Abstract
The further understanding of the mechanisms of gene regulatory networks requires comprehensive tools for both the representation of complicated signal transduction pathways and the in silico identification of genomic signals that govern the regulation of gene expression. Consequently, sophisticated notation must be developed to represent the signal transduction pathways in a form that can be readily processed by both computers and humans. We propose the regulator–reaction equations combined with detailed attributes including the associated cellular component, molecular function, and biological process and present the simulation-directed graphical notation that is derived from modification of Kohn’s method. We have developed the software suite, CADLIVE (Computer-Aided Design of LIVing systEms), which features a graphical user interface (GUI) to edit large-scale maps of complicated signal transduction pathways using a conventional XML-based representation. The regulator–reaction equations represent not only mechanistic reactions, but also semantic models containing ambiguous and incomplete processes. In order to demonstrate the feasibility of CADLIVE, we constructed a detailed map of the budding yeast cell cycle, which consists of 184 molecules and 152 reactions, in a really compact space. CADLIVE enables one to look at the whole view of a large-scale map, to integrate postgenomic data into the map, and to computationally simulate the signal transduction pathways, which greatly facilitates exploring novel or unexpected interactions.
INTRODUCTION
After the completion of the Human Genome Project, the next major challenge in biology is the detailed understanding of the dynamics of interactions between protein and DNA, through which biological cell functions ultimately emerge (1,2). A ‘signal transduction pathway’ is the common term used to describe the processes and interactions involved with a particular biochemical mechanism. Initial studies on signal transduction were focused mainly on growth factor signals that up- or down-regulate certain genes. Astonishingly rapid progress in this field, which covers model systems from bacteria to humans, demonstrates the importance of understanding various biological processes. The representation and understanding of signal transduction pathways has important applications for biotechnological, pharmaceutical and agricultural purposes.
Systems within a cell orchestrate biochemical interactions to form a signal transduction pathway that imparts a particular cellular function in each tissue or environment. Hence, to understand the processes that drive life, it is important to elucidate not only the function of each individual interaction, but also the role of the associated pathway as a whole. In order to make a complete catalog of substances and the interactions among them, as understood from experimental research data, powerful software for constructing the networks of biochemical interactions is required.
In the case of extensive genome knowledge databases, metabolic pathways are commonly represented by a graph-like structure in which nodes represent reactants, products or enzymes, and edges represent reactions. In signal transduction pathways, many types of molecule, which rarely appear in the metabolic pathway diagram, are involved, e.g. hormones, transmitters, membrane receptors, ion channels and transcription factors. They also contain other complicated intermolecular processes, such as the formation of multimeric complexes, protein modifications and the regulation of transcription. In addition, gene regulatory networks include aspects that are incomplete or are only partly understood.
The difficulties are not due merely to the large number of reactions, since large-scale diagrams of familiar metabolic pathways have been drawn in KEGG (3) and EcoCyc (4). The problem is to properly define the notation representing complicated and numerous signal transduction pathways, including known and unknown reactions (5–8). Although TRANSFAC is one of the most well known databases that focuses on describing signal transduction networks involved in the regulation of transcription factors and aims at furnishing a collection of data (9), more-defined notation is required for simulating a precise pathway map. In a traditional diagram including TRANSFAC, various semantics are used, such as arrows and nodes, which causes substantial confusion and requires reading large parts of the text to understand what the diagram really describes. Without consistent and unambiguous rules for representation, not only is information lost but also misinformation could be disseminated. In order to overcome the problem of representing signal transduction pathways, Kohn proposed a diagrammatical method that included novel symbols representing reactions and components (6). Maimon and Browning introduced link/like boxes to Kohn’s method and developed a new diagrammatic notation, which was targeted for use in the mathematical simulation of signal transduction pathways (10).
With the increase in the number of gene regulatory reactions, there is a great need for comprehensive tools for the in silico identification of genomic signals that regulate gene expression and the representation of detailed signal transduction pathway maps. In order to fulfill these requirements, sophisticated notation must be developed to represent the signal transduction pathway in a form that can be readily processed by both computers and humans, and to develop a user-friendly software suite that conveniently describes all possible signal transduction pathways and stores the detailed model pathways. Representation of the signal transduction pathways requires two features: one is a diagram-based database that can be readily understood by humans, and the other a text-based database that can be automatically processed by computers.
The objective of this study is to propose the regulator– reaction equations with precise attributes including the associated cellular component, molecular function and biological process using conventional XML-based representation, to present the simulation-directed graphical notation based on a modification of Kohn’s method (6), and to develop the software suite, CADLIVE (Computer-Aided Design of LIVing systEms) with a graphical user interface (GUI), which allows the editing of a large-scale map of complicated signal transduction pathways and generates the associated regulator–reaction equations automatically. We demonstrate that CADLIVE designs a large-scale map of the budding yeast cell cycle, which contains 184 molecules and 152 reactions and enables a computer to search or simulate its signal transduction pathways. CADLIVE can be an efficient platform for integrating postgenomic data such as DNA microarray and proteomics into a detailed biochemical map, which greatly facilitates exploring novel or unexpected interactions.
MATERIALS AND METHODS
Application program
CADLIVE is a software suite for constructing large-scale models of complicated biochemical reactions and for storing their regulator–reaction model in a database. Its GUI enables one to draw gene regulatory/metabolic maps in a simple manner, which eliminates the need for laborious, time-consuming and annoying activities typically involved in this process.
CADLIVE has improved on Kohn’s notation for network simulation, which describes complicated signal transduction pathways and metabolic circuits in a form that can be readily processed by both computers and humans. The regulator– reaction equations and their graphical notation are able to represent not only known interactions but also ambiguous reactions in the form of text and diagram. Using sophisticated notation draws complicated reactions compactly in the order of events, such as multicomplex formation, protein modification, regulation of transcription and transportation between organelles. Since our goal for constructing a signal transduction pathway is to infer a gene regulatory network based on various kinds of biological information obtained from high-throughput technology including microarrays and proteomics, or to simulate gene regulatory networks, the representation of the network needs to be processed with a computer automatically.
Several different kinds of database concerning gene regulatory networks have been developed, such as TRANSFAC (9) and KEGG (3), but a unified data format has not yet been established. Designing a large-scale molecular process for the whole cell requires data convertibility among various databases. Thus, the database of CADLIVE employs an architecture using a conventional XML-based representation, which provides the dynamic extensibility and configurability for CADLIVE, where components and reactions can be easily added, removed or exchanged among various systems.
Regulator–reaction equation
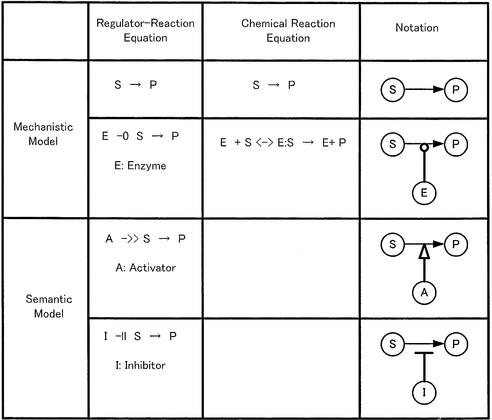
A formula is required to describe the molecular processes of a biological system. Chemical reaction equations are able to describe mechanistic pathways of signal transduction, but the chemical reaction equations are neither suitable for describing unknown pathways nor do they represent a meaningful flow of a signal transduction pathway. They cannot indicate the architecture of regulation, since they do not distinguish regulators (such as enzymes, activators and inhibitors) explicitly from their associated regulated processes. In order to overcome these problems, we propose the regulator– reaction equation, which describes not only mechanistic chemical reactions but also a semantic model of a meaningful flow of biopathways by specifying regulators and their regulated process.
