cao,L. and Kleckner,N. (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics, 116, 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]