Abstract
Fragile X syndrome, the most common cause of inherited mental retardation, is instigated by dynamic expansion of a d(CGG) trinucleotide repeat in the 5′-untranslated region of the first exon of the FMR1 gene, resulting in its silencing. The expanded d(CGG)n tract readily folds into hairpin and tetraplex structures which may contribute to the blocking of FMR1 transcription. In this work, we report that the cationic porphyrin 5,10,15,20-tetra(N-methyl-4-pyridyl)porphin (TMPyP4) effectively destabilizes in vitro the G′2 bimolecular tetraplex structure of d(CGG)n while it stabilizes the G′2 tetraplex form of the telomeric sequence d(TTAGGG)2. Similarly to TMPyP4, the hnRNP-related protein CBF-A also destabilizes G′2 tetrahelical d(CGG)n while binding and stabilizing tetraplex telomeric DNA. We report that relative to each agent individually, successive incubation of G′2 d(CGG)n with TMPyP4 followed by exposure to CBF-A results in a nearly additive extent of disruption of this tetraplex form of the repeat sequence. Our observations open up the prospect of unfolding secondary structures of the expanded FMR1 d(CGG)n tract of fragile X cells by their exposure to low molecular size drugs or to proteins such as TMPyP4 or CBF-A.
INTRODUCTION
Fragile X syndrome, the single most common inherited cause of mental impairment, is engendered by dynamic expansion of a d(CGG) trinucleotide repeat in the 5′-untranslated region of the first exon of the FMR1 gene (1). Expression of FMR1 is silenced and its replication is retarded in fully affected individuals who carry more than 200–2000 repeats of the d(CGG) triplet (2,3).
Hypermethylation of a promoter CpG island and of the fully expanded trinucleotide repeat sequence, as well as histone deacetylation, are major factors in the transcriptional silencing of FMR1 (4,5). However, FMR1 expression is only partially restored and FMRP is not detected in fragile X cells that are exposed to inhibitors of DNA methylation and of histone deacetylation (6,7). We and others demonstrated that the d(CGG)n tract spontaneously folds into hairpin structures (8–11). Two adjacent hairpins can associate to form G′2 bimolecular tetraplex structures. These structures are more stable than a monomeric hairpin. Indeed, the trinucleotide repeat sequence readily folds under physiological-like in vitro conditions and in the presence of alkali ions into stable bimolecular tetrahelical complexes (12–14). Hairpin and G′2 tetraplex forms of d(CGG)n have been shown to obstruct replicative DNA polymerases in vitro (15–17) and in vivo (18). Unwinding of these DNA secondary structures by WRN helicase alleviates the replicative block (17). It is conceivable that by impeding the transcription machinery, hairpin and tetraplex structures of an expanded d(CGG)n tract also contribute to the transcriptional silencing of FMR1 in fragile X cells. In addition, formation of secondary structures by the r(CGG)n tract in FMR1 mRNA molecules that escape the transcriptional block may obstruct its translation. Hence, in addition to DNA demethylation and histone hyperacetylation, full restoration of FMR1 expression and FMRP synthesis in fragile X cells might require destabilization of secondary structures of the genomic d(CGG)n tract and of the FMR1 mRNA r(CGG)n run.
Unfolding of secondary structures of d(CGG)n and r(CGG)n may be accomplished by over-expressing in fragile X cells proteins that destabilize folded forms of d(CGG)n and r(CGG)n. Alternatively, secondary structures of d(CGG)n might be resolved by exposing the cells to low molecular weight molecules that disrupt (CGG)n hairpin and tetraplex formation in DNA and RNA. We have identified previously three mammalian tetraplex d(CGG)n-destabilizing proteins (19,20) one of which is the heterogeneous nuclear ribonucleoprotein (hnRNP)-related protein CBF-A that destabilizes tetrahelical d(CGG)n while, paradoxically, it binds and stabilizes tetraplex telomeric DNA (20). The structural domains in CBF-A that mediate destabilization of tetraplex d(CGG)n are distinct from motifs which are responsible for the binding and stabilization of tetraplex telomeric DNA (21). Of special interest among tetraplex DNA-interacting drugs are three positional cationic porphyrin isomers: 5,10,15,20-tetra(N-methyl-2-pyridyl)porphin (TMPyP2); 5,10,15,20-tetra(N-methyl-3-pyridyl)porphin (TMPyP3); and 5,10,15,20- tetra(N-methyl-4-pyridyl)porphin (TMPyP4) (22–26). Simil arly to CBF-A, these drugs bind and stabilize tetraplex telomeric DNA. However, the three cationic porphyrins vary in their binding affinity for different tetraplex DNA molecules (24) and by their capacity to inhibit the activities of telomerase (27) or DNA helicase (25). A likely source for this variance is the divergent arrangement among tetraplex structures of different guanine-rich sequences of folded strands, groove sizes and base composition, and dimensions of loops (24,28).
In this work, we inquired whether in analogy with the CBF-A protein, cationic porphyrins also interact differentially with bimolecular tetraplex structures of telomeric DNA and of d(CGG)n. We find that while TMPyP4 stabilizes tetraplex telomeric DNA in vitro, it effectively disrupts tetraplex d(CGG)n. We also report that relative to the activity of each agent individually, the successive action of TMPyP4 followed by CBF-A enhances the destabilization of tetraplex d(CGG)n.