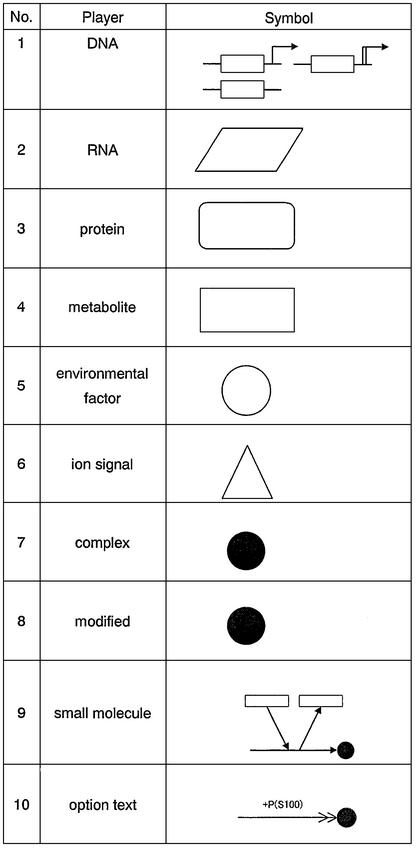
Table 1 shows the fundamental notation of the regulator– reaction equation, where a reaction process is divided into the regulator part and the regulated process. The regulator is classified as one of three different categories: enzyme (–○), activator (–>>) or inhibitor (–||). A regulator arrow acts on a reaction process shown with an arrow. When the enzyme is regarded as a regulator, it catalyzes the molecular reaction indicated by the arrow. In activation or inhibition processes, the regulator symbol acts on the arrow indicating a molecular reaction. Activator- or inhibitor-type regulators can regulate a semantic process that shows the meaningful flow of pathways without knowing any detailed mechanism, such as a shortcut for a complicated regulation or gene expression. On the other hand, the mechanistic model indicates the physical reality of molecular interactions such as enzyme–substrate complex formation and binding between molecules. The regulator– reaction model is able to represent both mechanistic and semantic models explicitly.
Table 1. Regulator–reaction equations and fundamental notation.
Map convention
A major consideration in design of the conventions of diagrams was the capability to trace all known interactions of any given molecular species. Accordingly, each molecular species should ideally appear only once in a diagram, and all interactions involving those species should emanate from a single symbolic object (6). Another important consideration was the presence of a concise method to represent multimolecular complexes. Multimeric proteins are common components of gene regulatory systems and sometimes function in large-scale multimolecular assemblies. Therefore, an extensible representation of such complexes is a fundamental requirement. A third major consideration was the representation of protein modifications, such as phosphorylations. One must be able to represent various modifications of a protein by unique graphical constructs. Often there are many interactions or modification sites that have diverse effects on function. The potential number of modification– multimerization combinations is tremendous, and the representation of all possible combinations of multimers and modifications in a single diagram is not practical. However, it is important to be able to represent combinations that may be significant.
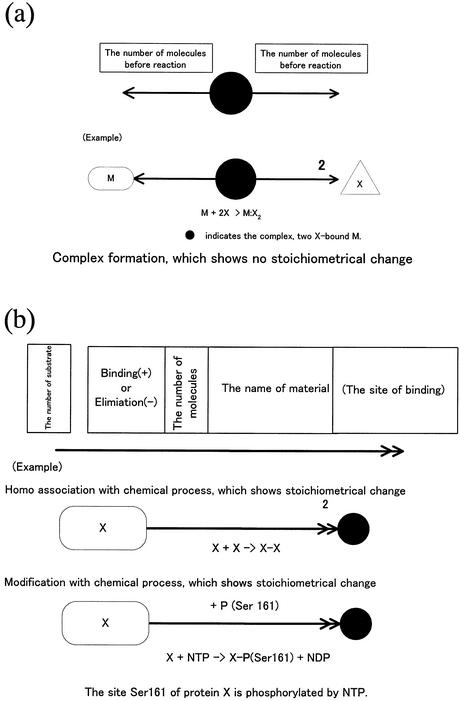
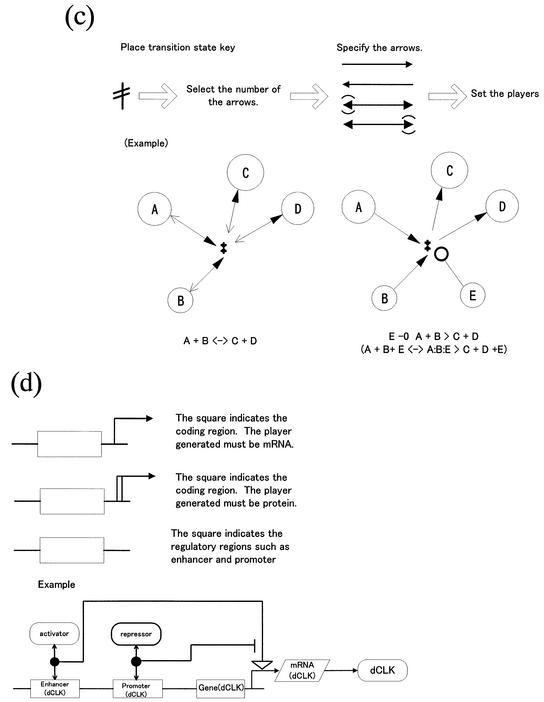
In CADLIVE, a molecule basically appears once in a diagram and its derivatives including modified ones and complexes are indicated by black circles without a name, shown in Figure 1 (Appendix 1: http://www.bse.kyutech.ac.jp/~kurata/NARwww/cadlive.html). This is one of the most important rules featured by the CADLIVE system. The name of a modified material is indicated using a bar (-), e.g. A + ATP → A-P + ADP. On the other hand, the name of a complex using a colon (:), e.g. A + B ↔ A:B. Modification indicates stoichiometric change, whereas complex formation shows no stoichiometric change. Table 2 provides a symbol for each variety of molecules so as to be easily identifiable by humans. The various arrows connecting molecular species must indicate the type of reactions. The proposed notation is established towards computer simulation, thus it emphasizes understanding the order of reaction events. We refer to the novel notations that we have proposed regarding modification, homoassociation, metabolic pathways including multi- substrate and multi-product enzyme reactions, regulation of gene expression, and reaction with cofactors. Most regulatory proteins are susceptible to a multitude of modifications, especially phosphorylation, which alter the function of the proteins. Such complicated modification presents a great challenge to any diagrammatic representation of regulatory processes. The difficulty increases further when various combinations of phosphorylations must also be considered. Kohn’s diagram accurately describes the detailed relationships among components, but it does not provide the step-wise view of specific biological processes, resulting in difficulties in understanding the event order. CADLIVE creates a unique diagram that satisfies both characteristics by employing a double barbed arrow attached by a text box, which points to various modification types and the amino acid positions susceptible to the modification. A complex containing multiple copies of the same monomer can be represented concisely by use of a number in the text box. Using the text box expresses multiple and complicated modifications to a protein according to the realistic order of events without the pill-shaped symbols employed by Kohn’s notation. The detailed notation is defined as shown in Figure 1a and b.
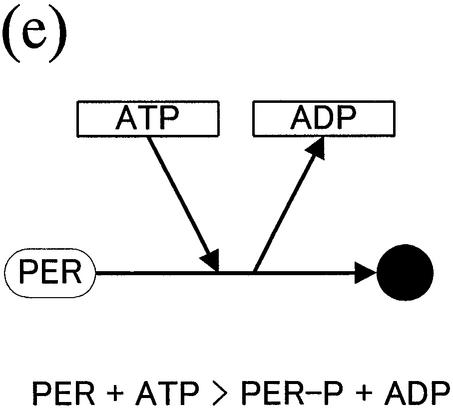
Figure 1.
(Opposite and above) Notations proposed for biochemical reaction networks. (a) Homoassociation and complex formation. (b) Homo association and modification with a chemical process. (c) Multi-substrate and multi-product reaction (metabolic pathways). (d) Gene expression control. (e) Reaction with cofactors.
Table 2. Symbols for players in CADLIVE.
Similar to problems encountered by describing a signal transduction pathway, it is also often hard to describe metabolic pathways definitively, since multiple substrates, cofactors and products participate in an enzymatic reaction, in which many reaction steps occur, and most of the detailed mechanisms are yet to be elucidated. Therefore, using a transition-state symbol is proposed that describes such multiple substrate and product reaction pathways (Fig. 1c). The notation for the regulation of gene expression, as shown in Figure 1d, is almost the same as Kohn’s method. The regulator–reaction model with activator or inhibitor accurately depicts the complexity of gene regulation using enhancer and promoter boxes. Finally, we explain the reaction example that does not follow the basic idea that a molecule appears once in a diagram. As shown in Figure 1e, a new notation is proposed that efficiently describes the reaction involving commonly-used small compounds such as cofactors, coenzymes and vitamins, which avoids increasing the number of arrows that emerge from the compounds.
