Abstract
The need to protect DNA from in vivo degradation is one of the basic tenets of therapeutic gene delivery and a standard test for any proposed delivery vector. The currently employed in vitro tests, however, presently provide no direct link between the molecular structure of the vector complexes and their success in this role, thus hindering the rational design of successful gene delivery agents. Here we apply atomic force microscopy (AFM) in liquid to visualise at the molecular scale and in real time, the effect of DNase I on generation 4 polyamidoamine dendrimers (G4) complexed with DNA. These complexes are revealed to be dynamic in nature showing a degree of mobility, in some cases revealing the addition and loss of dendrimers to individual complexes. The formation of the G4–DNA complexes is observed to provide a degree of protection to the DNA. This protection is related to the structural morphology of the formed complex, which is itself shown to be dependent on the dendrimer loading and the time allowed for complex formation.
INTRODUCTION
Despite much progress, there remains a number of well-known obstacles to the development of efficient gene delivery systems. To help overcome these challenges, it is clear that a deeper understanding of the detailed structure of proposed vector complexes and their response to environmental challenges would be of great benefit in the rational design of such systems. We demonstrate this philosophy here in a study of the effect of DNase I on the molecular structure of polyamidoamine (PAMAM) dendrimer–DNA complexes.
Such complexes form through electrostatic interactions between negatively charged phosphate groups of the DNA and protonated amino groups of the dendrimers (1–3). The complexes remain highly soluble, indicating that nuclease resistance may be achieved without forming insoluble complexes (3). In vitro experiments have shown that such dendrimers can chaperone DNA through cell membranes and promote efficient gene transfection (4). In parallel, atomic force microscopy (AFM) has demonstrated potential in studying processes involving DNA (5–8). For example, work imaging static DNA condensates induced by polylysine (6,7) and spermidine (9) has been reported. Liu et al. have also visualised the end point structure of a chitosan–DNA complex after exposure to DNase I (10). In these studies resolution has been achieved by imaging fixed samples in air and therefore information on process dynamics is limited, and structural morphology may contain drying artefacts. A limited number of studies have exploited AFM to visualise dynamical molecular processes involving DNA (5,11–15). Here our studies, which build upon this work, show the effect of the DNase I enzyme on DNA following complexation with increasing ratios and incubation times of generation 4 PAMAM dendrimers (G4).
DNase I is a pancreatic endonuclease, which catalyses the hydrolysis of double-stranded DNA (16). Structural analysis by X-ray crystallography shows that DNase I binds in the minor groove of B-type DNA (17–19). In the presence of divalent metal ions, DNase I cleaves DNA at the phosphodiester bonds that link adjacent nucleotides. In the presence of Mg2+, DNase I hydrolyses each strand of a duplex independently leading to random single strand breaks (SSBs). The purine–pyrimidine bonds are subsequently preferentially cleaved leading to double strand breaks (DSBs) leading to a final product of di- and higher oligonucleotides (16, 20–22). In the presence of Mn2+, the enzyme cleaves both strands of DNA at approximately the same site, leading directly to DSBs.
MATERIALS AND METHODS
G4, based on an ethylenediamine core (calculated formula weight 14 215) containing 64 surface primary amino groups, were used in this study (Dendritech, Midland, MI). Samples were obtained as methanol solutions. To generate aqueous solutions, a portion of the methanolic stock solution was dried under a stream of argon and subsequently placed in a vacuum oven (p ∼ 0.01 Torr) for several hours. The dried material was redissolved in deionised water obtained from ELGA MAXIMA system (Lane End, High Wycombe, Bucks, UK) with water of resistivity of 18.2 Ωcm to yield 20 µg ml–1 aqueous stock solutions, which were stored at 4°C for a maximum of a few days. This method follows previous reported work which avoids hydrolytic degradation of PAMAM dendrimers at room temperature (23). Immediately before use, a fraction of the stock solution was prepared to a concentration that allowed polymer and DNA solutions to be combined in equal volume. The DNA employed was a lyophilised plasmid pBR322, a well-characterised 4363 bp plasmid (Sigma-Aldrich, Poole, Dorset, UK), which was diluted to a stock solution of 10 µg ml–1 in deionised water and further diluted before use to 3.3 µg ml–1 in deionised water, if used for complex formation, or in 10% w/v phosphate buffered saline (PBS; 0.014 M NaCl, 0.001 M phosphate, pH 7.4, Sigma-Aldrich), 1 mM MgCl2 and 1 mM NiCl2, if used for the imaging of bare DNA on mica in the presence or absence of the DNase I enzyme. Water and buffers were filtered through a 0.2 µm pore size filter (Sartorius, Göettingen, Germany) prior to use. DNase I (Sigma-Aldrich, Poole, UK) was diluted with 1 mM PBS, containing 2 mM MgCl2 and 1 mM NiCl2, pH 7.4 just before use, except for the data shown in Figure 3b where the MgCl2 was replaced by MnCl2.
