Abstract
Replication protein A (RPA) is a heterotrimeric single-stranded DNA- (ssDNA) binding protein that can form a complex with the xeroderma pigmentosum group A protein (XPA). This complex can preferentially recognize UV-damaged DNA over undamaged DNA and has been implicated in the stabilization of open complex formation during nucleotide excision repair. In this report, nuclear magnetic resonance (NMR) spectroscopy was used to investigate the interaction between a fragment of the 70 kDa subunit of human RPA, residues 1–326 (hRPA701–326), and a fragment of the human XPA protein, residues 98–219 (XPA-MBD). Intensity changes were observed for amide resonances in the 1H–15N correlation spectrum of uniformly 15N-labeled hRPA701–326 after the addition of unlabeled XPA-MBD. The intensity changes observed were restricted to an ssDNA-binding domain that is between residues 183 and 296 of the hRPA701–326 fragment. The hRPA701–326 residues with the largest resonance intensity reductions were mapped onto the structure of the ssDNA-binding domain to identify the binding surface with XPA-MBD. The XPA-MBD-binding surface showed significant overlap with an ssDNA-binding surface that was previously identified using NMR spectroscopy and X-ray crystallography. Overlapping XPA-MBD- and ssDNA-binding sites on hRPA701–326 suggests that a competitive binding mechanism mediates the formation of the RPA–XPA complex. To determine whether a ternary complex could form between hRPA701–326, XPA-MBD and ssDNA, a 1H–15N correlation spectrum was acquired for uniformly 15N-labeled hRPA701–326 after the simultaneous addition of unlabeled XPA-MBD and ssDNA. In this experiment, the same chemical shift perturbations were observed for hRPA701–326 in the presence of XPA-MBD and ssDNA as was previously observed in the presence of ssDNA alone. The ability of ssDNA to compete with XPA-MBD for an overlapping binding site on hRPA701–326 suggests that any complex formation between RPA and XPA that involves the interaction between XPA-MBD and hRPA701–326 may be modulated by ssDNA.
INTRODUCTION
Nucleotide excision repair (NER) is a DNA repair mechanism specific for the bulky, helix-distorting adducts induced by exposing DNA to UV radiation or polycyclic aromatic hydrocarbons (1–7). The mechanism of NER requires the concerted action of at least 14 proteins that form a complex called the excision nuclease (3,6,8,9). One essential component of the excision nuclease is replication protein A (RPA) (10). RPA is a heterotrimeric, single-stranded DNA- (ssDNA) binding protein that is required for NER as well as replication and recombination (11). RPA forms a complex with the xeroderma pigmentosum group A protein (XPA), and this complex is thought to extend and stabilize the open complex that is formed around the site of damage by the ATP-dependent helicase activity of TFIIH (6,7,12). During the stabilization and extension of the open complex, it appears that XPA preferentially binds to the damaged strand and RPA binds to the undamaged strand (13). There is also evidence suggesting a role for the RPA–XPA complex in the damage recognition step of NER (9,14).
The RPA heterotrimer is composed of 70, 32 and 14 kDa subunits whose activities have been characterized extensively (11,15–19). XPA binding activity has been isolated to both the 70 and 32 kDa subunits of human RPA, referred to hereafter as hRPA70 and hRPA32, respectively (18,20,21). Previous studies of hRPA70 fragments localized the binding site for full-length XPA to hRPA70 residues 236–382 and 169–296 (18,21). Because hRPA70 residues 183–440 contain tandem ssDNA-binding domains, the XPA-binding site must be either in the middle of the tandem ssDNA-binding domains or in the more N-terminal ssDNA-binding domain (11,18,22).
Using a technique called chemical shift mapping, a previous investigation of the binding between an N-terminal fragment of hRPA70 (1–326; hRPA701–326) and a fragment of XPA containing the minimal DNA-binding domain residues (98–219; XPA-MBD) defined an hRPA701–326-binding surface on XPA-MBD (23). HRPA701–326 was originally designed as a stable fragment of hRPA70 capable of binding ssDNA. This fragment contains an N-terminal domain, residues 1–105 (NTD), that is important for hRPA70–protein interactions (11,24–26). The NTD is connected by a flexible linker to one of the tandem ssDNA-binding domains, residues 183–296 (SSB1), present in full-length hRPA70 (15,27). XPA-MBD was previously identified as the minimal fragment of XPA sufficient for damaged DNA binding (18,28).
Chemical shift mapping is well suited to study weak protein–protein interactions and can provide various levels of information about the structure and dynamics of the protein–protein complexes, depending upon the nature of the system under investigation (29). For systems with less than ideal solubility characteristics, it is often possible to identify binding surfaces or allostery (29). In addition, binding and kinetic constants can be measured for systems with more ideal solubility characteristics (29). In a mapping experiment, the chemical shifts can experience different perturbations: they can shift with or without broadening, they can remain in position and diminish in intensity, or they can remain unaffected. The specific behavior of the chemical shifts depends on the binding affinity and kinetics of the protein–protein complex that is formed.