Pathway search
In order to simulate the signal transduction pathways constructed by CADLIVE computationally, we employed a multiple search tree algorithm, which explores all possible relationships between a key component and a second messenger. The key components are nodes, and the interactions between the components are edges. The multiple tree algorithm searches all the possible second messengers following the key component, and determines the state of each reaction as activation or inhibition. When the increased concentration of a starting molecule causes that of a target molecule to increase, the pathway is defined as an activation pathway. Conversely, when an increase in the concentration of a starting material results in a decrease in the concentration of the target, it is defined as an inhibition pathway. Activation is annotated with a +1 sign, and inhibition with a –1 sign. Multiplying all the signs provides the state of the entire pathway. The multiple search tree determines the pathways and their state, as follows. (i) Search the reaction equations that contain key components, which are described as HEAD (–>>/–||/–○) LEFT → RIGHT, and determine where the key exists, the regulator part (HEAD), the substrate part (LEFT) or the product part (RIGHT). (ii) When the key is found in the HEAD part, the molecules in the substrate and product parts are selected as second messengers. If the pathway is an activation reaction, the concentration of molecules in the substrate decreases, and that in the product increases. If the key is found in the LEFT part, the molecules in the substrate part, with the exception of itself, are inhibited, and those in the product part are activated. (iii) The search is reiterated, setting the second messenger as a new key component.
Prediction of gene regulatory networks
To infer a gene regulatory network of the cell cycle of Saccharomyces cerevisiae, we employed the VoyaGene system (developed by Mitsui Knowledge Industry Co., Ltd.) that predicts the binary relationships of expressed mRNAs using a Bayesian network model. A Bayesian network model inferred the binary relation between mRNAs from the time course data for the mRNA expression, which were synchronized with respect to alpha, Cdc15, Cdc2 and eu (11) (http://genome-www.stanford.edu/cellcycle/). The numbers of the data point are 18 (alpha), 24 (cdc15), 17 (cdc28) and 14 (eu), and the total number is 73. The score value of the Bayesian network model, which varies from –1 to 1, verifies the presence of the binary relationship of mRNAs. The plus sign indicates that the expression of the binary mRNAs is correlated and the minus sign means there is no relationship. The coefficient indicates the regulation type, where the plus sign means activation regulation, and the minus sign shows inhibition regulation. In order to determine an optimal threshold value, the predicted relationship among G1 cyclin genes (Cln1, Cln2, Cln3, Swi4) was compared with those of the known network, changing the threshold value from 1.5 to 4.0. The threshold value of 2.0 showed the most similar network architecture to the known one. Detailed procedure is described elsewhere (Appendix 2: http://www.bse.kyutech.ac.jp/~kurata/NARwww/cadlive.html).
RESULTS AND DISCUSSION
Graphical user interface
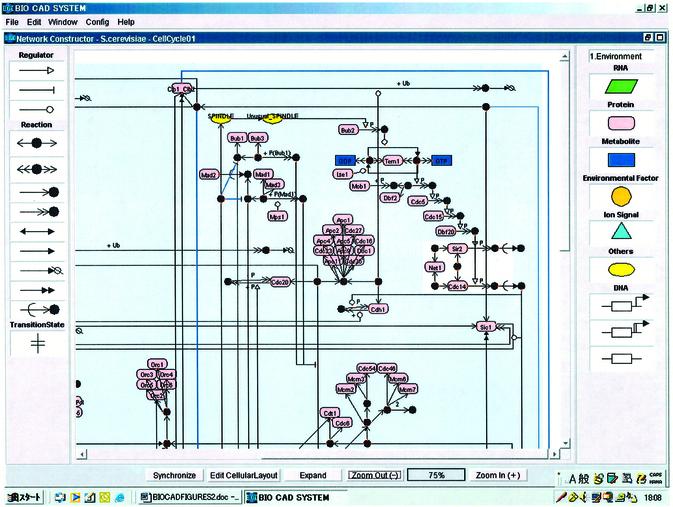
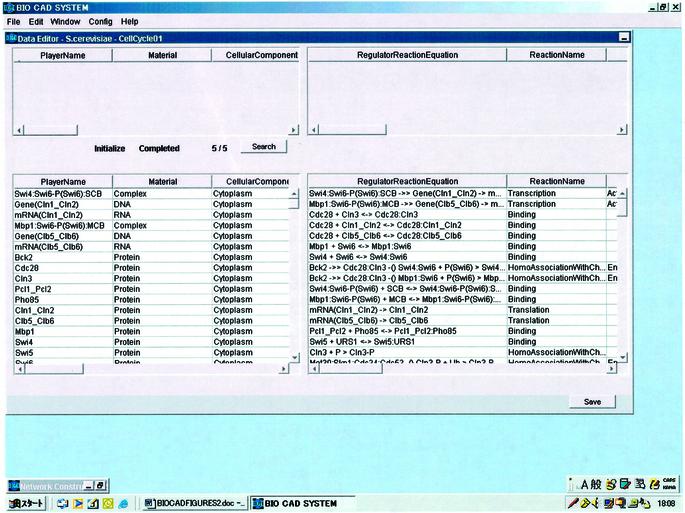
CADLIVE, which is written in Java, features a GUI that allows one to draw and describe a large-scale map of molecular networks. The editor consists of the Network Constructor for drawing a network graphically (Fig. 2) and the Data Editor (Fig. 3), which supports the GUI and can be used for searching the players of interest. The regulator–reaction equations are automatically generated in the Data Editor by drawing a graphical map. The player table describes detailed attributes containing the cellular component that shows where the player works and material class, and the reaction table describes the regulator–reaction equations with detailed tags including reaction types and regulator types, which are necessary for searching or simulating pathways computationally (Appendix 1: http://www.bse.kyutech.ac.jp/~kurata/NARwww/cadlive.html).
Figure 2.
Network editor for drawing a biochemical map graphically.
Figure 3.
Data editor for producing the tables of reactions and players.
Design of a yeast cell cycle map
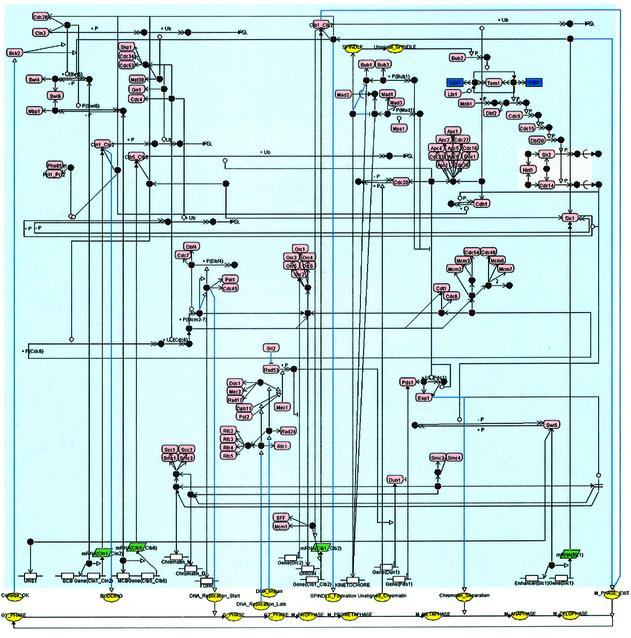
In order to demonstrate the feasibility of CADLIVE, we drew the budding yeast cell cycle map that consists of 184 players and 152 reactions by using CADLIVE, as shown in Figure 4. The diagram can be read from left to right, i.e. the cell cycle proceeds from G1 to M phase through S and G2 phases (12). From the bottom of the map, we draw a flow of the yeast cell cycle events, transcription regulation and protein signal transduction pathways. CADLIVE is able to describe not only reactions but also various events such as budding, DNA replication start and chromatid separation. This map is one of the most sophisticated images of the whole system of yeast cell cycle. The advantage of CADLIVE is not only to draw a large-scale biochemical map according to the well-defined rules, but also to automatically generate the regulator–reaction equations that correspond to the signal transduction pathways of the map.