MATERIALS AND METHODS
Preparation of tetraplex forms of DNA oligomers
Synthetic DNA oligomers 5′-tail TeR2, 5′-d[TAGACATG(TTAGGG)2TTA]-3′ that contains two copies of the telomeric repeat sequence d(TTAGGG); d(CGG)7, 5′-d(CGG)7-3′; 5′-tail d(CGG)7, 5′-d[GTCAGGTGC(CGG)7)]-3′; 3′-tail d(CGG)7, 5′-d[(CGG)7CGTGGACTG]-3′, each containing seven repeats of the d(CGG) trinucleotide; and d(5-meCGG)7, 5′-d(5-meCGG)7-3′ which is 5-methylated in each of its seven cytosine residues, were provided by Operon Technologies. The oligomers were purified by denaturing gel electrophoresis as we described (29) and were 5′-32P-end-labeled (30). The bimolecular G′2 tetraplex structure of the telomeric sequence was generated by incubating 85 µM 5′-[32P]5′-tail TeR2 DNA oligomer for 12–20 h at 37°C in TE buffer (10 mM Tris–HCl buffer pH 8.0, 1.0 mM EDTA) containing 1.0 M KCl. The bimolecular G′2 tetraplex structure of the fragile X expanded sequence was formed by incubating 55 µM 5′-[32P]d(CGG)7-containing oligomers for 12–20 h at 4°C in TE buffer containing 300 mM KCl. The tetraplex structures of the oligomers were resolved and purified by non-denaturing electrophoresis as previously described (31). The gel-purified tetraplex DNA structures were stored at –20°C until used. G′2 DNA structures existed in equilibrium with the single-stranded oligomer, constituting 40–70% of the total DNA upon storage at 4 or –20°C. Bimolecular stoichiometry of the tetraplex forms of both sequences was confirmed as detailed elsewhere (20). Resistance of the guanine residues to dimethyl sulfate attack (12) provided evidence that these tetraplex DNA structures were stabilized by guanine–guanine Hoogsteen hydrogen bonds, and circular dichroism (CD) spectroscopy indicated an antiparallel orientation of the DNA strands (20). Schemes of G′2 structures of 3′-tail d(CGG)7 and TeR2 DNA are shown in Figure 1.
Figure 1.
Schemes of G′2 bimolecular tetraplex forms of 3′-tail d(CGG)7 and 5′-tail TeR2 DNA. The bimolecular tetrahelices are both dimers of two hairpins bonded by guanine quartets. The single-stranded tails at the 3′ or 5′ ends of the respective oligomers are represented by dashed lines. Indicated are four of the seven guanine residues in each 3′-tail d(CGG)7 oligomer that participate in quartet formation in G′2 3′-tail d(CGG)7 and all six guanines of TeR2 DNA that form stacked guanine quartets in G′2 TeR2 DNA. The hairpins are aligned against each other in one of several possible orientations.
Cationic porphyrins and CBF-A protein
The three positional cationic porphyrin isomers TMPyP2, TMPyP3 and TMPyP4 (25) were dissolved in H2O to 5.0 mM and stored at –70°C until used. Recombinant CBF-A protein and its mutant variants were prepared as we recently described (21).
Tetraplex DNA stability assays
Assay mixtures for the measurement of G′2 tetraplex DNA stability in the presence of cationic porphyrins or CBF-A protein contained in a final volume of 10 µl of buffer D [20 mM Tris–HCl buffer, pH 8.0, 1.0 mM dithiothreitol (DTT), 0.5 mM EDTA, 20 mM KCl, 20% glycerol]: 21 nM 5′-[32P]G′2 5′-tail Ter2 DNA or 10–16 nM G′2 5′-[32P]3′-tail d(CGG)7 and the indicated amounts of a specified cationic porphyrin or recombinant CBF-A protein. Reaction mixtures were incubated for the indicated periods of time and temperatures and the reactions were terminated by rapid cooling of the mixtures to 4°C and addition of SDS to a final concentration of 1%. Intact and destabilized G′2 tetraplex forms of the oligomers were resolved from one another by electrophoresis at 4°C and 200–250 V in a non-denaturing 10% polyacrylamide gel in 0.5× TBE buffer (1.2 mM EDTA in 0.54 M Tris borate buffer pH 8.0, 1.0 mM EDTA) that contained 20 mM KCl. Electrophoresis was stopped after a bromophenol blue tracking dye migrated 7.0–7.5 cm into the gel. Proportions of intact and destabilized G′2 DNA were quantified by phosphorimaging of the dried gel.
RESULTS
TMPyP4 destabilizes G′2 3′-tail d(CGG)7 while stabilizing G′2 5′-tail TeR2 DNA
TMPyP4 was shown to bind to and stabilize both parallel and antiparallel tetraplex DNA structures (22,23) and to inhibit telomerase action by stabilizing tetraplex telomeric DNA (23,27,32). TMPyP4 and its two positional isomers, TMPyP2 and TMPyP3, differ in their capacity to promote parallel-stranded quadruplex DNA formation and to inhibit the unwinding of parallel and antiparallel tetraplex DNA molecules by the yeast Sgs1 helicase (25). We compared the effect of the three cationic porphyrins on the stability of antiparallel bimolecular G′2 structure of 3′-tail d(CGG)7. Neither TMPyP2 nor TMPyP3 affected the stability of this G′2 tetraplex DNA structure under a wide range of drug concentrations and temperatures (results not shown). In contrast, data shown in Figure 2 indicated that while TMPyP4 increased the thermal stability of G′2 5′-tail TeR2 DNA, it dramatically diminished the heat resistance of G′2 3′-tail d(CGG)7. The differential effect of TMPyP4 on the stabilities of G′2 structures of 5′-tail TeR2 DNA and 3′-tail d(CGG)7 were quantified by determining Tm values for pre-formed tetraplexes that were heated in the absence or presence of the drug. In addition, we determined the Tm value of the G′2 tetraplex structure of 3′-tail d(CGG)7 that was formed in the presence of TMPyP4. Typical G′2 DNA melting curves are shown in Figure 3. Linear plotting of DNA denaturation yielded sigmoid melting curves. To specifically calculate the melting temperature of the G′2 tetraplex form of 3′-tail d(CGG)7, we constructed semi-logarithmic plots of the descending portion of the curve of disappearance of its radioactive band. In this representative experiment, TMPyP4 elevated the Tm value of 5′-tail TeR2 DNA by 12.5°C (Fig. 3A). In contrast, addition of the drug to pre-formed G′2 3′-tail d(CGG)7 decreased its Tm by 14.0°C (Fig. 3B), and a similar decline in the Tm value was measured for G′2 3′-tail d(CGG)7 that was formed in the presence of TMPyP4 (Fig. 3C). Table 1 summarizes the results of a series of determinations of the effect of TMPyP4 on the melting temperatures of G′2 tetraplex forms of 5′-tail TeR2 and 3′-tail d(CGG)7. These results established that addition of TMPyP4 to pre-formed G′2 5′-tail TeR2 DNA significantly stabilized this tetraplex, increasing its average Tm value by 13.2°C. In clear contrast, TMPyP4 greatly destabilized G′2 3′-tail d(CGG)7, diminishing its Tm by 14.9°C on average. The Tm value of G′2 3′-tail d(CGG)7 that was generated in the presence of TMPyP4 was similarly lowered by 14.6°C relative to control DNA (Table 1). Hence, TMPyP4 exerted opposite effects on the thermal stabilities of bimolecular tetraplex structures of telomeric DNA and of the d(CGG) trinucleotide repeat.