In the previous nuclear magnetic resonance (NMR) study of the interaction between XPA-MBD and hRPA701–326, the 1H–15N chemical shift changes were monitored for a uniformly 15N-labeled sample of XPA-MBD before and after the addition of unlabeled hRPA701–326 (23). The observed chemical shift changes fell into the category of residue-specific intensity reductions for the amide 1H–15N chemical shifts of XPA-MBD after the addition of unlabeled hRPA701–326 (23). The XPA-MBD residues with the largest intensity reductions were in the zinc-binding core and the loop-rich subdomain, forming a mostly contiguous binding surface (23). This behavior was similar to a binding study of the cell–cell interaction proteins CD2 and CD48 (30). In the CD2–CD48 study, as the protein partner was added to the NMR sample, some resonances diminished in intensity without shifting frequency. For the CD2–CD48 complex, there was also a global decrease in resonance intensity at saturation due to slower molecular tumbling of the complex.
To further a molecular description of the interaction between hRPA70 and XPA, the reverse experiment has been performed. In this report, the amide 1H–15N chemical shifts of a uniformly 15N-labeled sample of hRPA701–326 were monitored before and after the addition of two forms of unlabeled XPA-MBD. In one of the forms of XPA-MBD, paramagnetic Co2+ was substituted for diamagnetic Zn2+ at the divalent metal-binding site (CoXPA-MBD) in an attempt to determine distance restraints for the hRPA701–326–XPA-MBD heterodimer based on paramagnetic pseudocontact chemical shifts induced by Co2+. The observed chemical shift changes for hRPA701–326, after the addition of either form of XPA-MBD, fell into the category of residue-specific intensity reductions for the amide 1H–15N chemical shifts with no significant changes in the 1H–15N chemical shift frequencies. The chemical shift intensity reductions observed for hRPA701–326 were all localized to the SSB1 domain. The residues with the largest intensity reductions were on the protein’s surface and showed significant overlap with the ssDNA-binding surface previously identified using X-ray crystallography and NMR spectroscopy (22,31). The presence of overlapping XPA-MBD and ssDNA-binding surfaces on SSB1 was consistent with our inability to form the ternary complex between hRPA701–326, XPA-MBD and ssDNA, and is discussed in context with the recent demonstration that XPA can inhibit the strand separation activity of RPA (32).
MATERIALS AND METHODS
Production and purification of XPA-MBD and hRPA701–326
XPA-MBD and CoXPA-MBD samples containing Zn2+ and Co2+, respectively, in the C4-type metal-binding domain were prepared as previously described (23,33). The following changes were made to the previously published expression and purification protocols for hRPA701–326: (i) for uniform 15N-labeling of hRPA701–326, standard minimal medium was used for bacterial growth that included 15N-enriched ammonium chloride purchased from CIL (Cambridge, MA); and (ii) a buffer containing 20 mM Tris–HCl pH 7.4, 50 mM KCl, 0.02% sodium azide and 5 mM dithiothreitol (DTT) was used for cell lysis and chromatography (27). This was also the final buffer used for the NMR experiments with XPA-MBD. For binding experiments with CoXPA-MBD, the DTT was replaced with Tris(2-carboxyethyl)phosphine (TCEP) to inhibit Co2+ precipitation. NMR spectroscopy and/or SDS–PAGE were used to verify the purity and stability of the hRPA701–326, XPA-MBD and CoXPA-MBD samples.
Oligonucleotide synthesis
An ssDNA nonamer, dCCAATAAC (d9), was synthesized using an Applied Biosystems DNA synthesizer model 392. After synthesis, d9 was lyophilized overnight to remove ammonia and incubated in 600 µl of 300 mM KCl for 1 h at 55°C. The nonamer was then purified by passage over a G25 desalting column using deionized, nanopure water as the eluent, lyophilized, and resuspended in 50 µl of the same buffer used for the NMR experiments. The final concentration of the purified sample was 10 mM.
NMR experiments
All NMR experiments were performed on Varian spectrometers operating at a 1H resonance frequency of 600 MHz. Two-dimensional, gradient-enhanced, 1H–15N heteronuclear single quantum coherence (HSQC) spectra were acquired on uniformly 15N-labeled hRPA701–326 samples in 90% H2O/10% D2O (34). The digital resolution for the HSQC experiments was 7.8 Hz/point in both the 1H and 15N dimensions, and the acquisition time was 128 ms. The amide 1H and 15N resonance assignments of hRPA701–326 were described previously and utilized in the current experiments (31). The interaction between hRPA701–326 and XPA-MBD was investigated by monitoring changes in the 1H–15N HSQC of a 0.5 mM uniformly 15N-labeled hRPA701–326 sample after a single addition of an equimolar amount of unlabeled XPA-MBD, unlabeled CoXPA-MBD, or a mixture of unlabeled XPA-MBD and d9. Prior to the NMR experiments, the sample volume was reduced from ∼1 ml to 300 µl using a Centricon centrifugal filtration device with a 3000 or 10 000 Da molecular weight cut-off (Millipore; Bedford, MA). All NMR spectra were processed and analyzed using the Felix software from the Accelerys Corporation (Cambridge, MA). Apodization was achieved in 1H and 15N dimensions using a squared sine bell function shifted by 90°. Apodization was followed by zero filling to twice the number of real data points and no linear prediction was used. Amide 1H–15N resonance intensity (peak heights) measurements were made using the Felix program. The intensity measurements were converted to tab-delimited ASCII files, and intensity ratio plots were generated using the KaleidaGraph software from Synergy (Reading, PA).