Figure 4.
Cell cycle map of S.cerevisiae constructed by CADLIVE.
In this section, we concretely explain the budding yeast cell cycle network that CADLIVE has described. Major cell cycle events in budding yeast are controlled by a single cyclin-dependent kinase (Cdc28) in conjunction with two families of cyclins, Cln1-3 and Clb1-6 (13). Since the roles of these cyclins overlap, it is sufficient to consider the interaction of Cdc28 with only four classes of cyclins: (i) Cln1 and Cln2, (ii) Cln3, (iii) Clb1 and Clb2, (iv) Clb5 and Clb6. Cln3:Cdc28 seems to govern the size at which newborn cells execute Start (14). Cln1:Cdc28 and Cln2:Cdc28 play major roles in budding. Clb5:Cdc28 and Clb6:Cdc28 are essential for timely DNA replication. Clb1:Cdc28 and Clb2:Cdc28 are responsible for completion of mitosis. Since the functions of Clb3:Cdc28 and Clb4:Cdc28 can be compensated by other Clbs, they are not essential.
At the beginning of the cycle, the cell has few cyclin molecules, because the transcription factor (SBF=Swi4:Swi6, MBF=Mbp1:Swi4, Mcm1) that regulates cyclin synthesis is inactive (15,16). Clb-dependent kinases are suppressed by a stoichiometric inhibitor (Sic1), and by efficient proteolyis of cyclin subunits by the APC (17,18). Cln3:Cdc28, which is present at low and nearly constant activity throughout the cycle, triggers a sequence of events leading ultimately to cell division (19). When the cell grows to a sufficiently large size, Cln3:Cdc28 and Bck2 activate SBF and MBF, causing Cln2 and Clb5 to accumulate. Clb5 accumulates in inactive trimers of Clb5:Cdc28:Sic1. Cln2:Cdc28 activity plays three important roles. First, it initiates bud formation. Second, it phosphorylates Sic1; subsequently the phosphorylated Sic1 is ubiquitinated and degraded by the SCF. Third, it inactivates Cdh1 by phosphorylation, which, in conjunction with the APC, is responsible for Clb2 degradation at mitotic exit (20). When Sic1 is destroyed, Clb5:Cdc28 activity rises abruptly and drives the cell into S phase (21).
When Sic1 and Cdh1 are inactivated, Clb2:Cdc28 can begin to rise, because Clb2:Cdc28 activates its own transcription factor, Mcm1 and SFF. Clb2:Cdc28 inactivates SBF, thus Cln2 synthesis shuts off and Cln2-dependent kinase activity begins to fall. Simultaneously, MBF is inactivated and the Clb5 level starts to fall. Rising Clb2:Cdc28 induces spindle formation and proceeds to mitosis.
When DNA is fully replicated and all chromosomes are aligned on the metaphase plate, Cdc20 is activated by dephosphorylation (22,23). The activated Cdc20 presents Pds1 to the APC for ubiquitination and degradation, resulting in activating Esp1 that induces chromatid separation (20,24). The activity of Esp1 is suppressed by a stoichiometric inhibitor (Pds1) in the form of Pds1:Esp1. Indirectly Cdc20 promotes dissociation of sister chromatids.
Cdc14 plays a critical role for the exit from mitosis by dephosphorylating Swi5, Sic1, and Cdh1. Cdc14 activates Swi5 by dephosphorylation to transcribe Sic1, the inhibitor for Clb2, and keeps Sic1 in an active form (25–27). The dephosphorylation-activated Cdh1 presents Clb2 to the APC for ubiquitination and degradation. When Sic1 makes a comeback to inhibit Clb2:Cdc28, and the action of Cdh1 destroys Clb2, the cell returns to G1. The activity of Cdc14 is regulated by reversible sequestration between nucleolus and nucleus. Cdc14, which is localized in nucleolus in the inactive form of RENT complex (Cdc14:Sir2:Net1), is released to function when it is necessary (26,28).
Briefly, two major transitions characterize wild-type cell cycles. At Start, a series of events is initiated in rapid succession: SBF turns on, Cln2 and Clb5 levels rise, Sic1 disappears, Cdh1 turns off and DNA synthesis and bud emergence commence. Shortly thereafter, Clb2 level rises and spindles start to form. At Finish, Cdc20 and Cdh1 turn on, Clb2 is destroyed and Sic1 makes a comeback. In simulations of various mutant strains, we will see how these chains of events can be dissociated.
DNA synthesis occurs just once in a cell cycle, which is strictly controlled by a licensing factor, Cdc6 (29,30). The proteins including Orc1-6, Mcm2-6, Cdt1 and Cdc6 form the complex at the replication origin of the chromosomes and are ready for DNA replication, but this complex does not start replication until Clb5:Cdc28 phosphorylates Cdc6. After the replication starts, the phosphorylated Cdc6 is presented to the SCF for degradation, and the complex of Dbf4 and Cdc7 phosphorylates Mcm proteins, which recruits the DNA polymerase to the replication fork to start DNA synthesis. Once Cdc6 disappears, DNA duplication does not occur any longer during this cell cycle. In addition, the transport of Mcm proteins between cytoplasm and nucleoplasm is involved in licensing of DNA synthesis.
Cell cycle implements the checkpoint system to monitor the state of the cycle and to solve problems that occur during the cycle, such as unreplicated DNA, impaired DNA, unaligned chromosome and unusual spindle assembly (31–33). In the DNA replication checkpoint, delay of DNA replication and impaired DNA transmit their signals to Rfc1 and Rad24, respectively, which form the complex with the tetramer of Rfc2,-3,-4,-5. The signals are converged to Mec1, which phosphorylates Rad53. The activated Rad53 stops or delays the cell cycle by suppressing Orc proteins at DNA synthesis level. In the spindle assembly checkpoint (34), the protein of Mad2, which binds to the kinetochore that is not captured by a spindle, is expected to sense the state of spindle assembly. Binding the spindle to the kinetochore releases Mad2, which forms the complex of Mad1:Mad2:Mad3 (35,36). In addition, the heterodimer of Bub1:Bub3 shows a relatively similar behavior of Mad2. Capturing the kinetochore by a spindle releases the dimer. Mad2, Bub1 and Bub3 are involved in sensing the unusual spindle assembly. Through an unknown mechanism, unusual formation of the spindle acts on Mps1 kinase, which phosphorylates Bub1 that is bound to Bub3. The modified complex of Bub1:Bub3 activates Mad1 within the complex Mad1:Mad2:Mad3 by phosphorylation, which results in suppressing the activity of Cdc20, leading to the inhibition of the degradation of Pds1. The deactivated Cdc20 seems to impinge on the APC degradation pathway (37). Consequently, chromatid separation is delayed or stopped. (Mad1, Mad2 and Mad3 form the complex with Cdc20 and APC.) Unusual spindles transmit their signal to Cdc14, enforcing the cell cycle to exit from mitosis, although the detailed mechanism is not clear.
Pathway search
Since the CADLIVE system implements sophisticated regulator–reaction equations with well-defined XML tags, it enables a computer to search signal transduction pathway efficiently. The pathway search program that implements a multiple search tree algorithm is helpful for users to follow the signal transduction pathway of their interest. Table 3 shows a part of the results of the searched pathways, which enhances understanding of the complicated cell cycle network. The Cdc28-cyclin engine drives the cell cycle by phosphorylation. The Cln3-associated Cdc28 phosphorylates MBF (Swi4: Swi6), which activates the transcription of the Cln1/2 genes, and the Cln1/2-associated Cdc28 controls the activity of Sic1 and Cdh1 by phosphorylation. The inhibitor of Sic1, whose activity is controlled by Cdc14 critically, regulates the activity of Clb5 or Clb2, resulting in the DNA replication start or the exit from mitotic phase, respectively. Cdc20 and Cdh1 carry the process of mitotic phase. The DNA replication late is sensed by a checkpoint system, and its signal is transmitted to Cdc6, stopping the cell cycle event. The signal of unusual spindles is transmitted to Cdc14 and Sic1, resulting in the exit from mitosis. Detailed pathways are searched elsewhere (Appendix 3: http://www.bse.kyutech.ac.jp/~kurata/NARwww/cadlive.html). Such a pathway search is very helpful for understanding a large-scale and complicated signal transduction network.