Figure 2.
TMPyP4 stabilizes a G′2 tetraplex form of the telomeric sequence 5′-tail TeR2 DNA and disrupts the G′2 structure of 3′-tail d(CGG)7. Bimolecular tetraplex structures of 5′-32P-labeled 5′-tail TeR2 DNA or 3′-tail d(CGG)7 oligomers were incubated for 10 min at the indicated temperatures in the absence or presence of 0.3 µM TMPyP4 under tetraplex DNA stability assay conditions (see Materials and Methods). Shown are representative phosphorimages of DNA resolved by non-denaturing electrophoresis. G′2, bimolecular tetraplex forms of the respective oligomers; SS, single-stranded oligomers.
Figure 3.
TMPyP4 conversely affects the thermal stabilities of tetraplex structures of telomeric and d(CGG)n sequences. 5′-32P-labeled G′2 5′-tail TeR2 DNA or G′2 3′-tail d(CGG)7 were incubated for 10 min at increasing temperatures and in the absence or presence of 0.3 µM TMPyP4. Single-stranded and G′2 forms of the oligomers were separated by non-denaturing electrophoresis and quantified by phosphorimaging analysis (see Materials and Methods). Shown are semi-logarithmic plots of the relative amounts of remaining tetraplex DNA structures as a function of increasing temperature. A relative initial value of 100% is denoted for unheated G′2 DNA structures that constituted 40–70% of the total incubated DNA. Melting temperatures, Tms, were those at which 50% of the initial amount of G′2 DNA was denatured. (A) Melting curves of G′2 5′-tail TeR2 DNA. Calculated Tm values for DNA incubated in the absence or presence of TMPyP4 are 42.5 and 55.0°C, respectively. (B) Melting curves of G′2 3′-tail d(CGG)7. Calculated Tm values for DNA incubated in the absence or presence of TMPyP4 are 45.0 and 30.5°C, respectively. (C) Melting curves of 5′-32P-labeled G′2 3′-tail d(CGG)7 that was generated under standard G′2 DNA formation conditions (see Materials and Methods) in the presence or absence of 0.3 µM TMPyP4. Both DNA preparations were heated without adding TMPyP4 to the mixtures. Calculated Tm values for G′2 3′-tail d(CGG)7 that was formed in the absence or presence of the drug are 45.5 and 32.0°C, respectively.
Table 1. Effect of TMPyP4 on the melting temperatures of G′2 tetraplex structures of 5′-tail TeR2 DNA and 3′-tail d(CGG)7.
| DNA |
Tm (n) |
Δ°C | |
|---|---|---|---|
| –TMPyP4 | +TMPyP4 | ||
| G′2 5′-tail TeR2 pre-formed | 44.6 ± 2.0 (4) | 57.8 ± 0.3 (3) | +13.2 |
| G′2 3′-tail d(CGG)7 pre-formed | 45.8 ± 0.7 (7) | 30.9 ± 1.0 (5) | –14.9 |
| G′2 3′-tail d(CGG)7 formed with TMPyP4 | 31.2 ± 2.3 (3) | – | –14.6a |
Tetraplex DNA molecules, 20 nM G′2 5′-[32P]5′-tail TeR2 DNA or 10 nM G′2 5′-[32P]3′-tail d(CGG)7 that were formed in the absence TMPyP4 were incubated for 10 min at various temperatures with or without 0.3 µM TMPyP4. In parallel, 10 nM G′2 5′-[32P]3′-tail d(CGG)7 that was generated in the presence of 0.3 µM TMPyP4 was similarly incubated without the drug. Values of the melting temperature, Tm, of the tetraplex DNA species were determined as described in Figure 3. Presented are average values ± SDs of the indicated number, n, of independent determinations.
aThe average decrease in the Tm of G′2 3′-tail d(CGG)7 that was formed in the presence of TMPyP4 was calculated relative to the Tm of control G′2 3′-tail d(CGG)7 (Tm = 45.8 ± 0.7°C).