Figure 3.
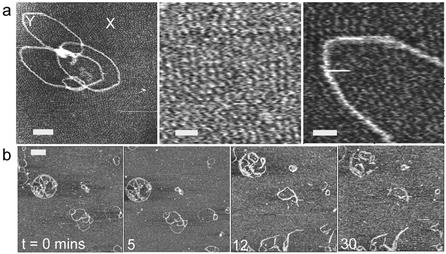
(a) AFM image of a ‘flower-like’ complex formed at 20:1 G4:DNA ratio. Higher resolution images (centred on regions marked X and Y) of the substrate background show features consistent with a densely packed layer of G4. Scale bar and z scale in nm, 50 and 5, 15 and 5, 13 and 5. (b) Time-lapse images showing the effect of DNase I on G4–DNA complex, 1:1 ratio, incubated for 15 min before deposition and imaged in 1 mM PBS, containing 2 mM MnCl2 and 1mM NiCl2, pH 7.4. Scale bar, 100 nm and z scale, 7 nm.
All AFM images were carried out on a Digital Instruments Nanoscope III MultiMode AFM with narrow oxide-sharpened silicon nitride tips (Veeco, Santa Barbra, CA). Imaging was conducted in tapping mode, with 256 × 256 pixel resolution, except for Figure 3a, which is 512 × 512 pixels. Selected fields were scanned at scan rates ranging from 6–7 Hz, each image was acquired within 36.6–43.6 s, except for Figure 3a where each image is aquired within 84 s, and tapping frequencies ranging from 8 to 10.5 kHz in liquid. Images were flattened to account for Z offsets and sample tilt. Setpoints were chosen close to the free oscillation amplitude to minimise forces exerted on the interfacial species. To verify the absence of adsorbed contaminant features, the mica (Agar Scientific, Essex, UK) was first scanned in the presence of deionised water to ensure no contamination was present. The DNA was imaged in the stated buffer so as to determine the configuration of the bare DNA in the absence of both the polymer and the enzyme. Polymer–DNA condensates were prepared by the addition of equal volumes (30 µl) of polymer solution to DNA solution. The quantity of polymer required to produce complexes of a given polymer to DNA ratio was calculated as the ratio between the phosphate groups of the DNA to the primary amino groups of the dendrimers. After a predetermined incubation time was achieved, 30 µl of each resulting solution was spotted as a single aliquot onto the freshly cleaved mica. For example, G4–DNA complexes (0.5:1 ratio) were prepared by adding 30 µl of polymer solution (1.05 µg ml–1) to 30 µl of DNA plasmid pBR322 solution (3.3 µg ml–1) in deionised water, a 30 µl aliquot of the obtained complex is then adsorbed immediately onto mica. Different G4:DNA ratios then were prepared in the same way and incubated for the times stated, at room temperature, adsorbed onto mica and then covered with the liquid cell. After stable imaging of complexes was obtained on freshly cleaved mica, in most cases it took less than a minute, images were recorded for a further 20 min. An aliquot of 15 µl of the fluid in the liquid cell was then exchanged with 15 µl of 2 U/ml DNase I. The time of this exchange was defined as time equals zero. Images were then captured according to the stated times and speeds. For moving features within images, we occasionally changed the x and y offsets to follow these features.
