Abstract
The heterogeneous nuclear ribonucleoprotein K protein is an RNA- and DNA-binding protein implicated in the regulation of multiple processes that comprise gene expression. We used chromatin immunoprecipitation (ChIP) assays to explore K protein interactions with serum-inducible, constitutively expressed and untranscribed gene loci in vivo. In the rat HTC-IR hepatoma cell line, serum treatment induced transient increases in the mRNA levels of two immediate-early genes, egr-1 and c-myc. ChIP analysis showed that the induction of egr-1 and c-myc genes was associated with a transient recruitment of K protein to multiple sites within each of these loci, including the promoter and transcribed regions. In contrast, recruitment of K protein to the constitutively transcribed β-actin locus and to randomly chosen non-transcribed loci was far weaker. In rat mesangial cells, c-myc was constitutively expressed while egr-1 remained serum responsive. In these cells, ChIP analysis showed serum-induced recruitment to the inducible egr-1 but not to the c-myc locus. Pre-treatment with the transcription inhibitor actinomycin D blocked the inducible but not the constitutive binding of K protein to these loci. Taken together, the results of this study suggest that the transient recruitment of K protein to serum-responsive loci depends on the inducible transcription of these immediate-early genes.
INTRODUCTION
Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is an evolutionarily conserved factor (1–3) found in multiple subcellular compartments including the nucleus (3), cytoplasm (4) and mitochondria (5). The finding that K protein is involved in a host of processes that comprise gene expression, such as chromatin remodeling (1), transcription (6,7), pre-mRNA splicing (8), mRNA export (9) and translation (10), has generated great interest in this factor (11–13). The involvement of K protein in these many processes probably reflects the interactions of its multiple domains (11,12) with a diversity of molecular partners including DNA (14,15), RNA (3), protein kinases (16–19), the GTP/GDP exchange factor Vav (20,21), and proteins involved in chromatin remodeling (22,23), transcription (6,24), mRNA splicing (23) and translation (12). Many of these interactions have been shown to be regulated by K protein phosphorylation induced either by changes in the extracellular environment (25) or by the activity of specific ligands (26). K protein is not only a kinase substrate (17) but it also regulates the activity of kinases (17,18). These findings are consistent with a model where K protein bridges signal transduction pathways to sites of nucleic acid-directed processes (11,12).
In the present study, we explored interactions of K protein with DNA targets in vivo. We show that K protein is transiently recruited to immediate-early gene loci in response to treatment of cells with serum, binding that appears to be transcription dependent.
MATERIALS AND METHODS
Cells
Rat HTC-IR hepatoma cells were grown in plastic cell culture flasks (26) in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, penicillin (100 U/ml) and streptomycin (0.01%), and humidified with a 7/93% CO2/air gas mixture. Rat mesangial cells were grown the same way except that RPMI 1640 medium was used (27).
Western blotting and immnuoprecipitations
Immunoprecipitations using anti-K protein antibody 54 directed against the C-terminus were carried out as previously described. Western blotting and immunostaining using anti-K protein antibody 54 were done by standard methods. Blots were developed using alkaline phosphatase colorimetric detection (26).
Chromatin immunoprecipitation (ChIP)
ChIP analysis was based on a previously published method (1,28). Cross-linking was done in the cold room, by adding 0.4 ml of 37% formaldehyde to 10 ml of overlaying media for 15 min. After cross-linking, cells were harvested and then washed twice with 1 ml of phosphate-buffered saline in Eppendorf tubes. Cells were lysed with 0.5 ml of IP buffer (150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.5% NP-40, 50 mM Tris–HCl, pH 7.5, 0.5 mM dithiothreitol) containing the following inhibitors: 10 µg/ml leupeptin, 0.5 mM phenylmethylsulfonyl fluoride, 30 mM p-nitrophenyl phosphate, 10 mM NaF, 0.1 mM Na3VO4, 0.1 mM Na2MoO4 and 10 mM β-glycerophosphate. After one wash with 1.0 ml of IP buffer, the pellet was resuspended in 1.0 ml of IP buffer (containing all inhibitors) and was sheared with a Bronson sonicator with two 10 s cycles, one pulsed and one continuous, at an output 3 and 80% duty cycle. Pull-downs were done using anti-K protein antibody either with or without blocking peptide (100 µM) and protein A beads (Pharmacia) (25). Beads were washed five times with 1 ml of IP buffer containing no inhibitors. DNA was eluted twice from the beads with 250 µl of elution buffer (1% SDS, 0.1 M NaHCO3) for 15 min with periodic vortexing (at room temperature). Cross-linking was reversed by adding 20 µl of 5 M NaCl and incubating the eluate overnight at 65°C. After adding 5 µg of linear acrylamide, DNA was precipitated with 1.0 ml of 100% ethanol. The pellet was washed with 1 ml of 70% ethanol, and then dissolved in 100 µl of TE pH 8.0. Proteins were digested by adding 11 µl of 10× proteinase K buffer (0.1 M Tris, pH 7.8, 50 mM EDTA, 5% SDS) and 1 µl of of 20 µg/µl proteinase K at 50°C for 30 min. DNA was extracted using phenol/chloroform, precipitated with ethanol and the final DNA pellet was dissolved in 20 µl of TE buffer.
PCR amplifications were done in 25 µl of 1× PCR buffer containing DNA, 0.5 µM primers (Table 1), 40 µM of each deoxynucleotide triphosphate (dNTP), 1.5 mM MgCl2 and 1.0 U of HotStart Taq DNA polymerase (Qiagen). One µCi of [α-32P]dCTP (NEN) was used in the reaction to label the PCR products. PCR products were resolved on native 5% polyacrylamide gels, the gels were dried and the PCR products were quantified using a phosphorimager (Cyclone, Packard). Densitometric analysis was performed using OptiQuant™ Image Analysis Software (Packard).
Table 1. List and sequences of PCR primers used.