TMPyP4 destabilizes G′2 forms of non-methylated and hypermethylated d(CGG)7 sequences
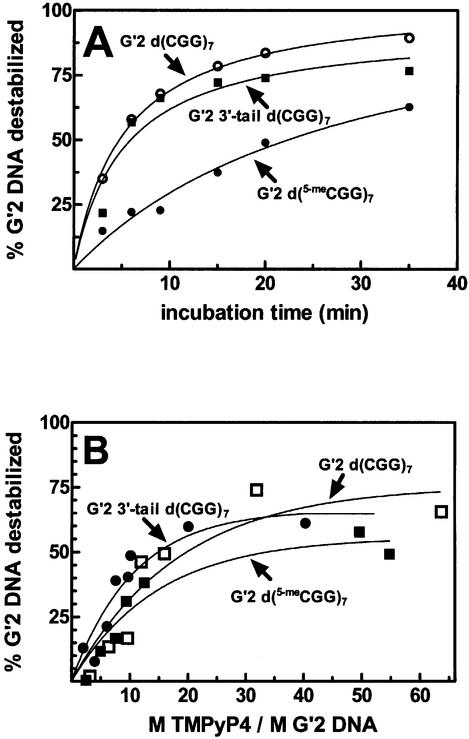
Next we inquired whether TMPyP4 destabilized G′2 tetraplex structures of d(CGG) repeat sequences other than 3′-tail d(CGG)7 and whether it was capable of disrupting a hypermethylated d(CGG)n sequence. We compared the kinetics and stoichiometry of destabilization by TMPyP4 of G′2 tetraplex forms of 3′-tail d(CGG)7, 5′-tail d(CGG)7, non-tailed d(CGG)7 and hypermethylated d(5-meCGG)7. As seen in Figure 4A, TMPyP4 similarly destabilized 65–75% of G′2 3′-tail d(CGG)7 or G′2 d(CGG)7 upon their incubation for 35 min at 30°C. Similar kinetics of destabilization by TMPyP4 were obtained for G′2 5′-tail d(CGG)7 (results not shown). However, <40% of G′2 d(5-meCGG)7 was destabilized following exposure to TMPyP4 for 35 min at 30°C (data not shown). This lower extent of disruption was plausibly due to the higher melting temperature of the tetraplex structure of the hypermethylated repeat sequence [Tm ∼49.0°C for G′2 d(5-meCGG)7 versus 45.8°C for G′2 3′-tail d(CGG)7]. Accordingly, incubation with TMPyP4 for 35 min at the higher temperature of 33°C resulted in destabilization of 60% of G′2 d(5-meCGG)7 (Fig. 4A). As seen in Figure 4B, molar excesses of 8–12 of TMPyP4 over tetraplex DNA were required to attain 50% of the maximum destabilization achieved under our experimental conditions for G′2 forms of d(CGG)7, G′2 3′-tail d(CGG)7 and d(5-meCGG)7. A progressively lower excess of drug was required for G′2 3′-tail d(CGG)7 destabilization with elevation of the incubation temperature (results not shown).
Figure 4.
Kinetics and stoichiometry of destabilization by TMPyP4 of G′2 tetraplex forms of d(CGG)7, 3′-tail d(CGG)7 and d(5-meCGG)7. (A) Kinetics of destabilization. 5′-32P-labeled G′2 3′-tail d(CGG)7 or d(CGG)7 at 45 nM each were incubated with 0.3 µM TMPyP4 for the indicated periods of time and at 30°C under tetraplex DNA stability assay conditions. G′2 d(5-meCGG)7 (50 nM) was similarly exposed to the drug, except that incubation was conducted at 33°C. Electrophoretic resolution of the DNA and quantification of G′2 tetraplex DNA destabilization were conducted as detailed in the legend to Figure 3. (B) Stoichiometry of destabilization. G′2 tetraplex forms of 5′-32P-labeled 3′-tail d(CGG)7 or d(CGG)7 at 45 nM each were incubated for 15 min at 30°C with increasing amounts of TMPyP4 under tetraplex DNA stability assay conditions. G′2 d(5-meCGG)7 (50 nM) was similarly incubated with increasing concentrations of TMPyP4 for 15 min and at 33°C. Single-stranded and G′2 forms of the DNA oligomers were separated by non-denaturing electrophoresis, and the relative amount of remaining tetraplex DNA was quantified as described in Figure 3.
TMPyP4 and CBF-A act in sequence to enhance destabilization of G′2 3′-tail d(CGG)7
In analogy with TMPyP4, the qTBP42/CBF-A protein binds and stabilizes mono- and bimolecular tetraplex forms of telomeric DNA (20,31) while it destabilizes bimolecular tetraplex structures of d(CGG)n oligomers (20,21). We inquired whether the extent of G′2 3′-tail d(CGG)7 disruption might be increased by the joint action of TMPyP4 and CBF-A.
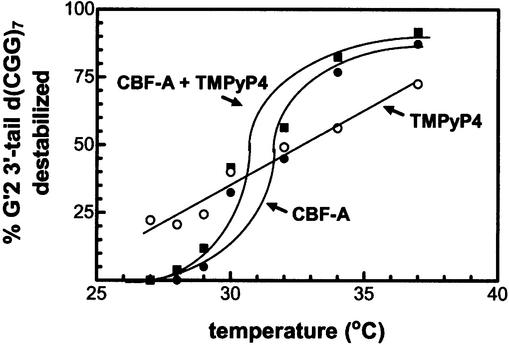
First, we measured the extent of 5′-32P-labeled G′2 3′-tail d(CGG)7 destabilization in reaction mixtures that were incubated at different temperatures in the presence of either TMPyP4 or CBF-A alone or a mixture thereof. As seen in Figure 5, whereas the rate of tetraplex DNA disruption by TMPyP4 increased linearly with increasing temperature, the temperature response of CBF-A-mediated destabilization was sigmoid. Destabilization in the presence of a mixture of TMPyP4 and CBF-A also displayed sigmoid temperature dependence, and the extent of G′2 3′-tail d(CGG)7 disruption by both agents was comparable with destabilization by CBF-A alone. These results implied that CBF-A might depress TMPyP4-mediated disruption of the tetraplex DNA.
Figure 5.
Destabilization of G′2 3′-tail d(CGG)7 by TMPyP4 and CBF-A. Reaction mixtures for the destabilization of 10 nM G′2 5′-[32P]3′-tail d(CGG)7 were incubated for 10 min at the indicated increasing temperatures in the presence of 0.35 µM TMPyP4 or 2.6 µM CBF-A, or a mixture thereof. Tetraplex and single-stranded forms of the DNA were resolved from one another by non-denaturing electrophoresis, and their relative amounts were quantified by phosphorimaging (see Materials and Methods).