RESULTS AND DISCUSSION
Figure 1a shows a topographical tapping mode AFM image of aggregates of G4 adsorbed at the water–mica interface and acquired upon exposing a mica surface to a 0.1 µg ml–1 aqueous solution of G4. In contrast to essentially featureless images of mica surfaces, a consistent set of small features can be seen which is attributed to adsorbed aggregates of G4 PAMAM dendrimers. Detailed cross-sectional measurements on a large number of such isolated features give an average height of 2.0 ± 0.6 nm and a full width at half-maximum height of 29.6 ± 3.3 nm. Figure 1a also shows a typical image of the plasmid DNA adsorbed to the mica surface, showing characteristic open ‘circle’ morphology. Taken together with other AFM data from this plasmid recorded within this study and also previously (24), morphologies consistent with linear, nicked-circular and supercoiled DNA were observed. This is supported by complementary agarose gel electrophoresis, which shows the pBR322 plasmid to consist of the three forms of DNA in the approximate ratio of 1:2.8:3.6 linear, nicked-circular and supercoiled, respectively (data not shown). Figure 1b depicts an example control AFM study of naked DNA adsorbed onto freshly cleaved mica and exposed to DNase I. This shows that fragmentation due to the accumulation of SSBs of the DNA molecules starts at ∼9 min after the addition of DNase I with complete fragmentation within 22 min (significantly shorter than time periods subsequently observed for G4–DNA complexes). Such data clearly demonstrate the accessibility of the DNase I to the cleavage site of the surface immobilised DNA.
Figure 1.
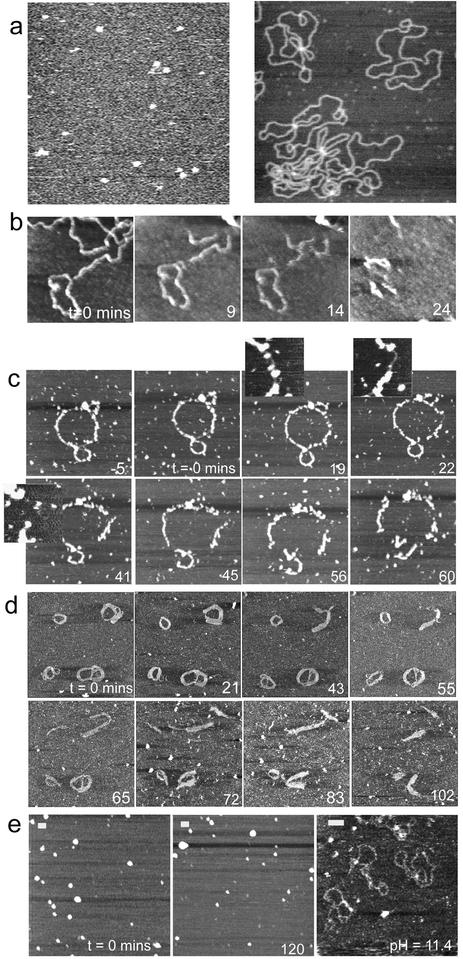
(a) Control AFM images of (left) aggregates of G4 adsorbed at the water–mica interface and acquired upon exposing a mica surface to a 0.1 µg ml–1 aqueous solution of G4 (z scale is 10 nm) and (right) an example AFM image of naked DNA adsorbed onto a freshly cleaved mica surfaces, recorded in 1 mM PBS containing 2 mM MgCl2 and 1 mM NiCl2. Examples of the effect of DNase I enzyme on (b) naked DNA and G4–DNA complexes on mica at (c) 0.5:1, (d) 1:1 and (e) 5:1 ratios. In (e), no degradation is observed within the time frame of the experiment, hence the images show different areas of the sample to illustrate this before pH is raised to disassemble the complexes. Scale bar and z scale in nm, (b) 100, 7, (c) 100, 10, (d) 100, 7 and (e) 100, 7.