| Primer name/accession number | Sequence | Direction | Figure |
|---|---|---|---|
| Rat egr1 promoter/J04154 | AAACACCATATAAGGAGCAGGAAG | Forward | 3A |
| TATTTCAAGGGTCTGAAACAGCAC | Reverse | ||
| Rat egr1 exon 1/J04154 | GGGGGCCCACCTACACTCC | Forward | 1A, 9A |
| CCACCAGCGCCTTCTCGTTATTCA | Reverse | ||
| Rat egr1 exon 2′/J04154 | ACTCCACTATCCACTATCAAAGCC | Forward | 3B, 8A, 8C |
| GTGGTCACTACGACTGAAATTACG | Reverse | ||
| Rat egr1 exon 2”/J04154 | TAGGACAATTGAAATTTGCTAAAGG | Forward | 3C |
| CACAGATGCTGTACAAAGATACAGG | Reverse | ||
| Rat c-myc promoter/Y00396 | TACTCTACTCCAGCTCTGGAACG | Forward | 2, 4A |
| ACCTACGACAGATGAGGTCTGAG | Reverse | ||
| Rat c-myc exon 2/Y00396 | GAGACATGGTGAATCAGAGCTTC | Forward | 4B, 8B, 8D |
| GTGTCTCTTCATGCAGCACTAGG | Reverse | ||
| Rat c-myc exon 3/Y00396 | GCAAATGCTCCAGCCCCAGGTC | Forward | 1B, 4C, 9B |
| AGTCCCAAAGCCCCAGCCAAGGTT | Reverse | ||
| Rat non-transcribed/AC120715 | AAGTTTGGGGCTGTTTAGTTC | Forward | 6A |
| GTTTCCACCACCAGGCACATA | Reverse | ||
| Rat non-transcribed/AC120715 | ACCAGCCAGTCAGAGTTCAGG | Forward | 6B |
| GGTTTCATCATCCCGAGAC | Reverse | ||
| Rat non-transcribed/AC120715 | GGGGCGATGCATACCTACAC | Forward | 6C |
| CTCCCTGCCATACCTTGACTCT | Reverse |
β-Actin PCR primers were purchased from Ambion, murine sequence accession X03672.
RNA isolation and quantitative RT–PCR
RNase inhibitor (1 U/µl) was used in all extractions and reactions involving RNA. Total cellular RNA was isolated using TRIzol reagent as per the manufacturer’s protocol (Invitrogen). RT–PCRs were carried out using SuperScript II reverse transcriptase (Invitrogen) and random primers in a 20 µl volume as per the manufacturer’s protocol. RT–PCRs were diluted 1:10 with water, and cDNAs were used in the PCR. Amplifications and analysis of 32P-labeled products were done as described above.
RESULTS AND DISCUSSSION
ChIP analysis of K protein interactions with serum-induced immediate-early gene loci
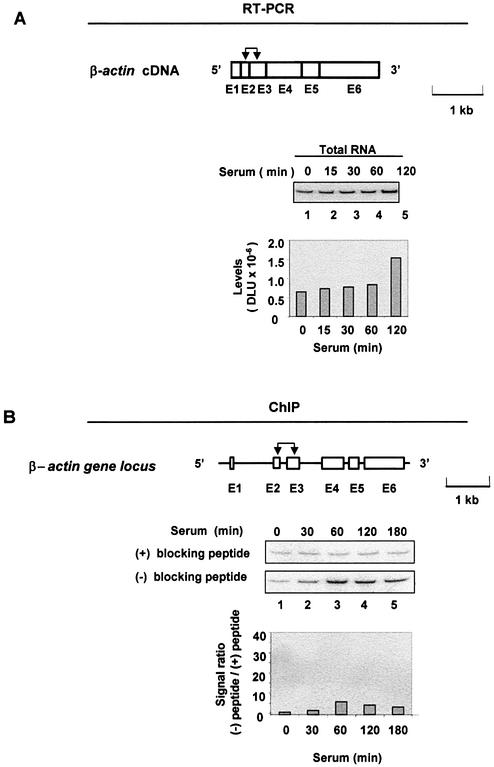
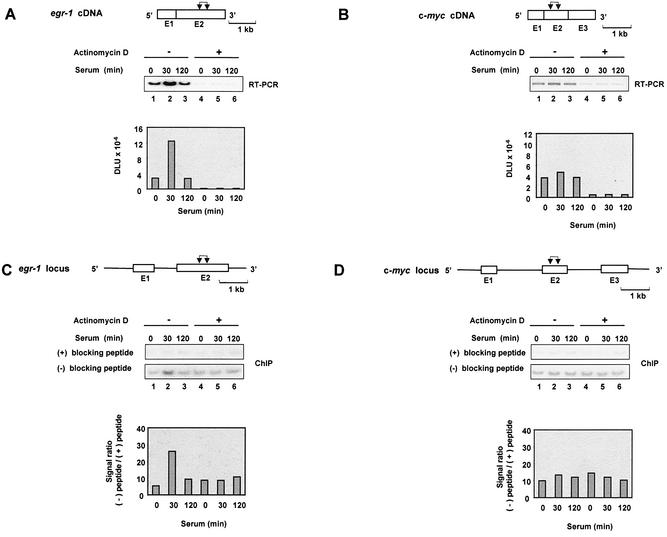
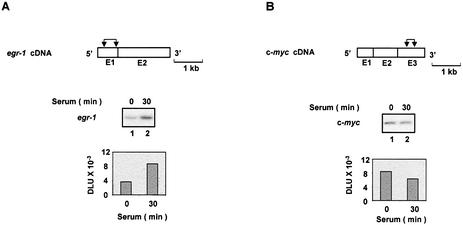
Treatment of cells with serum induces a large repertoire of genes, including the rapid induction of the immediate-early genes (29). Activation of these genes provides a good model to study the mechanisms of inducible gene expression (30,31). Serum-deprived rat HTC-IR cells were treated for given times with serum, whole-cell lysates were prepared and total RNA was purified. The levels of egr-1 and c-myc mRNAs were assessed using RT–PCR and gene-specific primers. Phosphorimaging of the 32P-labeled products separated by native PAGE revealed that the two transcripts were transiently induced by serum, with the peak levels seen 15–30 min following treatment (Fig. 1A and B, lanes 1–5). Compared with c-myc, a larger increase was seen in egr-1 mRNA, which also had lower constitutive expression in these serum-deprived cells.
Figure 1.