To explore this possibility, we compared the extent of G′2 3′-tail d(CGG)7 destabilization by TMPyP4 alone or in the presence of wild-type CBF-A, mutant CBF-A proteins that had lost their G′2 d(CGG)n-destabilizing activity, or proteins that do not interact with tetraplex DNA. Separately added TMPyP4 or wild-type CBF-A destabilized 43.1 or 64.5%, respectively, of the G′2 3′-tail d(CGG)7 substrate, and only 69.5% of this DNA was disrupted in the presence of both agents (Table 2). To examine whether the destabilizing activity of CBF-A is required for the inhibition of TMPyP4-mediated disruption of G′2 d(CGG)n, we used two CBF-A mutant proteins that are devoid of tetraplex DNA-destabilizing activity. These mutant proteins, T267K and ΔR11 contained, respectively, deactivating mutations in the ATP/GTP-binding box and the RNP11 motif (21). As demonstrated in Table 2, although both mutant proteins failed to disrupt the G′2 tetraplex DNA substrate, they effectively blocked its destabilization by TMPyP4. That this inhibition was specific to CBF-A was suggested by comparison with bovine serum albumin or ovalbumin that were incapable of binding or disrupting G′2 3′-tail d(CGG)7 (not shown). In contrast to wild-type CBF-A or its inactive mutants, these two proteins failed to significantly affect the extent of tetraplex 3′-tail d(CGG)7 destabilization by TMPyP4 (Table 2). Taken together, the results summarized in Figure 5 and Table 2 indicated that the interaction of active or inactive CBF-A protein with the G′2 3′-tail d(CGG)7 substrate specifically impeded its destabilization by TMPyP4.
Table 2. Effect of proteins on the G′2 3′-tail d(CGG)7-destabilizing capacity of TMPyP4.
| Agents added | % G′2 3′-tail d(CGG)7 destabilized |
|---|---|
| TMPyP4 | 43.1 |
| Wt CBF-A | 64.5 |
| Wt CBF-A + TMPyP4 | 69.6 |
| T267K CBF-A | 0.0 |
| T267K CBF-A + TMPyP4 | 0.0 |
| ΔR11 CBF-A | 0.0 |
| ΔR11 CBF-A + TMPyP4 | 3.9 |
| Bovine serum albumin | 0.0 |
| Bovine serum albumin + TMPyP4 | 30.7 |
| Ovalbumin | 0.0 |
| Ovalbumin + TMPyP4 | 37.5 |
Tetraplex DNA destabilization reaction mixtures containing 10 nM G′2 5′-[32P]3′-tail d(CGG)7 were incubated for 10 min at 30°C in the presence of the listed drug and/or proteins. Tetraplex and single-stranded forms of the DNA were resolved from one another by non-denaturing electrophoresis, and their relative amounts were quantified by phosphorimaging (see Materials and Methods). TMPyP4 was added at a 35-fold molar excess over G′2 3′-tail d(CGG)7 and proteins were present at molar excesses of 150–260-fold over tetraplex DNA.
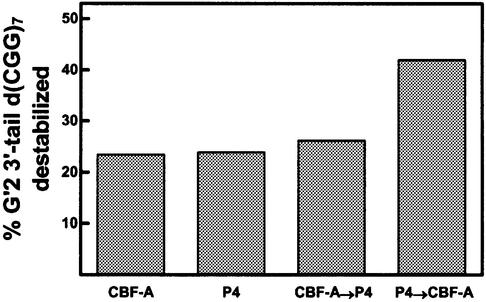
To substantiate that CBF-A blocked the action of TMPyP4 and to attempt to increase the extent of tetraplex DNA destabilization by both agents, G′2 3′-tail d(CGG)7 was exposed to TMPyP4 and CBF-A in succession. Reaction mixtures containing either TMPyP4 or CBF-A alone were incubated for 10 min at 30°C. CBF-A was added to the mixtures that were already incubated with TMPyP4, or TMPyP4 was added to mixtures that were incubated with CBF-A, whereas control mixtures were maintained with the single agent. Following incubation of all the mixtures for an additional period of 10 min at 30°C, the DNA was resolved by non-denaturing electrophoresis and the amounts of destabilized G′2 3′-tail d(CGG)7 were quantified. Results of a typical experiment, shown in Figure 6, indicated that 23.5 or 24.0% of the tetraplex DNA substrate were disrupted in mixtures that, respectively, contained CBF-A or TMPyP4 alone. A similar proportion, 26.2%, of the G′2 3′-tail d(CGG)7 was destabilized after being first exposed to CBF-A and then to TMPyP4. However, 42.0% of the tetraplex DNA substrate were disrupted when it was first exposed to TMPyP4 and subsequently to CBF-A. These results are in accord with the proposition that CBF-A inhibited TMPyP4 action by blocking its access to G′2 3′-tail d(CGG)7. This inhibitory activity could be circumvented by initially destabilizing the tetraplex DNA with TMPyP4 followed by additional unwinding by CBF-A. Such sequential action of the two agents resulted in an almost additive combination of their activities.
Figure 6.
TMPyP4 and CBF-A act in sequence to increase destabilization of G′2 3′-tail d(CGG)7. Tetraplex DNA destabilization reaction mixtures containing 0.13 µM TMPyP4 or 0.4 µM CBF-A were incubated for 10 min at 30°C. The mixtures were put on ice and CBF-A was added to 0.4 µM to the TMPyP4-containing mixtures, or TMPyP4 was added to 0.13 µM to mixtures that contained CBF-A. Control mixtures were maintained with the single original agent. All the assay mixtures were incubated for an additional period of 10 min at 30°C, the DNA was resolved by non- denaturing electrophoresis and amounts of destabilized G′2 3′-tail d(CGG)7 were quantified by phosphorimaging. Arrows indicate the order of sequential addition of TMPyP4 and CBF-A. P4 denotes TMPyP4.