Table 3. Pathway search for helping users understand the process of budding yeast cell cycle.
| Start | End | Pathway | Pathway length |
|---|---|---|---|
| Cln3 | Cln1_2 | Cln3(1) Cdc28:Cln3(1) Swi4:Swi6-P(1) SCB:Swi4:Swi6-P(1) mRNA(Cln1_2)(1) Cln1_2(1) | 6 A |
| Cln1_2 | Cdh1-P | Cln1_2(1) Cdc28:Cln1_2(1) Cdh1-P(1) | 3 A |
| Cln1_2 | Sic1-P | Cln1_2(1) Cdc28:Cln1_2(1) Sic1-P(1) | 3 A |
| Sic1 | DNA_ replication_Start | Sic1(1) Cdc28:Clb5_6(-1) Mcm:Cdc6-P:Cdt1: ORIGIN:Orc(1) Mcm:Cdc6-P-Ub:Cdt1:ORIGIN:Orc(1) Mcm:Cdt1:ORIGIN:Orc(1) Mcm-P:Cdt1:ORIGIN:Orc (1) FORK:DNA_Pol:Cdc45(1) DNA_Replication_Start(1) | 8 I |
| Sic1 | M phase exit | Sic1(1) Cdc28:Clb1_Clb2:Sic1(1) M_PHASE_EXIT(1) | 3 A |
| Cdc14-P(Cytoplasm) | Sic1 | Cdc14-P(Cytoplasm)(1) Sic1(1) | 2 A |
| Cdc14-P(Cytoplasm)(1) Swi5(1) Enhancer(Sic1):Swi5(1) mRNA(Sic1)(1) Sic1(1) | 5 A | ||
| Cdc20 | Chromatin_ separation | Cdc20(1) Apc:Cdc20(1) Esp1:Pds1-Ub(1) Esp1(1) Chromatin_Separation(1) | 5 A |
| Cdh1 | M phase exit | Cdh1(1) Apc:Cdh1(1) Clb1_Clb2(-1) M_PHASE_EXIT(-1) | 4 A |
| DNA_replication_ | DNA_ replication_ | DNA_Replication_Late(1) Rfc1:Rfc(1) Dpb11(1) Mec1(1) Rad53-P(1) Orc2(-1) Orc(1) | 16 I |
| late | start | ORIGIN:Orc(1) Cdc6:Cdt1:ORIGIN:Orc(1) Mcm:Cdc6:Cdt1:ORIGIN:Orc(1) | |
| Mcm:Cdc6-P:Cdt1:ORIGIN:Orc(1) Mcm:Cdc6-P-Ub:Cdt1:ORIGIN:Orc(1) Mcm:Cdt1:ORIGIN:Orc(1) | |||
| Mcm-P:Cdt1:ORIGIN:Orc(1) FORK:DNA_Pol:Cdc45(1) DNA_Replication_Start(1) | |||
| Unusual Spindle | Cdc14-P (cytoplasm) | Unusual_SPINDLE(1) Bub1-P(1) Tem1-GTP(1) (DDF2:MOB1-P)-P(1) Cdc5-P(1) Cdc15-P(1) Dbf20-P(1) Cdc14-P(Nucleosome)(1) Cdc14-P(Cytoplasm)(1) | 9 A |
(1), Activation; (-1), inhibition; A, activation; I, inhibition. Abbreviation: Apc = Apc1:Apc2:Cdc27:Apc4:Apc5:Cdc16:Cdc23:Apc9:Doc1:Apc11:Cdc26; Mcm = Mcm2:Mcm3:Cdc46:Cdc54:Mcm6:Mcm7; Orc = Orc1:Orc2:Orc3:Orc4:Orc5:Orc6; Rfc = Rfc2:Rfc3:Rfc4:Rfc5.
Integration of DNA microarray data into the map
To demonstrate that CADLIVE is an efficient platform to integrate DNA microarray data into a network map, we inferred gene regulations from DNA microarray data, and mapped them on the cell cycle network. Using VoyaGene, we inferred only 16 binary relationships between the mRNAs of Cln1, Clb5, Clb2 and Sic1 that appeared on the cell cycle map, because most of the detailed transcription regulations remain to be described on the map. Therefore, we searched the binary relationships between a protein and an mRNA (Cln1, Cln2, Cln3, Clb1, Clb2, Clb3, Clb4, Clb5, Clb6, Sic1), where the upstream of a protein regulates the downstream of an mRNA, and found 57 relationships on the map, which are shown in Table 4 (Appendix 3, http://www.bse.kyutech.ac.jp/~kurata/NARwww/cadlive.html). The cell cycle map contained the same binary relationships as inferred by VoyaGene, but most of the inferred gene regulations were missed on the map. The binary relationship between Swi4 and the mRNA of Cln1/2 was consistent with the pathway on the map. On the other hand, the relationships between Cln3 and Cln1/2, Mob1 and Cln2, Swi4 and Clb5/6, and Tem1 and Cln1/2 were recognized on the map, but each predicted regulation type did not correspond to that deduced from the map. Such inconsistency of the predicted regulations with the map is probably due to the incompleteness of the biochemical maps, indicating that many regulations, including transcription regulation, remain to be elucidated despite extensive studies in molecular biology.
Table 4. Binary relationship of gene regulations.
| Binary relation | Score value | Upstream process | Downstream process | Regulation type | Map |
|---|---|---|---|---|---|
| Cln3 -| Cln1 | 0.752187 | Initiation | Budding | FI | INC |
| Cln3 -| Cln2 | 0.675588 | Initiation | Budding | FI | INC |
| Swi4 → Cln1 | 0.631578 | Initiation | Budding | FA | CON |
| Swi4 → Cln2 | 0.654600 | Initiation | Budding | FA | CON |
| Swi4 → Clb5 | 0.540447 | Initiation | DNA replication | FA | INC |
| Swi4 → Clb6 | 0.761372 | Initiation | DNA replication | FA | INC |
| Swi4 → Sic1 | 0.634460 | Initiation | DNA replication | FA | |
| Swi4 -| Clb1 | 0.574502 | Initiation | Spindle formation | FI | |
| Swi4 -| Clb2 | 0.736314 | Initiation | Spindle formation | FI | |
| Dun1 → Cln1 | 0.777940 | DNA checkpoint | Budding | FA | |
| Dun1 → Cln2 | 0.787173 | DNA checkpoint | Budding | FA | |
| Rfc3 → Cln1 | 0.574689 | DNA checkpoint | Budding | FA | |
| Rfc5 → Cln1 | 0.542847 | DNA checkpoint | Budding | FA | |
| Ddc1 → Clb6 | 0.700711 | DNA checkpoint | DNA replication | FA | |
| Dpb11 → Clb6 | 0.574967 | DNA checkpoint | DNA replication | FA | |
| Dun1 → Clb5 | 0.520283 | DNA checkpoint | DNA replication | FA | |
| Dun1 → Clb6 | 0.806706 | DNA checkpoint | DNA replication | FA | |
| Rad17 → Clb6 | 0.568040 | DNA checkpoint | DNA replication | FA | |
| Dpb11 -| Sic1 | 0.685889 | DNA checkpoint | DNA replication | FA | |
| Rad24 -| Sic1 | 0.564004 | DNA checkpoint | DNA replication | FA | |
| Mec3 -| Clb5 | 0.716044 | DNA checkpoint | DNA replication | FI | |
| Mec3 -| Clb6 | 0.604183 | DNA checkpoint | DNA replication | FI | |
| Pol2 → Cln1 | 0.533419 | DNA replication | Budding | BA | |
| Pol2 → Cln2 | 0.612008 | DNA replication | Budding | BA | |
| Dbf4 → Cln1 | 0.721617 | DNA replication | Budding | BA | |
| Mcm2 -| Cln1 | 0.549940 | DNA replication | Budding | BI | |
| Mcm2 -| Cln2 | 0.541166 | DNA replication | Budding | BI | |
| Mcm6 -| Cln1 | 0.554601 | DNA replication | Budding | BI | |
| Pcl1 -| Clb1 | 0.621193 | DNA replication | Spindle formation | FI | |
| Pcl2 -| Clb1 | 0.506747 | DNA replication | Spindle formation | FI | |
| Pol2 -| Clb2 | 0.582428 | DNA replication | Spindle formation | FI | |
| Orc1 → Clb1 | 0.722655 | DNA replication | Spindle formation | FA | |
| Orc1 → Clb2 | 0.554201 | DNA replication | Spindle formation | FA | |
| Smc1 → Clb5 | 0.637312 | Chromatid condensation | DNA replication | BA | |
| Smc1 → Clb6 | 0.939309 | Chromatid condensation | DNA replication | BA | |
| Smc3 -| Clb2 | 0.549061 | Chromatid condensation | Spindle formation | FI | |
| Smc1 -| Clb2 | 0.553344 | Chromatid condensation | Spindle formation | FI | |
| Tem1 -| Cln1 | 0.809440 | Spindle checkpoint | Budding | BI | INC |
| Tem1 -| Cln2 | 0.829410 | Spindle checkpoint | Budding | BI | INC |
| Mob1 -| Cln2 | 0.539276 | Spindle checkpoint | Budding | BI | INC |
| Tem1 -| Clb5 | 0.701696 | Spindle checkpoint | DNA replication | BI | |
| Tem1 -| Clb6 | 0.888977 | Spindle checkpoint | DNA replication | BI | |
| Dbf20 -| Clb5 | 0.656491 | Spindle checkpoint | DNA replication | BI | |
| Mob1 -| Clb6 | 0.503613 | Spindle checkpoint | DNA replication | BI | |