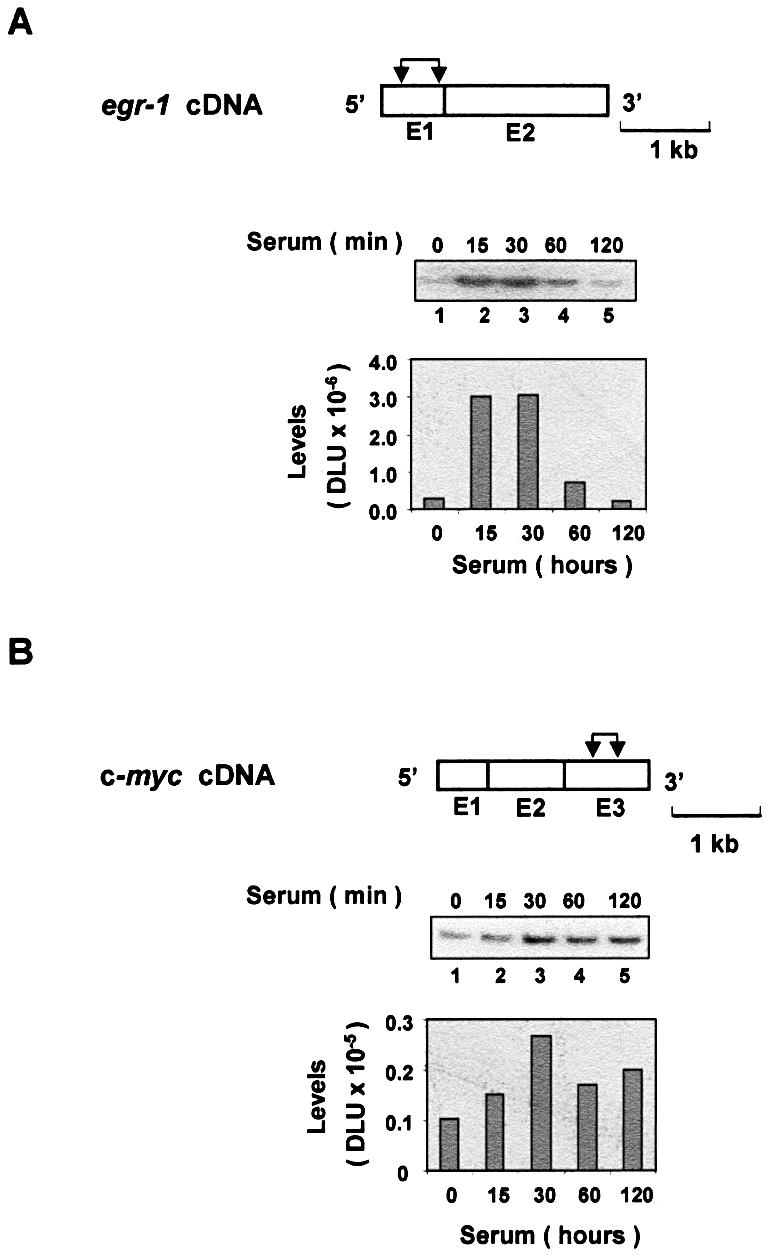
Serum-induced transcripts of the immediate-early genes. Serum-deprived HTC-IR cells were treated with 10% FBS for the indicated times. Cells were harvested for extraction of total RNA. Whole-cell RNA was used in RT–PCRs using oligo(dT) primer. PCR was carried out using primers for egr-1 (A) and c-myc (B) genes and [α-32P]dCTP. PCR products were resolved by non-denaturing 5% PAGE, and were analyzed using a phosphorimager. The diagram above the gels shows egr-1 (A) and c-myc (B) cDNAs; exons (E), which are numbered, are shown as boxes. The pairs of arrows designate the location of each pair of primers used to amplify the transcript fragments.
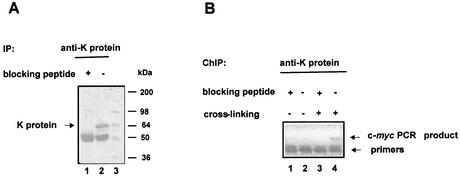
K protein is known to regulate c-myc gene expression (6,32). We used ChIP assays to test if K protein interacts with this and other serum-responsive gene loci in vivo. To ensure the specificity of the method, we used as a negative control anti-K protein antibody blocked with the peptide used to raise the serum. The western blot immunostained with anti-K protein antibody illustrates that pre-incubating the antibody with the peptide abrogates the ability of the antibody to precipitate K protein from cell lysates (Fig. 2A, compare lanes 1 and 2). Next we used these reagents in ChIP assays to assess binding of K protein to the c-myc locus, a gene whose transcription is thought to be regulated by K protein (6,33). HTC-IR cells grown in serum-containing medium were either treated (Fig. 2B, lanes 1 and 2) or not treated (Fig. 2B, lanes 3 and 4) with formaldehyde (1). Cells were harvested, washed and lysed, and the pellets were sheared using a sonicator. K protein–DNA adducts were mixed with either antibody alone or antibody pre-incubated with the blocking peptide; pulled-down DNA was purified from washed beads and used as a template in PCR using primers to the c-myc gene promoter. An ethidium bromide-stained agarose gel revealed a PCR product of the predicted size only in cells that were treated with the cross-liking agent and from beads that contained antibody that was not blocked with the peptide. These results validate this strategy to assess K protein interactions with gene loci in vivo.
Figure 2.
ChIP analysis using anti-K (αK) protein antibody. (A) Nuclear extracts from HTC-IR cells were mixed with anti-K protein antibody that was pre-incubated with (+, lane 1) or without (–, lane 2) the peptide used to generate the antibody in rabbits (48). Complexes were pulled-down with protein A beads and, after washing, proteins were eluted by boiling in SDS loading buffer. Proteins were resolved by SDS–PAGE and, after transfer to PVDF, the membranes were immunostained with anti-K protein antibody. Pre-stained molecular weight markers were run in lane 3, shown in kDa. (B) HTC-IR cells were treated with (+, lanes 3 and 4) or without (–, lanes 1 and 2) formaldehyde. Cells were lysed, nuclei were isolated and then sonicated. K protein was immunoprecipitated with anti-K protein antibody that was pre-incubated with either non-specific peptide (–, lanes 2 and 4) or a peptide that blocks K protein immunoprecipitation (blocking peptide, +, lanes 1 and 3) (A). Complexes were eluted, cross-linking was reversed and purified DNA was used as a template in a PCR using primers to the rat c-myc gene. DNA was separated by agarose gel electrophoresis and visualized with ethidium bromide. The arrow shows the predicted size fragment of the c-myc gene. The lower bands seen in every lane correspond to the PCR primers used.
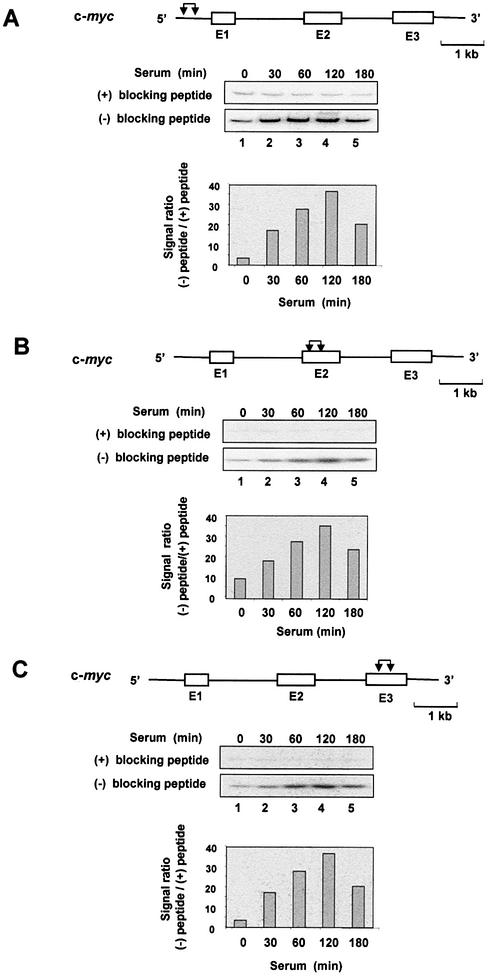
To explore the mechanisms of K protein interaction with egr-1 and c-myc loci, serum-deprived HTC-IR cells were treated with serum, and at given time points cells were cross-linked and ChIP analysis was done as above except that the number of cycles was decreased, PCR products were 32P-labeled and products were resolved on native PAGE to measure signals using a phosphorimager. Three sets of primers were used that span nearly the entire length of the egr-1 and c-myc loci, including the 5′-flanking promoter regions. The results of ChIP showed that K protein binds the egr-1 locus with similar kinetics for each set of primers used; in untreated cells, there was little K protein binding and the peak signal ratio [PCR signal (–) blocking peptide/(+) blocking peptide] was observed 60–120 min following treatment of cells with serum (Fig. 3, lanes 1–5). Similar kinetics of serum-induced recruitment of K protein were observed for the longer c-myc locus (Fig. 4). Because the chromatin was sheared to an average fragment size of 500 bp (assessed by agarose gel electrophoresis), the observation that similar signals were obtained with all three primers spanning the length of these loci suggests that K protein is recruited to multiple sites within the egr-1 and c-myc genes.