DISCUSSION
Oligomeric d(CGG)n in solution folds under physiological-like pH and temperature into hairpin structures that are stabilized by Watson–Crick and/or non-canonical Hoogsteen hydrogen bonds (8–11,33,34). Incubation of d(CGG)n in the presence of Li+, Na+ or K+ ions and under similar physiological-like conditions results in the formation of tetraplex structures of this DNA that are stabilized by Hoogsteen-bonded guanine quartets (12,13,35). Methylation of the deoxycytosine residues adds to the stability of these tetrahelices (12). When formed along a d(CGG)n-containing DNA template, hairpins and tetraplex structures of the repeat sequence obstruct the progression of DNA polymerases in vitro (15,17,36–38) and arrest DNA replication in vivo (18). Several models implicated secondary structures of the FMR1 d(CGG)n tract in its expansion in fragile X syndrome (18,39–43). In addition to their proposed contribution to d(CGG)n expansion, secondary structures of this sequence may also play a part in the transcriptional silencing of FMR1. First, guanine-rich DNA secondary structures are a preferred target for methyltransferase (10,44), and the ensuing hypermethylation of the repeat sequence and its upstream CpG island is a major (6,45–48), though not exclusive (49), factor in the silencing of FMR1. Secondly, similarly to their effective blocking of DNA polymerases (15,17,50), the thermodynamically stable d(CGG)n hairpins and tetraplexes might physically obstruct the transcription machinery.
The potentially detrimental consequences of secondary structures of d(CGG)n validate a search for agents that act to disrupt these formations. We have shown previously that proteins, WRN helicase (19) and the hnRNP-related proteins qTBP42/CBF-A (20,21) and uqTBP25 (20), destabilize G′2 bimolecular tetraplex forms of d(CGG)n. The principal observation of this report is that the cationic porphyrin TMPyP4 is also capable of disrupting a G′2 bimolecular tetraplex form of the d(CGG)n trinucleotide repeat. Similarly to qTBP42/CBF-A (20), TMPyP4 has a converse effect on the thermal stabilities of G′2 tetraplex structures of the telomeric sequence TeR2 DNA and of 3′-tail d(CGG)7. Results showed that whereas the melting temperature, Tm, of G′2 TeR2 DNA was ∼45°C in the absence of TMPyP4, it was elevated by 13°C to ∼58°C in the presence of the drug. In contrast, TMPyP4 effected a decrease of nearly 15°C in the Tm of G′2 3′-tail d(CGG)7, from ∼46°C to 31°C (Table 1, see also Figs 2 and 3). Likewise, the Tm of G′2 3′-tail d(CGG)7 that was formed in the presence of TMPyP4 was similarly diminished (Fig. 3C and Table 2). That the tetraplex DNA-destabilizing effect of TMPyP4 was not restricted to G′2 3′-tail d(CGG)7 was demonstrated by its capacity to effectively disrupt G′2 forms of non-tailed d(CGG)7, 5′-tailed d(CGG)7 and the hypermethylated d(5-meCGG)7 oligomer (Fig. 4 and Results). It was shown previously that the three cationic porphyrins, TMPyP2, TMPyP3 and TMPyP4, exhibit different binding affinities for different tetraplex DNA molecules (24) and that they vary in their capacity to inhibit the activities of DNA helicase (25) or telomerase (27). Data indicated that the different arrangement of folded strands, groove sizes, and length and base composition of loops in different tetrahelical structures of DNA dictate their dissimilar interaction with the cationic porphyrins (24,28). It is conceivable, therefore, that the opposite effect of TMPyP4 on the thermal stabilities of bimolecular tetraplex structures of telomeric and d(CGG)n sequences is also due to different structural features of the two tetrahelices. Indeed, similarly to TMPyP4, the hnRNP-related protein qTBP42/CBF-A also binds and stabilizes G′2 tetraplex telomeric DNA while disrupting G′2 tetraplex forms of d(CGG)n (20,21,31).
Although both CBF-A and TMPyP4 acted to destabilize G′2 3′-tail d(CGG)7, these two agents could act together to increase the extent of the destabilization of this tetraplex DNA. Results indicated that when CBF-A and TMPyP4 were added together to G′2 3′-tail d(CGG)7, only CBF-A acted to disrupt the tetraplex DNA substrate whereas TMPyP4 became inactive (Fig. 5 and Table 2). Data summarized in Table 2 strongly suggested that the CBF-A protein blocked the access of TMPyP4 to the tetraplex DNA substrate. This is probably due to the greater size and molar excess of the CBF-A protein which competes with TMPyP4 for the same tetraplex DNA target. However, initial destabilization of G′2 3′-tail d(CGG)7 by TMPyP4 alone, followed by exposure to CBF-A, resulted in an almost additive extent of disruption of the tetrahelical DNA structure (Fig. 6). Thus, enhanced destabilization of tetraplex d(CGG)n could be achieved by sequential action of drug and protein.
Clinical manifestations of fragile X syndrome might possibly be reversed by reactivation of FMR1 and synthesis of FMRP in neurons and in cells of other tissues of affected individuals. Together with DNA hypomethylation and histone hyperacetylation, destabilization of secondary structures of d(CGG)n might promote expression of the FMR1 gene in fragile X cells. Thus, an attractive extension of the reported observations is the possibility that secondary structures of the expanded d(CGG)n sequence might be destabilized in vivo. Exposure of fragile X cells to low molecular size drugs and/or proteins such as TMpyP4 or CBF-A, respectively, that disrupt d(CGG)n secondary structures might be instrumental in restoring FMR1 expression.
Acknowledgments
ACKNOWLEDGEMENTS
This study was supported by grants to M.F. from the US–Israel Binational Science Foundation, The Israel Science Foundation, the Conquer Fragile X Foundation Inc. and The Chief Scientist, Israel Ministry of Health.