RESULTS
Resonance intensity changes of hRPA701–326 bound to XPA-MBD
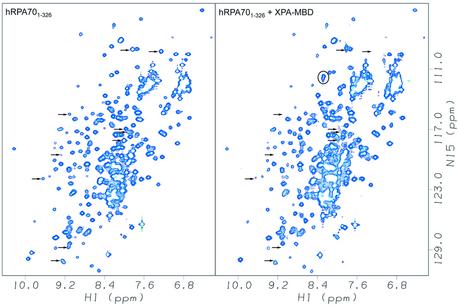
The amide 1H and 15N chemical shifts of a uniformly 15N-labeled sample of hRPA701–326 were monitored before and after the addition of an equimolar amount of unlabeled XPA-MBD. The changes observed for the hRPA701–326 amide 1H and 15N chemical shifts were residue-specific reductions in the resonance intensities, with no significant changes observed for the resonance frequencies. Figure 1 shows the amide region of the 1H–15N HSQC spectrum of hRPA701–326 before (left panel) and after (right panel) the addition of XPA-MBD. Arrows indicate the positions of assigned resonances that had a 2-fold or greater intensity reduction upon the addition of XPA-MBD. Circles indicate the appearance of new resonances upon the addition of XPA-MBD. Because of the poor solubility characteristics of the sample, these new resonances were not assignable. The signal losses observed are indicative of complex formation and result from the formation of either a stable, large molecular weight complex or a complex with an exchange rate between the free and bound forms that is similar to the chemical shift difference (35).
Figure 1.
Amide region of the 1H–15N HSQC spectra of hRPA701–326 on the left, and hRPA701–326 after the addition of XPA-MBD on the right. Arrows show the positions for assigned resonances that had a 2-fold or greater intensity reduction after the addition of XPA-MBD. New resonances that were not assignable are circled. Both spectra were collected at 25°C and at a 1H resonance frequency of 600 MHz.
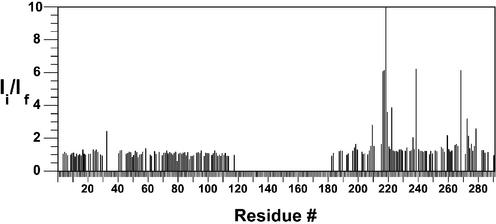
The amide resonance intensities in the 1H–15N HSQC spectrum of hRPA701–326 were measured before and after the addition of unlabeled XPA-MBD. The intensity ratio was determined by dividing the hRPA701–326 resonance intensities before the addition of XPA-MBD by the hRPA701–326 resonance intensities after the addition of XPA-MBD. Figure 2 shows the residue-specific intensity ratios for 88 residues from the NTD and 75 residues from SSB1. Due to spectral overlap in the 1H–15N HSQC spectra, unambiguous resonance intensity measurements were not possible for most of the linker residues. However, the position of these residues remains in the plot for clarity, and an inspection of the overlapped resonances from the linker residues in Figure 1 indicates only minor intensity changes.
Figure 2.
Plot of the hRPA701–326 intensity ratios versus residue number for binding to XPA-MBD. Ii refers to the resonance intensity before the addition of XPA-MBD and If refers to the resonance intensity after the addition of an equimolar amount of XPA-MBD.
The average intensity ratio for the NTD was 1.08 and for SSB1 was 2.67, indicating that XPA-MBD is interacting primarily with SSB1. In our previous NMR study of the interaction between XPA-MBD and an hRPA70 fragment containing the NTD and the flexible linker (1–168: hRPA701–168), no significant changes in the amide resonance intensities were observed in the 1H–15N HSQC spectrum of uniformly 15N-labeled XPA-MBD after the addition of an equimolar amount of unlabeled hRPA701–168 (23). Combining this previous observation with the current results is strong evidence that the interaction between hRPA701–326 and XPA-MBD is localized to SSB1. It is known that the flexible linker permits relatively independent rotational motion of the NTD and SSB1 in the hRPA701–326 fragment (31). The small amide resonance intensity changes observed for the NTD suggests that this is also the case for the complex that is formed between hRPA701–326 and XPA-MBD. If the rotational motion of the NTD is independent, the effective molecular weight for rotational tumbling of the complex between hRPA701–326 and XPA-MBD should be similar to the combined molecular weight of SSB1 and XPA-MBD, which is ∼25 kDa. An effective molecular weight for rotational tumbling of 25 kDa is within the detection range of the 1H–15N HSQC experiment. Because only a few new resonances were observed for the hRPA701–326–XPA-MBD complex and the effective molecular weight of the complex is ∼25 kDa, it is likely that the complex has not reached saturation (30). This is supported by the observation that the line widths for most hRPA701–326 amide 1H and 15N resonances are unaffected by the addition of XPA-MBD (data not shown).