Figure 3.
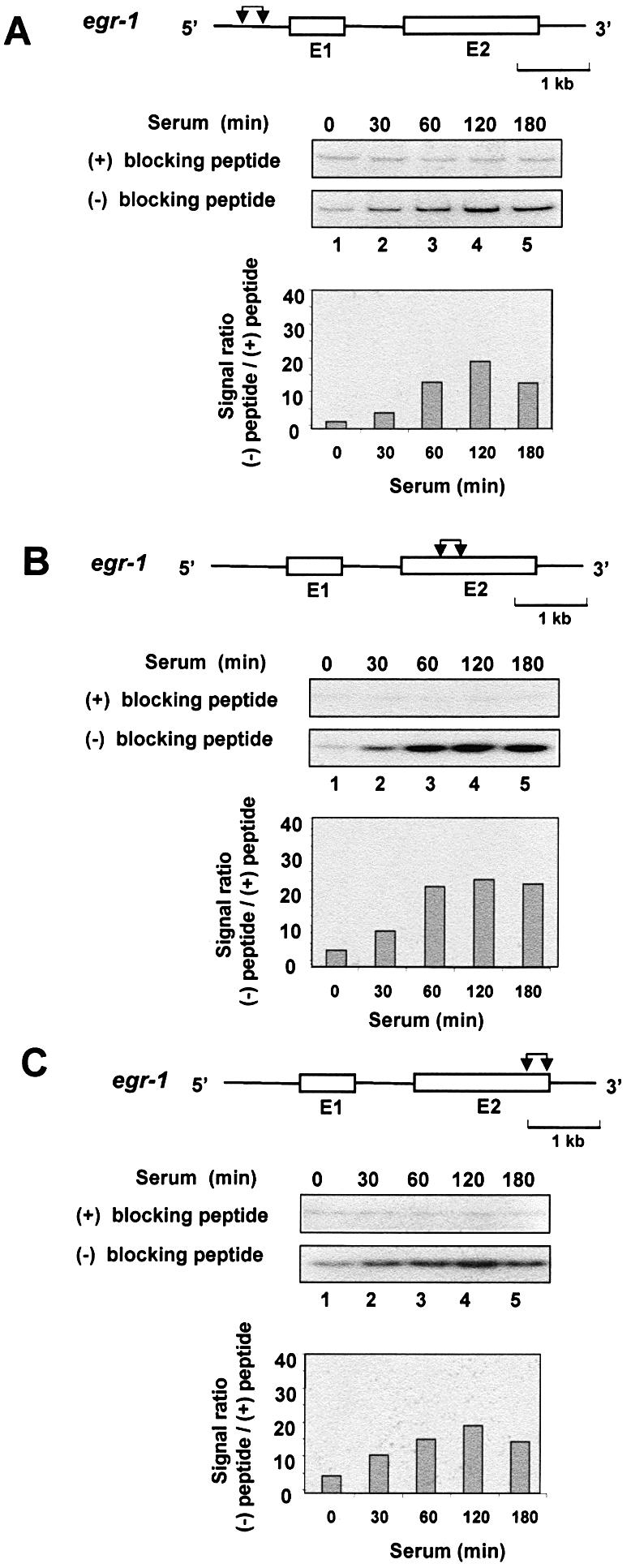
ChIP analysis of K protein interactions with the inducibly transcribed egr-1 gene locus. Serum-deprived HTC-IR cells were treated with 10% FBS for the indicated times. After cross-linking with formaldehyde, cells were lysed, pelleted and sonicated. For each time point, half of the nuclear DNA–protein sonicate was mixed with antibody blocked with the peptide [(+) blocking peptide] and the other half with antibody that was not blocked [(–) blocking peptide]. The rest of the ChIP assay was done as described in Figure 2, except that PCR was carried out using a series of primers spanning the length of the egr-1 locus. [α-32P]dCTP-labeled PCR products were resolved by non-denaturing 5% PAGE, and were analyzed using a phosphorimager. The graphs show signal ratios between the level of the 32P-labeled PCR product obtained without blocking the antibody [(–) blocking peptide] and the level obtained for the same time point with beads bearing blocked antibody [(+) blocking peptide]. The diagram above each gel shows the structure of the egr-1 gene; exons (E), which are numbered, are shown as boxes, and introns as lines. The pairs of arrows designate the location of the pair of primers used to amplify the fragments. The genes are drawn to the same scale; the length of a 1 kb unit is shown.
Figure 4.
ChIP analysis of K protein interactions with the inducibly transcribed c-myc gene locus. ChIP analysis of binding of K protein was done as described in Figure 3 using a series of primers spanning the c-myc gene locus. The diagram above each gel shows the structure of the c-myc gene; exons (E), which are numbered, are shown as boxes, and introns as lines. The pairs of arrows designate the location of the pair of primers used to amplify the fragments. The genes are drawn to the same scale; the length of a 1 kb unit is shown.
ChIP analysis of K protein interactions with constitutively expressed and non-transcribed loci
In cultured cells, the β-actin gene is constitutively expressed at high levels and it is not induced by serum treatment (Fig. 5A). Next, we examined the binding of K protein to the β-actin locus in serum-treated HTC-IR cells. Although there was apparent K protein recruitment to the β-actin locus, the level of binding was lower than that seen for the egr-1 and c-myc locus (signal ratio 4–5 for β-actin, compared with 20–40 for the immediate-early genes; compare Fig. 5B with Figs 3 and 4). The weak K protein recruitment may reflect the low affinity of K protein for the β-actin locus in these cells.
Figure 5.
Expression of the β-actin gene and its interaction with K protein. Serum-deprived HTC-IR cells were treated with 10% FBS for the indicated times. (A) RT–PCR of total β-actin mRNA was done as described in Figure 1. (B) ChIP analysis of the interaction of the β-actin locus with K protein was done as described in Figure 3. The diagram above each gel shows the structure of the β-actin cDNA and gene; exons (E), which are numbered, are shown as boxes, and introns as lines. The arrows designate the location of each pair of primers used. The gene and cDNA are drawn to the same scale; the length of a 1 kb unit is shown.