REFERENCES
- 1.Richards R.I. and Sutherland,G.R. (1997) Dynamic mutation: possible mechanisms and significance in human disease. Trends Biochem. Sci., 22, 432–436. [DOI] [PubMed] [Google Scholar]
- 2.Oostra B.A. and Chiurazzi,P. (2001) The fragile X gene and its function. Clin. Genet., 60, 399–408. [DOI] [PubMed] [Google Scholar]
- 3.O’Donnell W.T. and Warren,S.T. (2002) A decade of molecular studies of fragile X syndrome. Annu. Rev. Neurosci., 25, 315–338. [DOI] [PubMed] [Google Scholar]
- 4.Hansen R.S., Gartler,S.M., Scott,C.R., Chen,S.H. and Laird,C.D. (1992) Methylation analysis of CGG sites in the CpG island of the human FMR1 gene. Hum. Mol. Genet., 1, 571–578. [DOI] [PubMed] [Google Scholar]
- 5.El-Osta A. (2002) FMR1 silencing and the signals to chromatin: a unified model of transcriptional regulation. Biochem. Biophys. Res. Commun., 295, 575–581. [DOI] [PubMed] [Google Scholar]
- 6.Chiurazzi P., Pomponi,M.G., Willemsen,R., Oostra,B.A. and Neri,G. (1998) In vitro reactivation of the FMR1 gene involved in fragile X syndrome. Hum. Mol. Genet., 7, 109–113. [DOI] [PubMed] [Google Scholar]
- 7.Chiurazzi P., Pomponi,M.G., Pietrobono,R., Bakker,C.E., Neri,G. and Oostra,B.A. (1999) Synergistic effect of histone hyperacetylation and DNA demethylation in the reactivation of the FMR1 gene. Hum. Mol. Genet., 8, 2317–2323. [DOI] [PubMed] [Google Scholar]
- 8.Nadel Y., Weisman-Shomer,P. and Fry,M. (1995) The fragile X syndrome single strand d(CGG)n nucleotide repeats readily fold back to form unimolecular hairpin structures. J. Biol. Chem., 270, 28970–28977. [DOI] [PubMed] [Google Scholar]
- 9.Gacy A.M., Goellner,G., Juranic,N., Macura,S. and McMurray,C.T. (1995) Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell, 81, 533–540. [DOI] [PubMed] [Google Scholar]
- 10.Chen X., Mariappan,S.V., Catasti,P., Ratliff,R., Moyzis,R.K., Laayoun,A., Smith,S.S., Bradbury,E.M. and Gupta,G. (1995) Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc. Natl Acad. Sci. USA, 92, 5199–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitas M., Yu,A., Dill,J. and Haworth,I.S. (1995) The trinucleotide repeat sequence d(CGG)15 forms a heat-stable hairpin containing Gsyn·Ganti base pairs. Biochemistry, 34, 12803–12811. [DOI] [PubMed] [Google Scholar]
- 12.Fry M. and Loeb,L.A. (1994) The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA, 91, 4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F.M. (1995) Acid-facilitated supramolecular assembly of G-quadruplexes in d(CGG)4. J. Biol. Chem., 270, 23090–23096. [DOI] [PubMed] [Google Scholar]
- 14.Usdin K. (1998) NGG-triplet repeats form similar intrastrand structures: implications for the triplet expansion diseases. Nucleic Acids Res., 26, 4078–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodford K.J., Howell,R.M. and Usdin,K. (1994) A novel K+-dependent DNA synthesis arrest site in a commonly occurring sequence motif in eukaryotes. J. Biol. Chem., 269, 27029–27035. [PubMed] [Google Scholar]
- 16.Woodford K., Weitzmann,M.N. and Usdin,K. (1995) The use of K+-free buffers eliminates a common cause of premature chain termination in PCR and PCR sequencing. Nucleic Acids Res., 23, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamath-Loeb A.S., Loeb,L.A., Johansson,E., Burgers,P.M. and Fry,M. (2001) Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem., 276, 16439–16446. [DOI] [PubMed] [Google Scholar]
- 18.Samadashwily G.M., Raca,G. and Mirkin,S.M. (1997) Trinucleotide repeats affect DNA replication in vivo. Nature Genet., 17, 298–304. [DOI] [PubMed] [Google Scholar]
- 19.Fry M. and Loeb,L.A. (1999) Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem., 274, 12797–12802. [DOI] [PubMed] [Google Scholar]
- 20.Weisman-Shomer P., Naot,Y. and Fry,M. (2000) Tetrahelical forms of the fragile X syndrome expanded sequence d(CGG)n are destabilized by two heterogeneous nuclear ribonucleoprotein-related telomeric DNA-binding proteins. J. Biol. Chem., 275, 2231–2238. [DOI] [PubMed] [Google Scholar]
- 21.Weisman-Shomer P., Cohen,E. and Fry,M. (2002) Distinct domains in the CArG-box binding factor A destabilize tetraplex forms of the fragile X expanded sequence d(CGG)n. Nucleic Acids Res., 30, 3672–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anantha N.V., Azam,M. and Sheardy,R.D. (1998) Porphyrin binding to quadrupled T4G4. Biochemistry, 37, 2709–2714. [DOI] [PubMed] [Google Scholar]
- 23.Wheelhouse R.T., Sun,D., Han,H., Han,F.X. and Hurley,L.H. (1998) Cationic porphyrins as telomerase inhibitors: the interaction of tetra-(N-methyl-4-pyridyl)porphine with quadruplex DNA. J. Am. Chem. Soc., 120, 3261–3262. [Google Scholar]
- 24.Haq I., Trent,J.O., Chowdhry,B.Z. and Jenkins,T.C. (1999) Intercalative G-tetraplex stabilization of telomeric DNA by a cationic porphyrin1. J. Am. Chem. Soc., 121, 1768–1779. [Google Scholar]
- 25.Han H., Langley,D.R., Rangan,A. and Hurley,L.H. (2001) Selective interactions of cationic porphyrins with G-quadruplex structures. J. Am. Chem. Soc., 123, 8902–8913. [DOI] [PubMed] [Google Scholar]