Figure 1c–e shows examples of the effect of DNase I on G4–DNA complexes at (c) 0.5:1, (d) 1:1 and (e) 5:1 ratios (4); complexes were prepared in which DNA and G4 molecules were allowed to equilibrate in solution for a few seconds before being directly introduced to the mica and imaged for ∼20 min prior to the addition of the enzyme. The images observed before and at 0 min for all ratios demonstrate the influence of G4 loading upon the morphology of the complexes. For the 0.5:1 ratio (Fig. 1c), features consistent with individual molecules or small aggregates of G4 molecules are observed to decorate the DNA at –5 and 0 min. During the hour after addition of DNase I, a slight relaxation of the ring structure is initially observed via an extension in length of 66 nm to 1442 nm and eventually a fragmentation of the complex. At 0 min the measured thickness and height of the decorated DNA strands were 22 ± 5.5 and 3.7 ± 1.4 nm, respectively, while the measured thickness and height of the undecorated parts were 11.0 ± 0.5 and 0.5± 0.1 nm, respectively. This is consistent with some regions of the DNA plasmid having a high density of bound G4 whilst other regions remain relatively bare. Subsequent image data show this G4 coverage is in a constant state of flux. For example, the highlighted portion of the complex at 19 min, which is initially protected by G4 complexes, becomes bare after 22 min and is rapidly fragmented by 41 min. Figure 1d shows the effect of DNase I on a G4–DNA complex at 1:1 ratio. Here the observed complexes mostly appear as thickened ring-like structures (thickness ranging from 20 to 60 nm), similar to those previously reported for toroidal DNA condensates and their intermediates (10). The thickened regions are made up of parallel strands of condensed DNA molecules, consistent with DNA folding as a mechanism for DNA condensation. Features attributable to individual G4 molecules can no longer be seen to decorate the individual complexes, most likely due to their higher packing density along the DNA. The images recorded over 102 min, subsequent to enzyme addition, show dynamic changes in molecular conformation and a gradual degradation of the complexes. This fragmentation indicates that the enzyme is still able to access the DNA double-helix sufficiently to allow the accumulation of SSBs, and complete strand breakage. It is also interesting to note that regions of the complexes that appear to be composed of several laterally associated strands of DNA display more resistance to enzymatic degradation than isolated strands.
At the higher 5:1 G4:DNA ratio (Fig. 1e), the morphology of the observed condensates is markedly changed and consistent with ‘spheroid’ DNA condensates previously reported (25). Following the addition of DNase I, it can be seen that the complexes are still protected at 120 min without any significant degradation. The measured diameter and height of these features were 69 ± 15 and 10.6 ± 5.7 nm, respectively. Manipulation of the pH of the imaging medium to 11.4, in the absence of the enzyme, caused these complexes to rapidly disassemble, due to neutralisation of the charge on the dendrimers, and for free DNA molecules to be observed within 1 min. This confirms that the spheroids are made up of condensed plasmid molecules. The images displayed in Figure 1c–e thus demonstrate that the morphology of the G4–DNA complexes varies markedly with increasing G4 loading and that the degree of protection afforded by the complex is found to rise with increasing G4 loading, at least up to 5:1.
It is feasible that the complex architecture, and hence its ability to resist degradation, will also depend on the time allowed to form the system. Figure 2 displays images recorded for the 1:1 G4:DNA ratio, in which DNA and G4 molecules were allowed to equilibrate in solution for (a) 15 min and (b) 2 h prior to deposition onto mica. By consideration of the images in Figure 2 and Figure 1d (5 min incubation), it can be seen that for all incubations the morphology of the observed complexes is predominantly toroidal. The importance of the time allowed for complex formation is also apparent. For example, many of the features observed in Figure 1d appear to be composed of several loops, and may thus represent incompletely formed toroidal complexes or multimeric intermediates. In contrast, those complexes observed following longer incubation times are mostly ring-like, toroidal complexes of diameter 70–130 nm. From these and other data, we estimate an average for the observed lifetime until complete degradation of the naked DNA and the complexes at different ratios as 18 ± 8 min for naked DNA (n = 3), 66 ± 8 min for 0.5:1 G4:DNA ratio (n = 5) and 142 ± 37 min for 1:1 G4:DNA ratio (n = 46). The onset of degradation for both the 0.5:1 and the 1:1 ratio complexes was ∼30 min.
Figure 2.
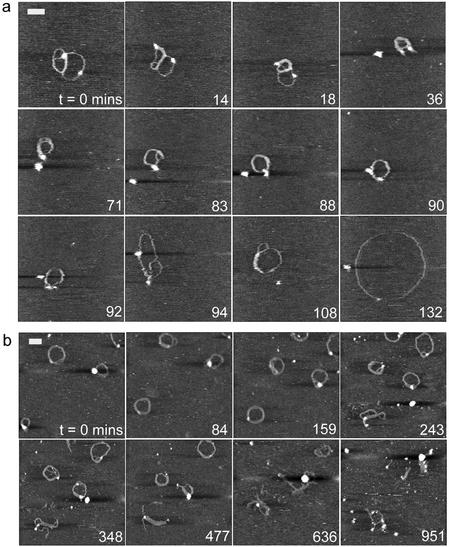
AFM image sequences for the 1:1 G4 to DNA ratio, in which DNA and dendrimer molecules were allowed to equilibrate in solution for (a) 15 min and (b) 2 h prior to deposition onto mica and subsequent exposure to the DNase I enzyme. Scale bar and z scale in nm, (a) 70 and 3, (b) 100 and 5.