Ten residues in SSB1 had a 2-fold or greater intensity reduction: R210, G217, E218, G219, K220, S223, N239, F269, K273 and T279. Of these, G219 had the largest intensity reduction (76-fold) and is the only residue off the scale in Figure 2. Large changes in amide resonance intensities during protein–protein binding provide evidence for the presence of an interaction (29). In this case, amino acid residues with the largest intensity changes were localized to a particular surface of the SSB1 structure. In some cases of chemical shift mapping, a global perturbation of resonances was found, indicating that an allosteric process occurred during binding and it is possible that even for localized perturbations, the mapping might report secondary structural changes that are remote from the direct site of interaction (29). However, the simplest interpretation of the current result is that the interaction site is localized on the surface of SSB1. From a recent ssDNA binding study with hRPA701–326, large chemical shift changes were observed for SSB1 resonances when hRPA701–326 was bound to ssDNA (31). The largest chemical shift changes observed for the SSB1 interaction with ssDNA included all the residues with the largest intensity changes observed when hRPA701–326 was bound to XPA-MBD, with the exception of N239 (31).
Resonance intensity changes of hRPA701–326 bound to Co2+-substituted XPA-MBD
In a previous NMR study of XPA-MBD, paramagnetic Co2+ was substituted for Zn2+ at the C4 site in a uniformly 15N-labeled sample of XPA-MBD (CoXPA-MBD) (23). Paramagnetic pseudocontact shifts were observed in the 1H–15N HSQC spectrum of CoXPA-MBD. The magnitude of the pseudocontact shifts was used to estimate distance restraints, and the distance restraints were used to refine the solution structure of XPA-MBD (23). A similar experiment was attempted here, where unlabeled CoXPA-MBD was added to uniformly 15N-labeled hRPA701–326, with the expectation that paramagnetic pseudocontact chemical shifts would be observed in the 1H–15N HSQC spectrum of hRPA701–326. However, no paramagnetic pseudocontact shifts were observed. Instead, a similar behavior was observed in the 1H–15N HSQC spectrum of hRPA701–326 when CoXPA-MBD was added as was observed after the addition of XPA-MBD. In general, the addition of CoXPA-MBD induced larger intensity reductions as well as a broader range of intensity reductions for the amide chemical shifts in the 1H–15N HSQC spectrum of hRPA701–326 than was observed after the addition of XPA-MBD.
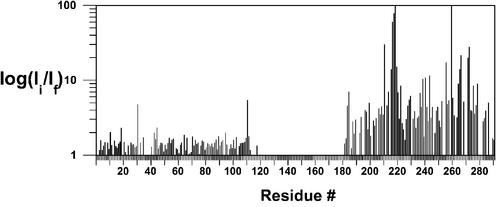
The resonance intensities from the amide region of the 1H–15N HSQC of hRPA701–326 were measured before and after the addition of unlabeled CoXPA-MBD. The intensity ratio was determined by dividing the hRPA701–326 resonance intensities before the addition of CoXPA-MBD by the hRPA701–326 resonance intensities after the addition of CoXPA-MBD. Figure 3 shows a log plot of the residue-specific intensity ratios for 99 residues from the NTD and 86 residues from SSB1. The average intensity ratio for the NTD was 1.5 and for SSB1 was 13.5. The small intensity reduction observed for the NTD is evidence that the linker remains flexible in the hRPA701–326–CoXPA-MBD complex and that the NTD has an average distance from the Co2+ center that is >20 Å (36). Over half of the SSB1 residues have an intensity reduction >2-fold. This is in contrast to the 2-fold intensity reduction observed for only 10 SSB1 residues in the hRPA701–326–XPA-MBD binding experiment. Further, the average intensity reduction for SSB1 is four times the average intensity reduction observed for the interaction with XPA-MBD and the difference in the average intensity change is almost 10-fold between the NTD and SSB1 when hRPA701–326 is bound to CoXPA-MBD. Because CoXPA-MBD contains a paramagnetic center, it is likely that the larger intensity reductions observed for SSB1 are the result of paramagnetic relaxation induced by the Co2+ (36).
Figure 3.
Log plot of the hRPA701–326 intensity ratios versus residue number for binding to CoXPA-MBD. Ii refers to the resonance intensity before the addition of CoXPA-MBD and If refers to the resonance intensity after the addition of an equimolar amount of CoXPA-MBD.
SSB1 residues with a ≥10-fold intensity reduction are T211, R216, G217, E218, G219, K220, L221, S223, N239, Q241, K244, Y256, G260, A265, N266, K267, V272, K273, Y276 and T279. G219 and G260 had the largest intensity reductions of 100- and 400-fold, respectively, and are close to or off the scale of Figure 3. For the interaction between hRPA701–326 and CoXPA-MBD, it is expected that the same regions will be perturbed as was observed for the interaction with XPA-MBD. Indeed, most of the residues with large intensity reductions from the hRPA701–326–XPA-MBD binding study also have large intensity reductions in the hRPA701–326–CoXPA-MBD complex. Exceptions to this trend were R210, with a 3.5-fold intensity reduction, and F269, with a 5-fold intensity reduction. However, both of these residues have intensity reductions that are greater than the average value observed for SSB1 in the hRPA701–326–XPA-MBD binding experiment. SSB1 residues with large intensity reductions that are unique to the hRPA701–326–CoXPA-MBD interaction are T211, R216, Q241, K244, Y256, G260, A265, N266, K267, V272 and Y276. It is reasonable to conclude that the SSB1 residues with large intensity reductions that are unique to the hRPA701–326–CoXPA-MBD interaction are within 20 Å of the Co2+ center (36).