| Sir2 → Clb5 | 0.666472 | Spindle checkpoint | DNA replication | BA | |
| Lte1 → Clb6 | 0.630102 | Spindle checkpoint | DNA replication | BA | |
| Tem1 → Clb1 | 0.868281 | Spindle checkpoint | Spindle formation | FA | |
| Tem1 → Clb2 | 0.886017 | Spindle checkpoint | Spindle formation | FA | |
| Dbf20 → Clb1 | 0.698233 | Spindle checkpoint | Spindle formation | FA | |
| Dbf20 → Clb2 | 0.819909 | Spindle checkpoint | Spindle formation | FA | |
| Mob1 → Clb1 | 0.571805 | Spindle checkpoint | Spindle formation | FA | |
| Dbf2 → Clb1 | 0.530131 | Spindle checkpoint | Spindle formation | FA | |
| Dbf2 → Clb2 | 0.500926 | Spindle checkpoint | Spindle formation | FA | |
| Bub1 -| Clb2 | 0.722445 | Spindle assembly | Spindle formation | BI | |
| Mps1 → Clb1 | 0.771476 | Spindle assembly | Spindle formation | BA | |
| Pds1 → Cln1 | 0.520284 | Chromatid separation | Budding | FA | |
| Pds1 → Cln2 | 0.603309 | Chromatid separation | Budding | FA |
Gene regulations among different processes are inferred by the Voyagene System. (→) means activation, and (-|) inhibition. Four kinds of fundamental regulations are shown as follows, FA, BA, FI and BI. (CON) indicates that the binary relation is consistent with the map, (INC) the relation is inconsistent with the map, and blank means no corresponding pathway on the map.
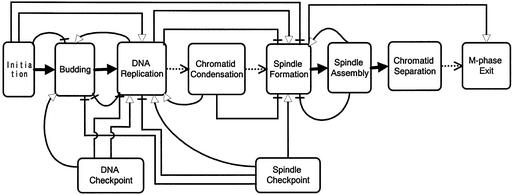
In order to clarify such unknown pathways on the CADLIVE map, the cell cycle was conveniently divided into ten processes, (i) initiation, (ii) budding, (iii) DNA replication, (iv) chromatid condensation, (v) spindle formation, (vi) spindle assembly, (vii) chromatid separation, (viii) M-phase exit, (ix) DNA replication checkpoint and (x) spindle assembly checkpoint. The flow of these processes is as shown in Figure 5, where each process proceeds with time in precise order. The detailed molecular network within each process has been studied intensively, whereas the causation among the processes has been hardly described at molecular level except the flows from initiation to DNA replication and from spindle formation to chromatid separation.
Figure 5.
A flow chart of cell cycle events. Various regulations among the processes are defined as follows, known activation pathway (solid arrow), unknown activation pathway (dotted arrow). The binary relationships inferred from DNA microarray data are defined as activation (open arrow head) and inhibition (no arrow head).
Integrating DNA microarray data into the CADLIVE map helps one understand causations among the processes. In Table 4, the binary relationships are sorted according to the processes in which the gene products function. We mapped the binary relations on the flow chart to specify the interactions among the processes. As shown in Figure 5, four kinds of regulations are presented, forward activation (FA) that a working process activates the subsequent one, forward inhibition (FI) that a working process inhibits the subsequent process such that the subsequent process does not start until the working process completes, backward activation (BA) that a working process activates the former one, and backward inhibition (BI) that a working process inhibits the former one such that the former process does not recur, enabling each event to occur once in a cell cycle. The regulations of FA, FI and BI, which seem to be suitable for processing cell cycle step by step, suggest the significant existence of unknown pathways that play critical roles in the event flow. In contrast, the BA seems to reverse the cycle, which suggests that complicated pathways are involved between the processes, or the components concerned might play another unexpected role in a different process.
Fifty-seven binary relationships of gene regulations between different processes, which are shown in Table 4, are summarized in Figure 5. Some of them indicate BA unexpectedly, but the other regulations seem reasonable for processing cell cycle events in order. Here, we deduce some interesting results out of them. Few pathways that connect chromatid condensation to the subsequent spindle formation have been found on the molecular network map, but DNA microarray data (Table 4) indicate that cohesins (Smc1, Smc3), which are responsible for linking chromatids, work as an inhibitor for the cyclin of Clb2, suggesting that the spindle formation is suppressed during the condensation process, and this inhibitory regulation helps the cell cycle processing its events step by step. Unexpectedly, the cohesins activate the former process (DNA replication), which seems to reverse the cell cycle. Although their detailed mechanism is not clear, these data present an interesting prediction that the structural proteins (cohesins) are involved in complicated communication between DNA replication and spindle formation, and the cohesins might have a regulatory function that has not been expected yet. In the same manner, the components (Mcm2, Mcm6) of the DNA replication process inhibit the former process of budding (Cln1, Cln2), whereas the proteins (Pol2, Dbf4) activate the former one unexpectedly, which seems to reverse cell cycle. The proteins (Pol2, Dbf4)-activated Cln1/Cln2 can be involved not in budding but in subsequent processes such as chromatid condensation and spindle formation. Actually, Cln2 is reported to be involved in spindle pole formation (14). This result suggests that Cln2 has another role in spindle formation, which requires further investigation
Some proteins (Ddc1, Dpb11, Dun1, Rad17) that belong to the DNA checkpoint system induce the process of DNA replication (Clb5, Clb6), which is consistent with our expectation that the checkpoint system should be activated before replication starts. In contrast, other proteins (Mec3, Rad24, Dpb11) in the checkpoint system inhibit genes regarding DNA replication, suggesting that they can be mediators to return signals of impaired DNAs or delay of replication to the replication process. The proteins (Tem1, Mob1, Dbf20, Sir2, Lte1) that belong to the spindle checkpoint system activate those (Clb1, Clb2) responsible for spindle formation, or inhibit the genes involved in budding and DNA replication (Cln1, Cln2, Clb5, Clb6), indicating that the spindle checkpoint system affects its neighboring processes. These regulations are quite reasonable, because the checkpoint system should be activated before the checked process starts, and suppress the former process so as not to occur reversibly. Novel gene regulatory pathways among these processes can be extended around the components.