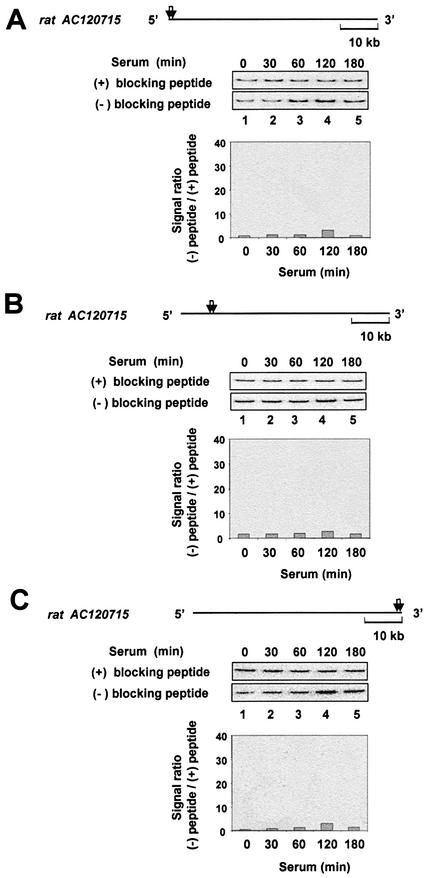
Because K protein appears to be recruited to the serum-induced immediate-early and β-actin loci (Figs 3–5), we wondered if this holds true for any stretch of DNA. To test such a possibility, we designed PCR primers to intragenic non-transcribed regions of two randomly chosen rat clones (NCBI) and used them to amplify fragments from the same immunoprecipitated DNA samples that were used above (Figs 3–5) to assess recruitment to the immediate-early gene loci. PCR results for these loci are shown in Figure 6. Compared with the signal ratios obtained with primers for the egr-1 and c-myc loci, the randomly chosen rat clones showed 10-fold lower signal ratios (in the range of 2–3 for the random sequence compared with 20–40 for the immediate-early genes). As for the immediate-early genes, some of these non-transcribed loci exhibited a weak transient increase (Fig. 6A and C), while with other primers the signal ratios were relatively unchanged by serum treatment (Fig. 6B). It is conceivable that the apparent weak recruitment of K protein to the randomly chosen loci (Fig. 6) represents background within the ChIP assay. For example, it may reflect binding of K protein to sites other than these untranscribed loci.
Figure 6.
ChIP analysis of K protein interactions with non-transcribed gene loci. ChIP was done as in Figure 3 using three pairs of PCR primers (A–C) to a randomly chosen rat clone (CH230-115D12; accession number AC120715). The diagram above each gel shows the 63 951 bp long fragment of the 182 396 bp rat CH230-115D12 clone. The arrows designate the location of each pair of primers used. The length of a 10 kb unit is shown.
Since for some loci K protein recruitment is transient while for others it appears to be constitutive, we wanted to address the global pattern of K protein–DNA interaction in vivo. To assess this, serum-deprived HTC-IR cells were pulse-labeled with [3H]thymidine and were then treated with 10% FBS for the indicated times (Fig. 7). After cells were cross-linked with formaldehyde, nuclei were sonicated and 3H-labeled DNA complexes were pulled-down with blocked and unblocked anti-K protein antibody. [3H]DNA eluted from the beads was measured using a scintillation counter. The counts measured from beads bearing peptide-blocked K protein antibody remained the same in untreated and treated samples; these 3H counts represent non-specific binding of DNA complexes to beads. There were twice as many 3H counts pulled-down with beads bearing anti-K protein antibody without blocking peptide and this did not change with serum treatment. Thus, while K protein recruitment to serum-responsive immediate-early gene loci was strongly inducible (Figs 3 and 4), the global DNA–K protein interaction was largely constitutive (Fig. 7). The signal ratio of the constitutive binding (2 U) was at least 10-fold below the peak signal ratio of the inducibly transcribed loci (Figs 2 and 3). K and other hnRNP proteins are components of nuclear matrix (34–36). Chromatin loops are attached to nuclear matrix through protein–DNA interactions. Thus, co-immunoprecipitation of 3H-labeled chromatin with K protein shown here (Fig. 7) may, in part, represent attachment of chromatin to the nuclear matrix. If so, the low level of binding seen in the untranscribed loci (Fig. 6) may also reflect these types of DNA–protein interactions.
Figure 7.
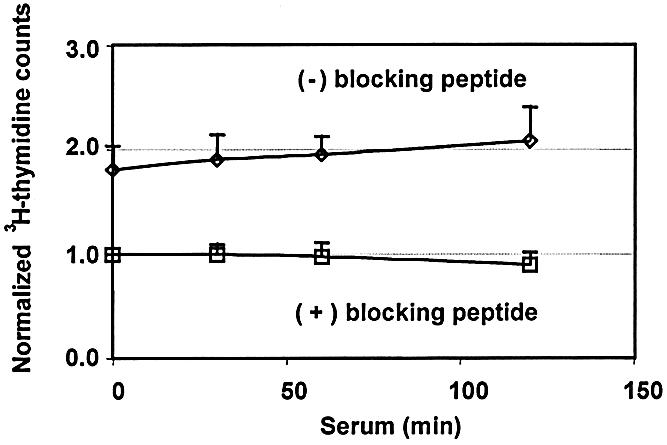
Assessment of global K protein–genome interactions in serum-treated cells. HTC-IR cells grown in DMEM supplemented with 10% FBS were labeled with [3H]thymidine (10 µCi in 10 ml of medium). After 12 h, the medium was replaced with serum-free DMEM for 24 h and serum- deprived cells were treated with 10% FBS for the indicated times. After cross-linking with formaldehyde, cells were lysed, and nuclei were isolated and sonicated. For each time point, half of the nuclear DNA–protein sonicate was mixed with antibody blocked with the peptide [(+) blocking peptide] and the other half with antibody that was not blocked [(–) blocking peptide]. After five washes with 1 ml of IP buffer, DNA was eluted from the beads and 3H counts were measured using a scintillation counter. The plot shows values for each time point normalized to the counts value of the zero time point for the (+) blocking peptide sample (mean ± SD, n = 4 separate experiments).