Comparison of the hRPA70 SSB1-binding surface for ssDNA and XPA-MBD
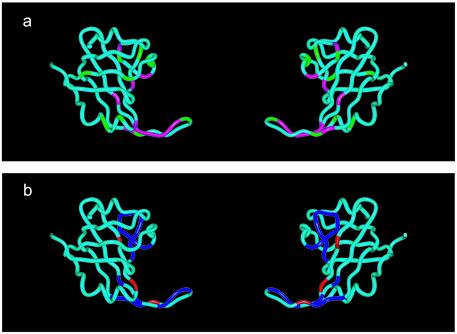
The crystal structure of the tandem ssDNA-binding domains of hRPA70 (hRPA70 residues 183–420) bound to ssDNA was solved recently (22). Figure 4 shows ribbon models of SSB1 made using the coordinates of hRPA70 residues 183–291 provided in the crystal structure (PBD ID: 1JMC) (22). The ssDNA has been deleted from the SSB1 structures shown in Figure 4. The SSB1 structure is shown in the same orientation on the left and rotated 180° around the vertices on the right. In Figure 4a, the SSB1 residues with the largest intensity reductions from the NMR analysis of the hRPA701–326–XPA-MBD complex are colored magenta and the SSB1 residues with the largest intensity reductions unique to the NMR analysis of the hRPA701–326–CoXPA-MBD complex are colored green. In Figure 4b, the SSB1 residues with the largest chemical shift changes in the NMR analysis of three different hRPA701–326–ssDNA complexes are colored blue and the SSB1 residues that are in contact with ssDNA in the crystal structure but were not assignable in the NMR analysis of the hRPA701–326–ssDNA complexes are colored red (22,31). An inspection of Figure 4a and b reveals that the XPA-MBD- and ssDNA-binding sites are not identical. However, very similar regions of the SSB1 structure are affected by the binding of both XPA-MBD and ssDNA.
Figure 4.
Ribbon models for residues 183–291 of SSB1 from hRPA70 adapted from the coordinates in PDB accession file 1jmc (22). (a) SSB1 residues with the largest resonance intensity reductions observed in the 1H–15N HSQC spectrum of hRPA701–326 when bound to XPA-MBD and CoXPA-MBD are colored magenta and green, respectively. (b) SSB1 residues with the largest chemical shift changes in the NMR analysis of three different hRPA701–326–ssDNA complexes are colored blue and SSB1 residues that are in contact with ssDNA in the crystal structure but were not assignable in the NMR analysis of the hRPA701–326–ssDNA complexes are colored red (22,31).
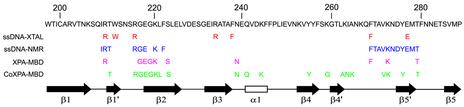
The linear sequence of SSB1 (hRPA70 residues 197–288) is shown in Figure 5, with a diagram of the secondary structure elements observed in the crystal structure for this region illustrated across the bottom (22). The first row under the SSB1 sequence, labeled ssDNA-XTAL and colored red, shows the residues that made direct contact with ssDNA in the crystal structure (22). The second row under the SSB1 sequence, labeled ssDNA-NMR and colored blue, shows residues that had the largest chemical shift changes in the presence of ssDNA (31). The third row under the SSB1 sequence, labeled XPA-MBD and colored magenta, shows residues that had the largest intensity reductions in the presence of XPA-MBD. The fourth row under the SSB1 sequence, labeled CoXPA-MBD and colored green, shows residues that had the largest intensity reductions in the presence of CoXPA-MBD.
Figure 5.
Diagram showing the amino acid sequence and secondary structure of SSB1 residues 197–288. SSB1 residues colored red under the linear sequence made direct contacts with ssDNA in the crystal structure (22). SSB1 residues colored blue under the linear sequence had the largest chemical shift changes when hRPA701–326 was bound to ssDNA (31). SSB1 residues colored magenta under the linear sequence had the largest resonance intensity reductions when hRPA701–326 was bound to XPA-MBD. SSB1 residues colored green under the linear sequence had the largest resonance intensity reductions when hRPA701–326 was bound to CoXPA-MBD.
According to Figures 4 and 5, there is extensive overlap between the regions of the SSB1 structure that are involved in binding both ssDNA and XPA-MBD. The exceptions to the trend were residues Q241, K244, Y256, G260, A265, N266 and K267. These exceptions were only observed in the NMR analysis of the hRPA701–326–CoXPA-MBD complex and may be explained by dipolar relaxation induced by the Co2+, suggesting they are within 20Å of the Co2+ quadrupole but are not part of the binding interface. The presence of overlapping, but non-identical, binding sites suggests one of two potential mechanisms for the interaction of hRPA701–326 with XPA-MBD and ssDNA. Similar binding regions could either interfere with or facilitate the formation of a ternary complex between hRPA701–326, XPA-MBD and ssDNA.