The data displayed in Figure 2a are noteworthy since the observed complex is still undergoing structural changes consistent with complexation before a clear enzymatic interaction takes place (71–94 min). Initially a multi-looped molecule is observed with a length of 861 nm, a width of 18.0 ± 1.2 nm and a maximum height at the marked high feature of 3.7 nm. The measured height of the remainder of the complex is 0.8 ± 0.2 nm. Over the next 36 min the DNA becomes increasingly condensed, as demonstrated by an increased width and height of the condensed complex to 27.6 ± 2.3 nm and 1.8 ± 0.3 nm, respectively, and a reduced length of 433 nm. Within the sequence a globular feature consistent with an enzyme molecule is apparent at 36 min. At 90–94 min an abrupt relaxation of the complex is coincident with the highlighted isolated feature ‘binding’ to the complex. The complex shows an immediate increase in observed length of ∼400 nm. The complex then opens out to a relaxed open circle morphology, reaching a maximum observed length of 1535 nm, a thickness of 11.0 ± 0.3 nm and a height of 0.5 ± 0.1 nm.
An increase in the time of incubation before deposition from 15 min to 2 h produces complexes that are consistently condensed into toroidal structures (Fig. 2b) and which do not display the continuing condensation seen in Figure 2a. It is evident that such complexes resist degradation for longer, showing extensive fragmentation only after ∼10 h through an opening of their structure with individual strands becoming partially detached.
It is not immediately apparent why, on some occasions, complexes are degraded to an open relaxed plasmid that is not fragmented as compared to the more common endpoint of complete fragmentation, although it may be related to the initial distribution in the DNA population of relaxed and supercoiled plasmids. From a total of 19 separate experiments, this former pathway was observed twice. In terms of the total number of individual complexes observed undergoing this degradation route, this corresponds to 2 complexes from 44, or ∼5%. We also note that this scenario always leads to a near perfect circular plasmid, which degrades no further, suggesting for this pathway that this is a minimum energy state of the DNA–dendrimer-substrate system, with relaxation of all writhe in the DNA. In this particular case, interactions between the plasmid and the substrate appear to dominate and that this acts to fully open the plasmid and inhibit further action of the DNase I beyond the inclusion of sufficient SSBs to relax the DNA.
Clearly, in interpreting the data, the effect of the substrate on DNA complex structure and behaviour must be considered. For example, one would expect free G4 to bind to the negatively charged mica surface and possibly play a role in condensing and protecting adsorbed DNA. However, control experiments adsorbing DNA to mica pre-treated with G4 showed no evidence of protection of DNA compared to adsorption on bare mica. It is worthy of note that, in experiments with a high excess of G4 to DNA, some evidence of the potential structural effects of the substrate was found. The data in Figure 3a show a ‘flower-like’ complex formed at 20:1 G4:DNA ratio following prior incubation for 2 h. Under such conditions we should expect to observe highly complexed spheroid-like structures (Fig. 1e). We propose that here the excess of dendrimer causes an artefactual structure to form through the immobilisation of the complex at the solid– solution interface. This is supported by higher resolution images of the substrate background in Figure 3a that reveal features consistent with a densely packed layer of the positively charged G4. In close proximity to the complex, this layer appears to order and align features within the complex, some of which now have dimensions consistent with exposed DNA. Hence, we believe that this process is driven by a reorganisation of the surface bound complex due to the attraction of the positive G4 to exposed portions of negative DNA. We propose that such exposed portions result from DNA that becomes transiently free of solution derived G4 (as observed dynamically in Figure 1c). These free regions are then preferentially attracted to the highly packed surface adsorbed dendrimers, causing the opening and distortion of the complex structure observed. Whilst such effects were not observed at lower G4:DNA ratios, they may play a role in the formation of the open loop plasmids such as observed in Figure 2a. Moreover, it is clear that since ‘open loop’ data were recorded in the presence of Mg2+, the action of enzyme leads to SSBs, and hence the open circular endpoint is a possibility. When carrying out similar experiments in Mn2+, in which the enzyme cleaves both strands of DNA, only fragmentation should be observed. This was indeed the case as evidenced by the data shown in Figure 3b.