Evidence that inducible recruitment of K protein to serum-responsive gene loci is transcription dependent
While studying different cell types, we have found that serum treatment of rat mesangial cells increased egr-1 but not c-myc mRNA levels (compare Fig. 8A, lanes 1–3 with Fig. 1B, lanes 1–3). Since this was different from the serum-responsive c-myc expression in hepatocytes, we used these cells to compare inducible recruitment of K protein to these loci. ChIP analysis revealed that there was serum-inducible recruitment of K protein to the transcriptionally active egr-1 locus (Fig. 8C, lanes 1–3). In contrast, there was no inducible recruitment to the unresponsive c-myc locus (Fig. 8D, lanes 1–3). These results suggest that the serum-responsive transient increase in K protein binding is dependent on the inducible transcription of the egr-1 target gene. To test this hypothesis further, we pre-treated (60 min) rat mesangial cells with the transcription inhibitor, actinomycin D (37,38). RT–PCR showed that this agent markedly decreased the constitutive levels of both egr-1 and c-myc mRNAs and blocked the serum-responsive induction of egr-1 (Fig. 8A and B, lanes 4–6). Both egr-1 and c-myc are immediate-early genes and their transcripts have short half-lives (20–30 min) (6). Thus, blocking transcription resulted in a rapid decline of transcript levels due to their degradation. In contrast, β-actin transcript has a longer half-life, and pre-treatment with actinomycin D did not alter the constitutive level of this mRNA (data not shown). ChIP analysis in mesangial cells pre-treated with actinomycin D showed that this agent blocked the inducible but not the constitutive binding of K protein to the egr-1 locus (Fig. 8C, compare lanes 1–3 with lanes 4–6). The constitutive binding of K protein to the c-myc locus was also not altered by pre-treatment of these cells with actinomycin D (Fig. 8D, compare lanes 1–3 with lanes 4–6). These results indicate that the transient recruitment of K protein to serum-inducible gene loci is transcription dependent. In contrast, the constitutive binding is not. This constitutive transcription-independent component may, in part, represent the genome-wide K protein binding pattern shown in the [3H]thymidine labeling experiments (Fig. 7).
Figure 8.
Evidence that transient recruitment of K protein to gene loci depends on gene-specific serum-inducible transcription. Serum-deprived rat mesangial cells were pre-incubated with or without 1 µg/ml actinomycin D (60 min) and then were treated with 10% FBS for the indicated times. Total RNA was isolated for RT–PCR, and ChIP analysis was done using K protein antibody as before (Figs 1 and 3). RT–PCR and ChIP analysis were done using primers for the indicated regions within the egr-1 and the c-myc loci.
K protein was first identified as a component of the hnRNP particle (3,39), and presumably can bind to mRNAs as they are synthesized. Thus, the serum-responsive recruitment to transcribed regions may represent binding of K protein to the nascent transcripts. To test if K protein binds to immediate-early gene transcripts in vivo, serum-deprived HTC-IR cells were either untreated or were treated with 10% serum for 30 min. Cells were then incubated with formaldehyde (28) and RNA bound to K protein was immunoprecipitated with anti-K protein antibody. This RNA was used as a template in RT–PCR using c-myc and egr-1 primers. Predicted PCR products were obtained with egr-1 (Fig. 9A) and c-myc (Fig. 9B) primer pairs. Without reverse transcriptase, no PCR product was obtained, indicating no DNA contamination (data not shown). These results show that both transcripts were immunoprecipitated with K protein and provide evidence that K protein associates with these transcripts in vivo. The binding of K protein to these RNAs is consistent with the possibility that the recruitment to the transcribed sites may, in part, be mediated by binding of K protein to these immediate-early nascent transcripts (40). However, K protein binds many proteins, DNA and RNA (12). Some of the K protein partners are recruited to transcribed loci by interacting with DNA, transcriptionally active complexes or nascent RNA. One or more of these K protein interactions could be responsible for the recruitment of K protein to the transcribed loci by an indirect mechanism.
Figure 9.
Evidence for the association of egr-1 (A) and c-myc (B) mRNA with K protein in vivo. Serum-deprived HTC-IR cells were treated with 10% FBS for the indicated times After cross-linking with formaldehyde, cells were lysed with IP buffer. K protein was immunoprecipitated with anti-K protein antibody, beads were washed and complexes were eluted. RNA was precipitated with ethanol, washed with 70% ethanol, and then dissolved in TE pH 8.0. Proteins were then digested by proteinase K. RNA was extracted using phenol/chloroform and the final RNA pellet was dissolved in TE buffer. RNA pulled-down with K protein was used in an RT–PCR, and products were analyzed as above (Fig. 1).
The level of egr-1 mRNA co-immunoprecipitated with K protein increased with serum treatment, while the amount of c-myc mRNA decreased (Fig. 9, lanes 1 and 2). Although serum treatment increased the levels of c-myc mRNA, the amount bound to K protein decreased (Figs 1B and 9B). Similarly decreased K protein–RNA binding was observed for β-actin RNA in response to serum treatment of these cells (data not shown). The protein composition of mRNPs is dynamic and depends on subcellular compartmentalization of the protein–RNA complex (40) and extracellular signals (18). For example, a signal-dependent release of K protein from the 15-lipoxygenase (LOX) mRNA in erythroid precursors is thought to activate translation of the LOX transcript by allowing its shift to polysomes (18). The RNA co-immunoprecipitation results (Fig. 9, and data not shown) may represent the net effect of the K protein–c-myc mRNA interactions that occur predominantly in the cytoplasmic compartment. Unlike egr-1, there is a relatively high level of constitutive expression of c-myc mRNA (Fig. 1A and B). Thus, the serum-induced decrease of c-myc mRNA bound to K protein may represent reduced K protein binding to this constitutive and presumably cytoplasmic pool. Similarly to the LOX mRNA (18), this decrease may reflect a shift of c-myc mRNA from mRNPs to the polysome-bound fraction. In contrast, it is conceivable that in the nucleus, the level of binding of K protein to nuclear c-myc RNA follows the pattern determined by serum-induced transcription (40).
In the above studies, we demonstrate inducible and constitutive binding of K protein to immediate-early gene loci that respond to serum (Figs 3, 4 and 8B). The results of the ChIP analysis suggest that both inducible and constitutive binding of K protein occur at multiple sites including the promoter and transcribed regions (Figs 3 and 4). Although novel, the observation that K protein binds to multiple sites within target loci is not unexpected. K protein has been shown to regulate promoter activity including that of the c-myc gene (6). In vitro K protein binds the c-myc promoter CT element directly, and in reporter gene assays mutation of this element abrogates the ability of K protein to activate the c-myc promoter (41). The in vivo interaction of K protein with the c-myc promoter shown here (Fig. 4) is consistent with the previous suggestions that K protein regulates endogenous c-myc promoter (6,32,41). K protein has also been shown to stimulate transcription of reporter genes even if the promoters do not contain CT elements (42). This may reflect effects of K protein on steps downstream of transcription initiation, such as transcription elongation and pre-RNA processing. We have shown that K protein interacts in vivo with mRNAs encoded by target loci including egr-1 and c-myc (Fig. 9) (26), suggesting that it regulates RNA-directed processes. Indeed, K protein has recently been shown to regulate splicing of the β-tropomyosin pre-mRNA (8). Thus, recruitment to sites within both the promoter and transcribed regions (Figs 3 and 4) is consistent with the suggestion that K protein is involved in multiple processes that comprise gene expression, including transcription and pre-mRNA processing.