No ternary complex forms between hRPA701–326, XPA-MBD and ssDNA
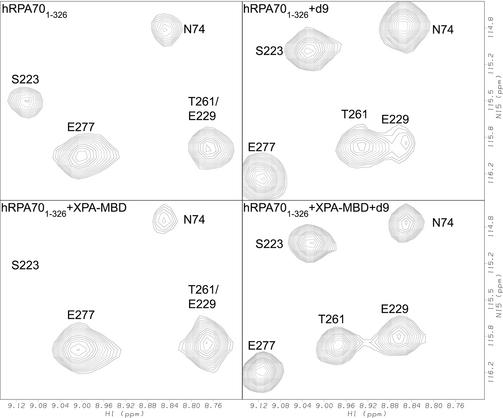
The formation of a ternary complex between hRPA701–326, XPA-MBD and ssDNA was investigated by mixing equimolar amounts of unlabeled XPA-MBD and ssDNA with a 15N-labeled sample of hRPA701–326 followed by monitoring the amide 1H and 15N chemical shifts. Figure 6 shows a selected region from the 1H–15N HSQC spectrum of hRPA701–326 with resonances labeled for residues N74, S223, E277 and T261/E229. The top left panel of Figure 6 shows the spectrum of hRPA701–326 in the absence of XPA-MBD and d9. The top right panel of Figure 6 shows the spectrum of hRPA701–326 after the addition of an equimolar amount of d9. The bottom left panel of Figure 6 shows the spectrum of hRPA701–326 after the addition of an equimolar amount of XPA-MBD. The bottom right panel of Figure 6 shows the spectrum of hRPA701–326 after the addition of an equimolar amount of XPA-MBD and d9. The bottom right panel of Figure 6 clearly demonstrates that it is the spectrum of the hRPA701–326–d9 complex that is recovered when both XPA-MBD and d9 are added to hRPA701–326. Under these conditions, the chemical shift changes observed in the rest of the hRPA701–326 1H–15N HSQC spectrum are identical to those observed in the presence of d9 alone (31). Based on this result, it appears that a ternary complex between hRPA701–326, XPA-MBD and the ssDNA cannot form. The same resonance intensity reductions were not observed for hRPA701–326 residues in the presence of XPA-MBD and d9 as was observed in the presence of XPA-MBD alone. This is further evidence for overlapping XPA-MBD- and ssDNA-binding sites on hRPA701–326. Although it is not detectable in this experiment, XPA-MBD has previously been shown to bind to d9, hence, it is possible that some d9 is bound to XPA-MBD in the mixture of all three compounds (37). Taken together, these results suggest that the affinity that both protein fragments have for ssDNA is greater than their affinity for one another, and are consistent with a recent study showing that XPA did not effect the kon of the RPA heterotrimer binding to a 40-base oligomer of ssDNA (32).
Figure 6.
Selected regions from the 1H–15N HSQC spectra for hRPA701–326. The figure shows the spectrum of hRPA701–326 in the absence of XPA-MBD and d9 (top left), in the presence of an equimolar amount of d9 (top right), in the presence of an equimolar amount of XPA-MBD (bottom left), and in the presence of an equimolar amount of XPA-MBD and d9 (bottom right).
DISCUSSION
Controversy exists over the details of the interaction between RPA and XPA. Two independent groups have demonstrated that 33 amino acids near the C-terminus of the hRPA32 subunit are necessary for the interaction with full-length XPA (18,21). These two groups did not reach a consensus on whether hRPA70 plays a significant role in the interaction with XPA. Here, we have shown that the minimal fragment of XPA necessary for DNA binding has an interaction with SSB1 from hRPA70. Furthermore, it was concluded that the XPA-MBD-binding interface overlaps the ssDNA-binding interface on the surface of SSB1.
Due to the sensitivity of chemical shift mapping to weak binding, it is possible that the observed interaction between hRPA701–326 and XPA-MBD is too weak to be physiologically relevant. It is important to emphasize that the RPA and XPA fragments used in this study were selected to investigate the structural details of just one of the many interactions between the two full-length proteins. In the cell, RPA and XPA will bind with some intrinsic affinity that will include the contributions from the different interactions that have been identified. Previous studies have described various details about the binding affinities between RPA, XPA and ssDNA (12,18,20,28). For instance, surface plasmon resonance was used to investigate the binding between full-length RPA and XPA (20). In this study, the binding was too fast to reliably determine rate constants, but a KD of 1.9 × 10–8 M was determined from a Scatchard analysis of the steady-state binding levels (20). This study also showed that the XPA interaction with RPA70 was localized to the XPA-MBD fragment. In another study, enzyme-linked immunosorbent assays (ELISAs) showed that ∼80% of the binding affinity between RPA and XPA is with the hRPA32 subunit and ∼20% with the hRPA70 subunit (18).
The affinity of RPA and XPA for ssDNA has also been investigated (12,18). The hRPA701–326 fragment had an apparent KA for a 30-base thymine oligomer, dT30, of 4.2 × 107 M–1. This value was 50- to 100-fold lower than the apparent KA of hRPA70 for dT30 (18). The KD of XPA for a short (octamer) piece of ssDNA was 1.3 × 10–8 M, and this binding affinity appears to be localized to the XPA-MBD fragment (12,28). Based on these data, the KA for the interaction between hRPA701–326 and XPA-MBD should be of the order of 1 × 107 M–1, so it is unclear why the chemical shift changes observed for 15N-labeled hRPA701–326 did not show more evidence for saturated binding. It is likely that the fragments used in this study, especially hRPA701–326, are missing amino acid residues that form part of the binding interface. However, the observation that ssDNA can compete with XPA-MBD for an overlapping binding site on hRPA701–326 is consistent with the published binding constants.