Clearly there are many factors that delineate the ability of a DNA condensing agent to form stable condensates capable of resisting enzymatic degradation for a sufficient period to facilitate transfection. We have demonstrated an approach based upon direct molecular observation that reveals the effect of structural factors and the process of degradation itself. In the example of a G4 system, we have shown that increasing polymer loading and incubation time can facilitate increased DNA protection. The data reveal the dynamic motion associated with latter stages of condensation and structural changes thereafter, and the effect of introducing DNase I into the environment. As yet it is difficult to completely separate these two processes as even at equilibrium dynamic changes are seen in complex structure (13), and indeed such conformational shifts in complex structure may facilitate the degradation caused by the enzyme. Nevertheless, this approach offers an accessible means of studying a range of complex biomolecular processes at interfaces such as, for example, membrane environments. In this context, a future combination of dynamic imaging with molecular scale sample manipulation and single molecule force spectroscopy approaches (26) would represent a significant step.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Prof. X. Chen, School of Pharmaceutical Sciences, The University of Nottingham, for his valuable discussion on the AFM work. H.G.A. thanks the Egyptian Government and The University of Nottingham for funding a PhD scholarship. S.A. thanks Pfizer Global Research and Development for funding of her lectureship.
REFERENCES
- 1.Hill I.R.C., Garnett,M.C., Bignotti,F. and Davis,S.S. (2001) Determination of protection from serum nuclease activity by DNA–polyelectrolyte complexes using an electrophoretic method. Anal. Biochem., 291, 62–68. [DOI] [PubMed] [Google Scholar]
- 2.Bielinska A.U., Kukowska-Latallo,J.F. and Baker,J.R.,Jr (1997) The interaction of plasmid DNA with polyamidoamine dendrimers: mechanism of complex formation and analysis of alteration induced in nuclease sensitivity and transcriptional activity of the complexed DNA. Biochim. Biophys. Acta, 1353, 180–190. [DOI] [PubMed] [Google Scholar]
- 3.Kukowska-Latallo J.F., Bielinska,A.U., Johnson,J., Spindler,R., Tomalia,D.A. and Baker,J.R.,Jr (1996) Efficient transfer of genetic material into mammalian cells using starburst polyamidoamine dendrimers. Proc. Natl Acad. Sci. USA, 93, 4897–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang M.X., Redemann,C.T. and Szoka F.C. (1996) In vitro gene delivery by degraded polyamidoamine dendrimers. Bioconjugate Chem., 7, 703–714. [DOI] [PubMed] [Google Scholar]
- 5.Bezanilla M., Drake,B., Nudler,E., Kashlev,M., Hansma,P.K. and Hansma,H.G. (1994) Motion and enzymatic degradation of DNA in the atomic force microscope. Biophys. J., 67, 2454–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golan R., Pietrasanta,L.I., Hsieh,W. and Hansma,H.G. (1999) DNA toroids: stages in condensation. Biochemistry, 38, 14069–14076. [DOI] [PubMed] [Google Scholar]
- 7.Hansma H.G., Golan,R., Hsieh,W., Lollo,C.P., Mullen-Ley,P. and Kwoh,D. (1998) DNA condensation for gene therapy as monitored by atomic force microscopy. Nucleic Acids Res., 26, 2481–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umemura K., Nagami,F., Okada,T. and Kuroda,R. (2000) AFM charactarization of single strand-specific endonuclease activity on linear DNA. Nucleic Acids Res., 28, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Y. and Hoh,J.H. (1998) Early intermediates in spermidine induced DNA condensation on the surface of mica. J. Am. Chem. Soc., 120, 8903–8909. [Google Scholar]