A class of factors recently identified appears to bridge signal transduction pathways to sites of nucleic acid-directed processes (25,43,44). In addition to K protein (16,18,25, 45,46), this class of factors includes YB-1 (43), Sam68 (47) and others (44). Here we demonstrate for the first time (Figs 3 and 4) that a member of this class of signaling molecules is recruited to multiple sites within an inducible gene locus. Because K protein is a substrate and interacts with several inducible kinases (16,18,25,45,46), these observations suggest that K protein participates in signaling processes at the target gene loci. The role of K protein recruitment to these inducible gene loci remains to be defined.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the NIH GM45134, DK45978, Fogarty International Research Collaboration Award (FIRCA) TW05685, the Juvenile Diabetes Research Foundation and the Northwest Kidney Foundation (K.B.). Y.K. was supported by a postdoctoral fellowship from the American Heart Association Northwest Affiliate.
REFERENCES
- 1.Denisenko O.N. and Bomsztyk,K. (2002) Yeast hnRNP K-like genes are involved in regulation of the telomeric position effect and telomere length. Mol. Cell. Biol., 22, 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charroux B., Angelats,C., Fasano,L., Kerridge,S. and Vola,C. (1999) The levels of the bancal product, a Drosophila homologue of vertebrate hnRNP K protein, affect cell proliferation and apoptosis in imaginal disc cells. Mol. Cell. Biol., 19, 7846–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swanson M.S. and Dreyfuss,G. (1988) Classification and purification of proteins of heterogenous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol. Cell. Biol., 8, 2237–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostrowski J., Sims,J.E., Sibley,C.H., Valentine,M.A., Dower,S.K., Meier,K.E. and Bomsztyk,K. (1991) A serine/threonine kinase activity is closely associated with a 65-kDa phosphoprotein specifically recognized by the kappa B enhancer element. J. Biol. Chem., 266, 12722–12733. [PubMed] [Google Scholar]
- 5.Ostrowski J., Wyrwicz,L., Rychlewski,L. and Bomsztyk,K. (2002) Heterogeneous nuclear ribonucleoprotein K protein associates with multiple mitochondrial transcripts within the organelle. J. Biol. Chem., 277, 6303–6310. [DOI] [PubMed] [Google Scholar]
- 6.Michelotti E.F., Michelotti,G.A., Aronsohn,A.I. and Levens,D. (1996) Heterogenous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol., 16, 2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Q., Melnikova,I.N. and Gardner,P.D. (1998) Differential effects of heterogeneous nuclear ribonucleoprotein K on Sp1 and Sp3-mediated transcriptional activation of neuronal nicotinic acetylcholine receptor promoter. J. Biol. Chem., 273, 19877–19883. [DOI] [PubMed] [Google Scholar]
- 8.Expert-Bezancon A., Le Caer,J.P. and Marie,J. (2002) HnRNP K is a component of an intronic splicing enhancer complex that activates the splicing of the alternative exon 6A from chicken-beta-tropomyosin pre-mRNA. J. Biol. Chem., 277, 16614–16623. [DOI] [PubMed] [Google Scholar]
- 9.Michael W.M., Eder,P.S. and Dreyfuss,G. (1997) The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J., 16, 3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostareck D.H., Ostareck-Lederer,A., Wilm,M., Thiele,B.J., Mann,M. and Hentze,M.W. (1997) mRNA silencing in erythroid differentiation: hnRNP K nad hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell, 89, 597–606. [DOI] [PubMed] [Google Scholar]
- 11.Makeyev A.V. and Liebhaber,S.A. (2002) The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA, 8, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bomsztyk K., Van Seuningen,I., Suzuki,H., Denisenko,O. and Ostrowski,J. (1997) Diverse molecular interactions of the hnRNP K protein. FEBS Lett., 403, 113–115. [DOI] [PubMed] [Google Scholar]
- 13.Ostareck-Lederer A., Ostareck,D.H. and Hentze,M.W. (1998) Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem. Sci., 23, 409–411. [DOI] [PubMed] [Google Scholar]
- 14.Ostrowski J., Van Seuningen,I., Seger,R., Rouch,C.T., Sleath,P.R., McMullen,B.A. and Bomsztyk,K. (1994) Purification, cloning and expression of a murine phosphoprotein that binds the κB motif in vitro identifies it as the homolog of the human hnRNP K protein. Description of a novel DNA-dependent phosphorylation process. J. Biol. Chem., 269, 17626–17634. [PubMed] [Google Scholar]
- 15.Ito K., Sato,K. and Endo,H. (1994) Cloning and characterization of a single-stranded DNA binding protein that specifically recognizes a deoxycytidine stretch. Nucleic Acids Res., 22, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schullery D.S., Ostrowski,J., Denisenko,O.N., Stempka,L., Shnyreva,M., Suzuki,H., Gschwendt,M. and Bomsztyk,K. (1999) Regulated interaction of protein kinase Cd with the heterogeneous nuclear ribonucleoprotein K protein. J. Biol. Chem., 274, 15101–15109. [DOI] [PubMed] [Google Scholar]
- 17.Van Seuningen I., Ostrowski,J., Bustelo,X.R., Sleath,P.R. and Bomsztyk,K. (1995) The K protein domain that recruits the interleukin 1-responsive K protein kinase lies adjacent to a cluster of c-Src and Vav SH3-binding sites. Implications that K protein acts as a docking platform. J. Biol. Chem., 270, 26976–26985. [DOI] [PubMed] [Google Scholar]
- 18.Ostareck-Lederer A., Ostareck,D.H., Cans,C., Neubauer,G., Bomsztyk,K., Superti-Furga,G. and Hentze,M.W. (2002) c-Src mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol., 22, 4535–4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wadd S., Bryant,H., Filhol,O., Scott,J.E., Hsieh,T.Y., Everett,R.D. and Clements,J.B. (1999) The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J. Biol. Chem., 274, 28991–28998. [DOI] [PubMed] [Google Scholar]
- 20.Hobert O., Jallal,B., Schlessinger,J. and Ullrich,A. (1994) Novel signaling pathway suggested by SH3 domain-mediated p95vav/heterogeneous ribonucleoprotein K interaction. J. Biol. Chem., 269, 20225–20228. [PubMed] [Google Scholar]