As mentioned above, the DNA microarray data predict the existence of gene regulatory pathways among different processes. Many interactions have been provided as activation or inhibition. Especially, lots of regulations have been detected in both DNA checkpoint and spindle checkpoint processes, providing a clue to elucidating signal transduction pathways between the checkpoint system and its neighboring processes. Such predicted regulation tells scientists what to do next, which leads to understanding the whole image of the cell cycle network. CADLIVE is demonstrated to be an efficient platform to integrate postgenomic data into the detailed map constructed based on molecular biology.
Mapping protein interaction data on the network
To demonstrate that CADLIVE is useful for integrating proteomics data into the cell cycle map, we mapped the protein–protein interaction data on it. We detected hundreds of interactions between the proteins that appeared on the cell cycle map from BIND database (http://www.bind.ca/). As shown in Table 5, lots of binding relations are verified from the map directly. For example, the DNA checkpoint system (Rfc2, Rfc3, Rfc4, Rfc5, Mec1, Mec3, Ddc1, Dun1, Rad17, Rad24, Rad53) and DNA replication system (Orc1, Orc2, Orc3, Orc4, Orc5) seems to exist as a macromolecular complex as has been expected. In contrast, some relations have not been described on the map. The binding relations that are shown in the database but not found on the map help one finding novel interactions. Especially, the use of protein interactions among different processes can predict unknown signal transduction pathways that connect distant cell cycle processes.
Table 5. Mapping protein–protein interaction data (http://www.bind.ca/) on the cell cycle network.
| Protein 1 | Protein 2 | ||
|---|---|---|---|
| Y | R | N | |
| Cdc28 | Cln1, Cln2, Clb5, Clb2 | Mbp1, Cdh1, Sic1 | Net1 |
| Cln1 | Cln2 | Sic1 | Clb5, Clb2 |
| Cln2 | Sic1 | ||
| Cln3 | Mad3 | ||
| Pcl2 | Pho85 | Swi5 | |
| Clb5 | Clb2 | ||
| Mbp1 | Swi6 | Net1 | |
| Cdc4 | Skp1 | Cdc16, Rad53 | |
| Grr1 | Skp1 | Rad53 | |
| Cdc7 | Cdc53 | ||
| Pol1 | Rfc1, Rfc3, Rfc4, Rfc5 | ||
| Orc1 | Orc2, Orc3, Orc4, Orc5 | ||
| Orc2 | Orc3, Orc4, Orc5 | ||
| Orc3 | Orc4, Orc5 | ||
| Orc4 | Orc5 | ||
| Mcm3 | Mcm2, Cdc46 | Skp1, Cdc53 | |
| Cdc46 | Mcm2, Mcm6 | Skp1, Cdc53 | |
| Rfc2 | Rfc1, Rfc3, Rfc4, Rfc5, Rad24 | Dun1 | |
| Rfc3 | Rfc1, Rfc4, Rfc5, Rad24 | Dun1 | |
| Rfc4 | Rfc1, Rfc5 | Pol2, Mec3, Ddc1 | |
| Rfc5 | Rfc1, Rad24 | Dun1 | |
| Rad24 | Dun1 | ||
| Mec3 | Rad17, Ddc1 | ||
| Rad53 | Dun1, Mec1 | Swi4, Cdc16, Smc3 | |
| Srl2 | Met30, Mcm1, Tem1, Srl2 | ||
| Clb2 | Cdh1 | ||
| Apc1 | Apc2, Cdc27, Cdc16, Cdc23 | ||
| Apc2 | Cdc27, Cdc16, Cdc23, Doc1 | ||
| Cdc16 | Cdc27, Cdc23, Doc1 | ||
| Cdc23 | Doc1 | ||
| Smc1 | Smc3 | Cdc5 | |
| Smc3 | Cdc5 | ||
| Bub3 | Bub1 | Mad3 | |
| Cdc20 | Mad2, Mad3 | ||
| Tem1 | Cdc15 | Pho85 | |
| Mob1 | Dbf2 | Dbf20 | |
| Sir2 | Net1 |
Protein 2 are searched that bind to protein 1. Duplicated data are removed. Binding interactions are recognized on the map (Y), or not detected on the map (N). Proteins seem to interact on the map indirectly (R).
In this study, the protein–protein interaction data present novel relationships that cannot be obtained from DNA microarray data. Out of them, we refer to several binding relationships between the proteins located at distant processes on the map. The protein Rad53 in the DNA checkpoint system interacts with several proteins (Swi4, Smc3, Cdc16) that function in different processes, suggesting that Rad53 is a hub for coordinating various processes such as initiation, chromatid condensation and chromatid separation. For example, the binding between Swi4 and Rad53 suggests that the initiation process (Swi4) is involved in the DNA checkpoint process (Rad53) and may be a key to elucidating the mechanism of how the DNA checkpoint system follows the initiation process. It is interesting to investigate whether Swi4 activates Rad53 through their binding interaction. The binding between Cln3 and Mad3 suggests that the Cln3 cyclin kinase, whose level is kept over the entire period of cell cycle, plays another role in the spindle assembly process. Finally, the binding interactions between Srl2 and Tem1, and between Srl2 and Mcm1 indicate that the processes of DNA checkpoint (Srl2), spindle checkpoint (Tem1) and spindle formation (Mcm1) interact with one another. Srl2 is known to suppress the function of Rad53 (DNA replication checkpoint). These binding interactions can support the DNA microarray data of Tem1→Clb2, assuming that Tem1 binds to Srl2, releasing Mcm1 from the complex Mcm1:Srl2, which results in the induction of Clb2. Srl2 seems a bridge among the processes of DNA checkpoint and spindle formation/assembly. Although there are various possible mechanisms that explain the CADLIVE map with the protein interaction data, we show one possible pathway. Assuming that the binding of Srl2 to Mcm1 and Tem1 causes Mcm1 and Tem1 to reduce their functions, i.e. deactivation of Srl2, suppressed induction of spindle formation, and suppression of the spindle checkpoint, Rad53 becomes active for enhancing the DNA checkpoint system, whereas the spindle formation and its checkpoint process are suppressed. Srl2 can be a switch of the DNA checkpoint system, watching and controlling the spindle formation and spindle checkpoint processes, where Srl2 controls the activities of the regulatory proteins (Tem1 and Mcm1) by sequestering them away. Such sequestration-dependent regulation is seen in other cell cycle systems, e.g. Esp1 is sequestered by Pds1, and Clb2:Cdc28 is captured by Sic1 until they are required to proceed cell cycle events.
CADLIVE employs the idea that one molecule appears once in a map, thereby enabling one not only to draw a detailed network in a really compact space, which shows the whole view of a large-scale map, but also to integrate postgenomic data into it. In addition, the capability of CADLIVE to generate regulator–reaction equations enables one to explore biochemical pathways computationally. These are great advantages, which have not been achieved by other systems, to explore novel or unexpected interactions among the components that are located at distant processes from using postgenomic data. In the postgenomic era, biological predictions would be facilitated by exploring the interaction among the components that are located at distant processes on a map rather than by focusing on local signal transduction pathways intensively.
Towards simulation
The completion of the draft of the human genome sequence reminds one again that the understanding of gene regulatory networks is an important target of genomic research. However, in order to construct the entire gene regulatory network, a huge amount of biological data is required, such as regulation of gene expression, protein–protein interactions, sequence similarity and so on. The magnitude of these data is too large for biologists to deal with by hand. Thus, we propose a simulation-directed notation and the regulator–reaction models that are readable by both humans and computers. We developed CADLIVE with GUI that constructs a genetic network graphically, generates the regulator–reaction equations, and stores both types of information in an XML format. The notation represents a higher-level description for genetic and metabolic networks, and enables one to draw a compact diagram and to generate a regulator–reaction model.
The conventional XML-based representation format provides the dynamic extensibility and configurability of CADLIVE, where components and reactions can be easily added, removed, or exchanged among various databases. It is important to be able to import constructed models separately, since actual biochemical reactions within a cell can be constructed by combining various modules of biological processes. CADLIVE has the capability of importing or exporting various models and features a high compatibility among pathway databases, thus enabling one to build a comprehensive model of all biological processes.