- 10.Liu W.G., Yao,K.D. and Liu,Q.G. (2001) Formation of a DNA/N-dodecylated chitosan complex and salt-induced gene delivery. J. Appl. Polym. Sci., 82, 3391–3395. [Google Scholar]
- 11.Kasas S., Thomson,N.H., Smith,B.L., Hansma,H.G., Zhu,X.S., Guthold,M., Bustamante,C., Kool,E.T., Kashlev,M. and Hansma,P.K. (1997) E.coli RNA polymerase activity observed using atomic force microscopy. Biochemistry, 36, 461–468. [DOI] [PubMed] [Google Scholar]
- 12.Pope L.H., Davies,M.C., Laughton,C.A., Roberts,C.J., Tendler,S.J.B. and Williams,P.M. (1999) Intercalation-induced changes in DNA supercoiling observed in real-time by atomic force microscopy. Anal. Chim. Acta, 400, 27–32. [Google Scholar]
- 13.Martin A., Davies,M.C., Roberts,C.J., Stolnik,S., Tendler,S.J.B. and Williams,P.M. (2000) Observation of DNA–polymer condensate formation in real time at a molecular level. FEBS Lett., 480, 106–112. [DOI] [PubMed] [Google Scholar]
- 14.Ono M.Y. and Spain,E.M. (1999) Dynamics of DNA condensates at the solid-liquid interface by atomic force microscopy. J. Am. Chem. Soc., 121, 7330–7334. [Google Scholar]
- 15.Ellis D.J., Dryden,D.T.F., Berge,T., Edwardson,I.M. and Henderson,R.M. (1999) Direct observation of DNA translocation and cleavage by EcoK1 endonuclease using atomic force microscopy. Nature Struct. Biol., 6, 15–17. [DOI] [PubMed] [Google Scholar]
- 16.Pan C.Q., Ulmer,J.S., Herzka,A. and Lazarus,R.A. (1998) Mutational analysis of human DNase I at the DNA binding interface: implications for DNA recognition, catalysis and metal ion dependence. Protein Sci., 7, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suck D. (1997) DNA recognition by structure-selective nucleases. Biopolymers, 44, 405–421. [DOI] [PubMed] [Google Scholar]
- 18.Lahm A. and Suck,D.J. (1991) DNase I-induced DNA conformation 2Å structure of a DNase I-octamer complex. J. Mol. Biol., 221, 645–667. [DOI] [PubMed] [Google Scholar]
- 19.Paul T., Young,M.J., Hill,I.E. and Ingold,K.U. (2000) Strand cleavage of supercoiled DNA by water-soluble peroxyl radicals. the overlooked importance of peroxyl radical charge. Biochemistry, 39, 4129–4135. [DOI] [PubMed] [Google Scholar]
- 20.Weston S.A., Lahm,A. and Suck,D.J. (1992) X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 Å resolution. J. Mol. Biol., 226, 1237–1256. [DOI] [PubMed] [Google Scholar]
- 21.Pan Q.C. and Lazarus,R.A. (1999) Ca2+-dependent activity of human DNase I and its hyperactive variants. Protein Sci., 8, 1780–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fojta M., Kubièárová,T. and Paleèek,E. (1999) Cleavage of supercoiled DNA by deoxyribonuclease I in solution and at the electrode surface. Electroanalysis, 11, 1005–1012. [Google Scholar]
- 23.Hill I.R.C., Garnett,M.C., Bignotti,F. and Davis,S.S. (2001) Determination of protection from serum nuclease activity by DNA–polyelectrolyte complexes using an electrophoretic method. Anal. Biochem., 291, 62–68. [DOI] [PubMed] [Google Scholar]
- 24.Pope L.H., Davies,M.C., Laughton,C.A., Roberts,C.J., Tendler,S.J.B. and Williams,P.M. (2000) AFM studies of intercalation induced changes in plasmid DNA tertiary structure. J. Microsc., 199, 68–78. [DOI] [PubMed] [Google Scholar]
- 25.Rackstraw B.J., Martin,A.L., Stolnik,S., Roberts,C.J., Garnett,M.C., Davies,M.C. and Tendler,S.J.B. (2001) Microscopic investigations into PEG-cationic polymer-induced DNA condensation. Langmuir, 17, 3185–3193. [Google Scholar]
- 26.Scheuring S., Stahlberg,H., Chami,M., Houssin,C., Rigaud,J.L. and Engel,A. (2002) Charting and unzipping the surface layer of Corynebacterium glutamicum with the atomic force microscope. Mol. Microbiol., 44, 675–684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.