The presence of overlapping XPA-MBD- and ssDNA-binding sites in hRPA701–326 has significant implications for the role of the RPA–XPA complex in NER. Recent studies on the function of XPA found that it stabilizes the duplex DNA structure around the damaged site by inhibiting the strand separation activity of RPA, suggesting some transient interference with the ability of RPA to bind ssDNA (32). According to the data presented herein, this interference may result from the interaction between XPA-MBD and SSB1 and is most easily explained if the RPA–XPA complex is formed prior to open complex formation. A recent study demonstrated strand-specific binding by XPA and RPA to a damaged duplex DNA (13). According to this study, XPA binds to the damaged strand and RPA binds to the undamaged strand. Based on the evidence presented in this report, it is likely that the RPA–XPA complex undergoes a structural rearrangement to accommodate both SSB1 binding to ssDNA on the undamaged strand, enhancing the stability of open complex formation, and the binding of XPA-MBD to the damaged strand (13). This hypothesis is supported by the fact that complementary structural studies of ssDNA and RPA binding to XPA-MBD also showed significant overlap in the binding interfaces (23).
Acknowledgments
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Pamela Vise and Lee Fortunato for helpful discussions during the preparation of this manuscript, and Nancy Isern for the preparation of d9. This work was performed under the auspices of the US Department of Energy (Contract DE-AC06-76RLO1830) and was supported by the Department of Energy Office of Biological and Environmental Research Program under Grant 249311 KP11-01-01.
REFERENCES
- 1.Friedberg E.C. (1997) Correcting the Blueprint of Life: An Historical Account of the Discovery of DNA Repair Mechanisms. Cold Springs Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 2.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC. [Google Scholar]
- 3.deBoer J. and Hoeijmakers,J.H. (2000) Nucleotide excision repair and human syndromes. Carcinogenesis, 21, 453–460. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T., Karran,P. and Wood,R.D. (1997) DNA excision repair pathways. Curr. Opin. Genet. Dev., 7, 158–169. [DOI] [PubMed] [Google Scholar]
- 5.Wallace S.S., Van Houten,B. and Kow,Y.W. (1994) DNA damage: effect on DNA structure and protein recognition. Ann. NY Acad. Sci., 276, 236–292. [Google Scholar]
- 6.de Laat W.L., Jaspers,N.G. and Hoeijmakers,J.H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev., 13, 768–785. [DOI] [PubMed] [Google Scholar]
- 7.Wood R.D. (1997) Nucleotide excision repair in mammalian cells. J. Biol. Chem., 272, 23465–23468. [DOI] [PubMed] [Google Scholar]
- 8.Sancar A. (1996) DNA excision repair. Annu. Rev. Biochem., 65, 43–81. [DOI] [PubMed] [Google Scholar]
- 9.Wakasugi M. and Sancar,A. (1999) Order of assembly of human DNA repair excision nuclease. J. Biol. Chem., 274, 18759–18768. [DOI] [PubMed] [Google Scholar]
- 10.Mu D., Hsu,D.S. and Sancar,A. (1996) Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem., 271, 8285–8294. [DOI] [PubMed] [Google Scholar]
- 11.Wold M.S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem., 66, 61–92. [DOI] [PubMed] [Google Scholar]
- 12.Wang M., Mahrenholz,A. and Lee,S.H. (2000) RPA stabilizes the XPA-damaged DNA complex through protein–protein interaction. Biochemistry, 39, 6433–6439. [DOI] [PubMed] [Google Scholar]
- 13.Hermanson-Miller I.L. and Turchi,J.J. (2002) Strand-specific binding of RPA and XPA to damaged duplex DNA. Biochemistry, 41, 2402–2408. [DOI] [PubMed] [Google Scholar]
- 14.He Z., Henricksen,L.A., Wold,M.S. and Ingles,C.J. (1995) RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature, 374, 566–569. [DOI] [PubMed] [Google Scholar]
- 15.Gomes X.V., Henricksen,L.A. and Wold,M.S. (1996) Proteolytic mapping of human replication protein A: evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry, 35, 5586–5595. [DOI] [PubMed] [Google Scholar]
- 16.Lao Y., Gomes,X.V., Ren,Y., Taylor,J.S. and Wold,M.S. (2000) Replication protein A interactions with DNA. III. Molecular basis of recognition of damaged DNA. Biochemistry, 39, 850–859. [DOI] [PubMed] [Google Scholar]
- 17.Lao Y., Lee,C.G. and Wold,M.S. (1999) Replication protein A interactions with DNA. 2. Characterization of double-stranded DNA-binding/helix-destabilization activities and the role of the zinc-finger domain in DNA interactions. Biochemistry, 38, 3974–3984. [DOI] [PubMed] [Google Scholar]