- 26.Siddiqui-Jain A., Grand,C.L., Bearss,D.J. and Hurley,L.H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA, 99, 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi D.F., Wheelhouse,R.T., Sun,D. and Hurley,L.H. (2001) Quadruplex-interactive agents as telomerase inhibitors: synthesis of porphyrins and structure–activity relationship for the inhibition of telomerase. J. Med. Chem., 44, 4509–4523. [DOI] [PubMed] [Google Scholar]
- 28.Arthanari H., Basu,S., Kawano,T.L. and Bolton,P.H. (1998) Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res., 26, 3724–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fry M., Perrino,F.W., Levy,A. and Loeb,L.A. (1988) Factor D is a selective single-stranded oligodeoxythymidine binding protein. Nucleic Acids Res., 16, 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J. and Russel,D.W. (2001) Molecular Cloning: A Laboratory Manual, Vol. 2, 3rd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 31.Sarig G., Weisman-Shomer,P., Erlitzki,R. and Fry,M. (1997) Purification and characterization of qTBP42, a new single-stranded and quadruplex telomeric DNA-binding protein from rat hepatocytes. J. Biol. Chem., 272, 4474–4482. [DOI] [PubMed] [Google Scholar]
- 32.Han F.X., Wheelhouse,T.T. and Hurley,L.H. (1999) Interaction of TMPyP4 and TMPyP2 with quadruplex DNA. Structural basis for the differential effects on telomerase inhibition. J. Am. Chem. Soc., 121, 3561–3570. [Google Scholar]
- 33.Mariappan S.V., Catasti,P., Chen,X., Ratliff,R., Moyzis,R.K., Bradbury,E.M. and Gupta,G. (1996) Solution structures of the individual single strands of the fragile X DNA triplets (GCC)n·(GGC)n. Nucleic Acids Res., 24, 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitas M., Yu,A., Dill,J., Kamp,T.J., Chambers,E.J. and Haworth,I.S. (1995) Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucleic Acids Res., 23, 1050–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kettani A., Kumar,R.A. and Patel,D.J. (1995) Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol., 254, 638–656. [DOI] [PubMed] [Google Scholar]
- 36.Usdin K. and Woodford,K.J. (1995) CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res., 23, 4202–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang S., Ohshima,K., Shimizu,M., Amirhaeri,S. and Wells,R.D. (1995) Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem., 270, 27014–27021. [DOI] [PubMed] [Google Scholar]
- 38.Weitzmann M.N., Woodford,K.J. and Usdin,K. (1996) The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J. Biol. Chem., 271, 20958–20964. [DOI] [PubMed] [Google Scholar]
- 39.Jeffreys A.J., Barber,R., Bois,P., Buard,J., Dubrova,Y.E., Grant,G., Hollies,C.R., May,C.A., Neumann,R., Panayi,M., Ritchie,A.E., Shone,A.C., Signer,E., Stead,J.D. and Tamaki,K. (1999) Human minisatellites, repeat DNA instability and meiotic recombination. Electrophoresis, 20, 1665–1675. [DOI] [PubMed] [Google Scholar]
- 40.Buard J., Shone,A.C. and Jeffreys,A.J. (2000) Meiotic recombination and flanking marker exchange at the highly unstable human minisatellite CEB1 (D2S90). Am. J. Hum. Genet., 67, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells R.D. (1996) Molecular basis of genetic instability of triplet repeats. J. Biol. Chem., 271, 2875–2878. [DOI] [PubMed] [Google Scholar]
- 42.Chen X., Mariappan,S.V., Moyzis,R.K., Bradbury,E.M. and Gupta,G. (1998) Hairpin induced slippage and hyper-methylation of the fragile X DNA triplets. J. Biomol. Struct. Dyn., 15, 745–756. [DOI] [PubMed] [Google Scholar]
- 43.Gordenin D.A., Kunkel,T.A. and Resnick,M.A. (1997) Repeat expansion—all in a flap? Nature Genet., 16, 116–118. [DOI] [PubMed] [Google Scholar]
- 44.Smith S.S., Laayoun,A., Lingeman,R.G., Baker,D.J. and Riley,J. (1994) Hypermethylation of telomere-like foldbacks at codon 12 of the human c-Ha-ras gene and the trinucleotide repeat of the FMR-1 gene of fragile X. J. Mol. Biol., 243, 143–151. [DOI] [PubMed] [Google Scholar]
- 45.Sutcliffe J.S., Nelson,D.L., Zhang,F., Pieretti,M., Caskey,C.T., Saxe,D. and Warren,S.T. (1992) DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet., 1, 397–400. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y.H. and Griffith,J. (1996) Methylation of expanded CCG triplet repeat DNA from fragile X syndrome patients enhances nucleosome exclusion. J. Biol. Chem., 271, 22937–22940. [PubMed] [Google Scholar]
- 47.deVries B.B., Jansen,C.C., Duits,A.A., Verheij,C., Willemsen,R., van Hemel,J.O., van den Ouweland,A.M., Niermeijer,M.F., Oostra,B.A. and Halley,D.J. (1996) Variable FMR1 gene methylation of large expansions leads to variable phenotype in three males from one fragile X family. J. Med. Genet., 33, 1007–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pietrobono R., Pomponi,M.G., Tabolacci,E., Oostra,B., Chiurazzi,P. and Neri,G. (2002) Quantitative analysis of DNA demethylation and transcriptional reactivation of the FMR1 gene in fragile X cells treated with 5-azadeoxycytidine. Nucleic Acids Res., 30, 3278–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tassone F., Hagerman,R.J., Chamberlain,W.D. and Hagerman,P.J. (2000) Transcription of the FMR1 gene in individuals with fragile X syndrome. Am. J. Med. Genet., 97, 195–203. [DOI] [PubMed] [Google Scholar]
- 50.Grabczyk E. and Usdin,K. (2000) The GAA·TTC triplet repeat expanded in Friedreich’s ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res., 28, 2815–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]