The XML-based regulator–reaction model documentation enables one to process signal transduction pathways by computer, and thus allows the search of pathways. In the near future, to simulate the dynamic features of a large-scale biological system, CADLIVE will represent a powerful tool for constructing these complicated networks using GUI. The regulator–reaction model is very useful for describing semantic and mechanistic models, and it would be feasible to parse and convert the data into a mathematical model automatically. At present, we are developing a simulator with various conversion methods. The combination of CADLIVE with such a simulator will form a powerful system for analyzing the dynamic molecular architecture of a biological system.
The CADLIVE system not only constructs a large-scale map of complicated signal transduction pathways based on the information provided by molecular biology, but also has the capability to map the heterogeneous experimental data derived from DNA microarrays and proteomics studies on a biochemical network of interest. This is because the data produced by DNA microarrays are converted into a meaningful flow between mRNAs, and proteomics data are converted into binding reactions. CADLIVE is expected to become a core system for integrating all the network information in order to elucidate the architecture of a large-scale biological system.
Acknowledgments
ACKNOWLEDGEMENTS
This study was carried out as a part of The Project for Development of a Technological Infrastructure for Industrial Bioprocesses on R&D of New Industrial Science and Technology Frontiers by Ministry of Economy, Trade & Industry (METI), and entrusted by New Energy and Industrial Technology Development Organization (NEDO). We are grateful to Mitsui Knowledge Industry Co., Ltd. for predicting the gene regulatory network of budding yeast cell cycle.
REFERENCES
- 1.Abbott A. (1999) Alliance of US labs plans to build map of cell signalling pathways. Nature, 402, 219–220. [DOI] [PubMed] [Google Scholar]
- 2.Kitano H. (2002) Systems biology: a brief overview. Science, 295, 1662–1664. [DOI] [PubMed] [Google Scholar]
- 3.Kanehisa M., Goto,S., Kawashima,S. and Nakaya,A. (2002) The KEGG databases at GenomeNet. Nucleic Acids Res., 30, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karp P.D., Riley,M., Saier,M., Paulsen,I.T., Collado-Vides,J., Paley,S.M., Pellegrini-Toole,A., Bonavides,C. and Gama-Castro,S. (2002) The EcoCyc Database. Nucleic Acids Res., 30, 56–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirson I., Fortemaison,N., Jacobs,C., Dremier,S., Dumont,J.E. and Maenhaut,C. (2000) The visual display of regulatory information and networks. Trends Cell Biol., 10, 404–408. [DOI] [PubMed] [Google Scholar]
- 6.Kohn K.W. (1999) Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Mol. Biol. Cell, 10, 2703–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voit E.O. (2000) Computational Analysis of Biochemical Systems. Cambridge University Press. [Google Scholar]
- 8.Ichikawa K. (2001) A-Cell: graphical user interface for the construction of biochemical reaction models. Bioinformatics, 17, 438–484. [DOI] [PubMed] [Google Scholar]
- 9.Wingender E., Chen,X., Fricek,E., Geffers,R., Hehl,R., Liebich,I., Krull,M., Matys,V., Michael,H., Ohnhauser,R. et al. (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res., 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maimon R. and Browning,S. (2001) In Yi,T.-M., Hucka,M., Morohashi,M. and Kitano,H. (eds), Proceedings of the Second International Conference on Systems Biology, Pasadena, pp. 311–317.
- 11.Spellman P.T., Sherlock,G., Zhang,M.Q., Iyer,V.R. Anders,K., Eisen,M.B., Brown,P.O., Botstein,D. and Futcher,B. (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell, 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasmyth K. (1996) At the heart of the budding yeast cell cycle. Trends Genet., 12, 405–412. [DOI] [PubMed] [Google Scholar]
- 13.Mendenhall M.D. and Hodge,A.E. (1998) Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 62, 1191–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K.C., Csikasz-Nagy,A., Gyorffy,B., Val,J., Novak,B. and Tyson,J.J. (2000) Kinetic analysis of a molecular model of the budding yeast cell cycle. Mol. Biol. Cell, 11, 369–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasmyth K. and Dirick,L. (1991) The role of SWI4 and SWI6 in the activity of G1 cyclins in yeast. Cell, 66, 995–1013. [DOI] [PubMed] [Google Scholar]
- 16.Koch C., Moll,T., Neuberg,M., Ahorn,H. and Nasmyth,K. (1993) A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science, 261, 1551–1557. [DOI] [PubMed] [Google Scholar]
- 17.Donovan J.D., Toyn,J.H., Johnson,A.L. and Johnston,L.H. (1994) P40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev., 8, 1640–1653. [DOI] [PubMed] [Google Scholar]
- 18.Schwob E., Bohm,T., Mendenhall,M.D. and Nasmyth,K. (1994) The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S.cerevisiae. Cell, 79, 233–244. [DOI] [PubMed] [Google Scholar]
- 19.Wittenberg C., Sugimoto,K. and Reed,S.I. (1990) G1-specific cyclins of S.cerevisiae: cell cycle periodicity, regulation by mating pheromone and association with the p34CDC28 protein kinase. Cell, 62, 225–237. [DOI] [PubMed] [Google Scholar]
- 20.Visintin R., Prinz,S. and Amon,A. (1997) CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science, 278, 460–463. [DOI] [PubMed] [Google Scholar]
- 21.Verma R., Annan,R.S., Huddleston,M.J., Carr,S.A., Reynard,G. and Deshaies,R.J. (1997) Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science, 278, 455–460. [DOI] [PubMed] [Google Scholar]
- 22.Shirayama M., Zachariae,W., Ciosk,R. and Nasmyth,K. (1998) The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J., 17, 1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto A., Guacci,V. and Koshland,D. (1996) Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J. Cell Biol., 133, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciosk R., Zachariae,W., Michaelis,C., Shevchenko,A., Mann,M. and Nasmyth,K. (1998) An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell, 93, 1067–1076. [DOI] [PubMed] [Google Scholar]
- 25.Visintin R., Craig,K., Hwang,E.S., Prinz,S., Tyers,M. and Amon,A. (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell, 2, 709–718. [DOI] [PubMed] [Google Scholar]
- 26.Visintin R., Hwang,E.S. and Amon,A. (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature, 398, 818–823. [DOI] [PubMed] [Google Scholar]
- 27.Jaspersen S.L., Charles,J.F. and Morgan,D.O. (1999) Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr. Biol., 9, 227–236. [DOI] [PubMed] [Google Scholar]
- 28.Shou W., Seol,J.H., Shevchenko,A., Baskerville,C., Moazed,D., Chen,Z.W., Jang,J., Charbonneau,H. and Deshaies,R.J. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell, 97, 233–244. [DOI] [PubMed] [Google Scholar]
- 29.Botchan M. (1996) Coordinating DNA replication with cell division: current status of the licensing concept. Proc. Natl Acad. Sci. USA, 93, 9997–10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishitani H. and Lygerou,Z. (2002) Control of DNA replication licensing in a cell cycle. Genes Cells, 7, 523–534. [DOI] [PubMed] [Google Scholar]
- 31.Burke D.J. (2000) Complexity in the spindle checkpoint. Curr. Opin. Genet. Dev., 10, 26–31. [DOI] [PubMed] [Google Scholar]
- 32.Hartwell L.H. and Weinert,T.A. (1989) Checkpoints: controls that ensure the order of cell cycle events. Science, 246, 629–634. [DOI] [PubMed] [Google Scholar]
- 33.Weinert T. (1997) Yeast checkpoint controls and relevance to cancer. Cancer Surv., 29, 109–132. [PubMed] [Google Scholar]
- 34.Amon A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69–75. [DOI] [PubMed] [Google Scholar]
- 35.Nicklas R.B., Ward,S.C. and Gorbsky,G.J. (1995) Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J. Cell Biol., 130, 929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X. and Nicklas,R.B. (1995) Mitotic forces control a cell-cycle checkpoint. Nature, 373, 630–632. [DOI] [PubMed] [Google Scholar]
- 37.Hwang L.H., Lau,L.F., Smith,D.L., Mistrot,C.A., Hardwick,K.G., Hwang,E.S., Amon,A. and Murray,A.W. (1998) Budding yeast Cdc20: a target of the spindle checkpoint. Science, 279, 1041–1044. [DOI] [PubMed] [Google Scholar]