- 18.Walther A.P., Gomes,X.V., Lao,Y., Lee,C.G. and Wold,M.S. (1999) Replication protein A interactions with DNA. 1. Functions of the DNA-binding and zinc-finger domains of the 70-kDa subunit. Biochemistry, 38, 3963–3973. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y.L., Chen,C., Keshav,K.F., Winchester,E. and Dutta,A. (1996) Dissection of functional domains of the human DNA replication protein complex replication protein A. J. Biol. Chem., 271, 17190–17198. [DOI] [PubMed] [Google Scholar]
- 20.Saijo M., Kuraoka,I., Masutani,C., Hanaoka,F. and Tanaka,K. (1996) Sequential binding of DNA repair proteins RPA and ERCC1 to XPA in vitro. Nucleic Acids Res., 24, 4719–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stigger E., Drissi,R. and Lee,S.H. (1998) Functional analysis of human replication protein A in nucleotide excision repair. J. Biol. Chem., 273, 9337–9343. [DOI] [PubMed] [Google Scholar]
- 22.Bochkarev A., Pfuetzner,R.A., Edwards,A.M. and Frappier,L. (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature, 385, 176–181. [DOI] [PubMed] [Google Scholar]
- 23.Buchko G.W., Daughdrill,G.W., de Lorimier,R., Rao,B.K., Isern,N.G., Lingbeck,J.M., Taylor,J.S., Wold,M.S., Gochin,M., Spicer,L.D. et al. (1999) Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies. Biochemistry, 38, 15116–15128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braun K.A., Lao,Y., He,Z., Ingles,C.J. and Wold,M.S. (1997) Role of protein–protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry, 36, 8443–8454. [DOI] [PubMed] [Google Scholar]
- 25.Gomes X.V. and Wold,M.S. (1996) Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry, 35, 10558–10568. [DOI] [PubMed] [Google Scholar]
- 26.Jacobs D.M., Lipton,A.S., Isern,N.G., Daughdrill,G.W., Lowry,D.F., Gomes,X. and Wold,M.S. (1999) Human replication protein A: global fold of the N-terminal RPA-70 domain reveals a basic cleft and flexible C-terminal linker. J. Biomol. NMR, 14, 321–331. [DOI] [PubMed] [Google Scholar]
- 27.Henricksen L.A., Umbricht,C.B. and Wold,M.S. (1994) Recombinant replication protein A: expression, complex formation and functional characterization. J. Biol. Chem., 269, 11121–11132. [PubMed] [Google Scholar]
- 28.Kuraoka I., Morita,E.H., Saijo,M., Matsuda,T., Morikawa,K., Shirakawa,M. and Tanaka,K. (1996) Identification of a damaged-DNA binding domain of the XPA protein. Mutat. Res., 362, 87–95. [DOI] [PubMed] [Google Scholar]
- 29.Zuiderweg E.R. (2002) Mapping protein–protein interactions in solution by NMR spectroscopy. Biochemistry, 41, 1–7. [DOI] [PubMed] [Google Scholar]
- 30.McAlister M.S., Mott,H.R., van der Merwe,P.A., Campbell,I.D., Davis,S.J. and Driscoll,P.C. (1996) NMR analysis of interacting soluble forms of the cell–cell recognition molecules CD2 and CD48. Biochemistry, 35, 5982–5991. [DOI] [PubMed] [Google Scholar]
- 31.Daughdrill G.W., Ackerman,J., Isern,N.G., Botuyan,M.V., Arrowsmith,C., Wold,M.S. and Lowry,D.F. (2001) The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res., 29, 3270–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patrick S.M. and Turchi,J.J. (2002) Xeroderma pigmentosum complementation group A protein (XPA) modulates RPA–DNA interactions via enhanced complex stability and inhibition of strand separation activity. J. Biol. Chem., 277, 16096–16101. [DOI] [PubMed] [Google Scholar]
- 33.Buchko G.W., Ni,S., Thrall,B.D. and Kennedy,M.A. (1997) Human nucleotide excision repair protein XPA: expression and NMR backbone assignments of the 14.7 kDa minimal damaged DNA binding domain (Met98–Phe219). J. Biomol. NMR, 10, 313–314. [DOI] [PubMed] [Google Scholar]
- 34.Kay L.E., Keifer,P. and Saarinen,T. (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc., 114, 10663–10665. [Google Scholar]
- 35.Kay L.E., Muhandiram,D.R., Farrow,N.A., Aubin,Y. and Forman-Kay,J.D. (1996) Correlation between dynamics and high affinity binding in an SH2 domain interaction. Biochemistry, 35, 361–368. [DOI] [PubMed] [Google Scholar]
- 36.Gochin M. and Roder,H. (1995) Protein structure refinement based on paramagnetic NMR shifts: applications to wild-type and mutant forms of cytochrome c. Protein Sci., 4, 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchko G.W., Tung,C.S., McAteer,K., Isern,N.G., Spicer,L.D. and Kennedy,M.A. (2001) DNA–XPA interactions: a (31)P NMR and molecular modeling study of dCCAATAACC association with the minimal DNA-binding domain (M98–F219) of the nucleotide excision repair protein XPA. Nucleic Acids Res., 29, 2635–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]