- 21.Bustelo X.R., Suen,K.-I., Michael,W.M., Dreyfuss,G. and Barbacid,M. (1995) Association of the vav proto-oncogene product with poly(rC)-specific RNA-binding proteins. Mol. Cell. Biol., 15, 1324–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denisenko O.N. and Bomsztyk,K. (1997) The product of murine homolog of the Drosophila extra sex comb gene displays transcriptional repressor activity. Mol. Cell. Biol., 17, 4707–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shnyreva M., Schullery,D.S., Suzuki,H., Higaki,Y. and Bomsztyk,K. (2000) Interaction of two multifunctional proteins. Heterogeneous nuclear ribonucleoprotein K and Y-box binding protein. J. Biol. Chem., 275, 15498–15503. [DOI] [PubMed] [Google Scholar]
- 24.Denisenko O.N., O’Neill,B., Ostrowski,J., Van Seuningen,I. and Bomsztyk,K. (1996) Zik, a transcriptional repressor that interacts with the heterogenous nuclear ribonuclear particle K protein. J. Biol. Chem., 271, 27701–27706. [DOI] [PubMed] [Google Scholar]
- 25.Ostrowski J., Schullery,D.S., Denisenko,O.N., Higaki,Y., Watts,J., Aebersold,R., Stempka,L., Gschwendt,M. and Bomsztyk,K. (2000) Role of tyrosine phosphorylation in the regulation of the interaction of heterogenous nuclear ribonucleoprotein K protein with its protein and RNA partners. J. Biol. Chem., 275, 3619–3628. [DOI] [PubMed] [Google Scholar]
- 26.Ostrowski J., Kawata,Y., Schullery,D., Denisenko,O.N., Higaki,Y., Abrass,C.K. and Bomsztyk,K. (2001) Insulin alters heterogeneous ribonucleoprotein K protein binding to DNA and RNA. Proc. Natl Acad. Sci. USA, 98, 9044–9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki H., O’Neill,B.C., Suzuki,Y., Denisenko,O.N. and Bomsztyk,K. (1996) Activation of a nuclear DNA-binding protein recognized by a transcriptional element, bcn-1, from the laminin B2 chain gene promoter. J. Biol. Chem., 271, 18981–18988. [DOI] [PubMed] [Google Scholar]
- 28.Kuo M.H. and Allis,C.D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods, 19, 425–433. [DOI] [PubMed] [Google Scholar]
- 29.Iyer V.R., Eisen,M.B., Ross,D.T., Schuler,G., Moore,T., Lee,J.C.F., Trent,J.M., Staudt,L.M., Hudson,J.,Jr, Boguski,M.S. et al. (1999) The transcriptional program in the response of human fibroblasts to serum. Science, 283, 83–87. [DOI] [PubMed] [Google Scholar]
- 30.Jhun B.H., Haruta,T., Meinkoth,J.L., Leitner,W., Draznin,B., Saltiel,A.R., Pang,L., Sasaoka,T. and Olefsky,J.M. (1995) Signal transduction pathways leading to insulin-induced early gene induction. Biochemistry, 34, 7996–8004. [DOI] [PubMed] [Google Scholar]
- 31.Spencer C.A. and Groudine,M. (1990) Transcription elongation and eukaryotic gene regulation. Oncogene, 5, 777–785. [PubMed] [Google Scholar]
- 32.Mandal M., Vadlamudi,R., Nguyen,D., Wang,R., Costa,L., Bagheri-Yarmand,R., Mendelsohn,J. and Kumar,R. (2001) Growth factors regulate heterogeneous nuclear ribonucleoprotein K expression and function. J. Biol. Chem., 276, 9699–9704. [DOI] [PubMed] [Google Scholar]
- 33.Michelotti E.F., Tomonaga,T., Krutzsch,H. and Levens,D. (1995) Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J. Biol. Chem., 270, 9494–9499. [DOI] [PubMed] [Google Scholar]
- 34.Nickerson J.A., Krochmalnic,G., Wan,K.M. and Penman,S. (1989) Chromatin architecture and nuclear RNA. Proc. Natl Acad. Sci. USA, 86, 177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattern K.A., van der Kraan,I., Schul,W., de Jong,L. and van Driel,R. (1999) Spatial organization of four hnRNP proteins in relation to sites of transcription, to nuclear speckles and to each other in interphase nuclei and nuclear matrices of HeLa cells. Exp. Cell Res., 246, 461–470. [DOI] [PubMed] [Google Scholar]
- 36.Samuel S.K., Spencer,V.A., Bajno,L., Sun,J.M., Holth,L.T., Oesterreich,S. and Davie,J.R. (1998) In situ cross-linking by cisplatin of nuclear matrix-bound transcription factors to nuclear DNA of human breast cancer cells. Cancer Res., 58, 3004–3008. [PubMed] [Google Scholar]
- 37.Sussman M. (1966) Protein synthesis and the temporal control of genetic transcription during slime mold development. Proc. Natl Acad. Sci. USA, 55, 813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinol-Roma S. and Dreyfuss,G. (1992) Shuttling of pre-RNA binding proteins between nucleus and cytoplasm. Nature, 355, 730–732. [DOI] [PubMed] [Google Scholar]
- 39.Matunis M.J., Xing,J. and Dreyfuss,G. (1994) The hnRNP K protein: unique primary structure, nucleic acid-binding properties and subcellular localization. Nucleic Acids Res., 22, 1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mili S., Shu,H.J., Zhao,Y. and Pinol-Roma,S. (2001) Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol. Cell. Biol., 21, 7307–7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takimoto M., Tomonaga,T., Matunis,M., Avigan,M., Krutzsch,H., Dreyfuss,G. and Levens,D. (1993) Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J. Biol. Chem., 268, 18249–18258. [PubMed] [Google Scholar]
- 42.Lee M.-H., Mori,S. and Raychaudhuri,P. (1996) Trans-activation by the hnRNP K protein involves an increase in RNA synthesis from the reporter genes. J. Biol. Chem., 271, 3420–3427. [DOI] [PubMed] [Google Scholar]
- 43.Chen C.Y., Gherzi,R., Andersen,J.S., Gaietta,G., Jurchott,K., Royer,H.D., Mann,M. and Karin,M. (2000) Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev., 14, 1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 44.Vernet C. and Artzt,K. (1997) STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet., 13, 479–484. [DOI] [PubMed] [Google Scholar]
- 45.Habelhah H., Shah,K., Huang,L., Burlingame,A., Shokat,K. and Ronai,Z. (2001) Identification of new JNK substrate using ATP pocket mutant JNK and a corresponding ATP analogue. J. Biol. Chem., 276, 18090–18095. [DOI] [PubMed] [Google Scholar]
- 46.Habelhah H., Shah,K., Huang,L., Ostareck-Lederer,A., Burlingame,A., Shokat,K., Hentz,M. and Ronai,Z. (2001) ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nature Cell Biol., 3, 325–330. [DOI] [PubMed] [Google Scholar]
- 47.Matter N., Herrlich,P. and Konig,H. (2002) Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature, 420, 691–695. [DOI] [PubMed] [Google Scholar]
- 48.Van Seuningen I., Ostrowski,J. and Bomsztyk,K. (1995) Description of an IL-1-responsive kinase that phosphorylates the K protein. Enhancement of phosphorylation by sequence-selective DNA and RNA motifs. Biochemistry, 34, 5644–5650. [DOI] [PubMed] [Google Scholar]