Abstract
The binding of the novel cytotoxic acridine derivative, 1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea (ACRAMTU) to various self-complementary oligonucleotide duplexes has been studied by combined high-resolution NMR spectroscopy/restrained molecular dynamics and equilibrium binding assays to establish the sequence and groove specificity of intercalation. The binding mode in the sequences d(GGACGTCC)2 and d(GGAGCTCC)2 was deduced from chemical shift changes and intermolecular NOEs between the ligand and the oligonucleotides. ACRAMTU intercalated into the 5′-CG/CG and 5′-GA/TC base steps, and penetration of the duplexes occurred from the minor groove. Intercalation of ACRAMTU in d(GGTACC)2 occurs at the central TA/TA step, based on the absence of the internucleotide A4H8–T3H1′ and A4H8–T3H3′ cross-peaks in the 1:1 complex of this sequence. An energy- minimized AMBER model of the 1:2 complex, [d(GGAGCTCC)2(ACRAMTU)2], was generated, which was based on restricted molecular dynamics/ mechanics calculations using 108 NOE distance restraints (including 11 DNA–drug distances per ligand). Equilibrium dialysis experiments were performed using octamers containing various base steps present in the ‘NMR sequences’. The highest affinity for ACRAMTU was observed in d(TATAT ATA)2, followed by d(CGCGCGCG)2 and d(GAG ATCTC)2. The binding levels for CG/CG and GA/TC were virtually the same. The unusual tolerance of the GA/TC intercalation site and the pronounced groove specificity of ACRAMTU play a significant role in the molecular recognition between the corresponding platinum conjugate, Pt-ACRAMTU, and DNA.
INTRODUCTION
DNA-directed rational anticancer drug development has made extensive use of the acridine pharmacophore (1). The 9-anilinoacridine derivative amsacrine, 4′-(acridin-9-ylamino)methanesulfon-m-anisidide, has been in clinical use since the 1970s as an investigational drug in the treatment of adult leukemia (2). The cytotoxic effect of amsacrine (m-AMSA) and its second-generation analogues is closely associated with the ability of these DNA intercalators to adversely interfere with the DNA strand cleavage and resealing activity of type II topoisomerase enzymes (3). More recent developments in acridine research include polycyclic acridines (4) and acridine-4-carboxamides (5) that show promising cell kill in a wide range of human cancer cell lines. DACA, a representative of the latter class of 4-substituted acridines currently in clinical trials, has demonstrated dual topoisomerase I and II specificity (6). Such compounds are potential candidates for the treatment of multi-drug resistant cancers (7). Acridine derivatives and related chromophores have also been used as components in drug conjugates to enhance the DNA affinity and efficacy of known clinical and experimental therapeutics, such as cisplatin (8–10), nitrogen mustards (11,12) and groove binders (13).
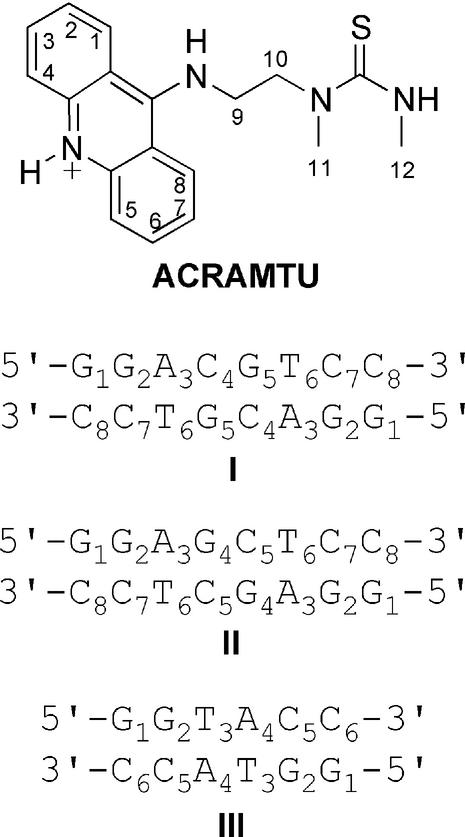
In following our interest in therapeutically useful metal–acridine conjugates exhibiting a spectrum of antitumor activity different from that of established clinical agents, we have developed the cytotoxic complex [PtCl(en)(ACRAMTU-S)]2+ (Pt-ACRAMTU) (14). This conjugate was generated by substituting one of the two chloro leaving groups in [PtCl2(en)] (en = ethane-1,2-diamine), a cisplatin derivative, with sulphur of the novel 9-aminoacridine derivative, 1-[2-(acridin-9-ylamino)ethyl]-1,3-dimethylthiourea (ACRAMTU, acridinium cation, Fig. 1). The presence of only one substitution-labile chloro ligand in Pt-ACRAMTU (thiourea sulphur and diamine are not replaced by DNA nucleophiles) renders this compound a monofunctional covalent DNA modifier. The unwinding of negatively supercoiled DNA by 21°/adduct detected in a gel mobility shift assay indicated a ‘pseudobifunctional’ covalent/intercalative binding mode (15,16) and confirmed the persistence of the sulphur-mediated platinum–acridine linkage in covalent DNA adducts of Pt-ACRAMTU (17). The inability to induce cross-links, we argued, should not only prevent platinum from forming cisplatin-type adducts, but may also enhance binding to sequences dictated by the intercalator rather than the metal moiety. In contrast, in virtually all platinum–acridine conjugates containing the classical cis-diaminedichloroplatinum(II) moiety the metal dominates the binding selectivity leading to adducts in runs of adjacent purine bases in double-stranded DNA (8). The 1,2-intrastrand cross-link is the most abundant adduct and principle cytotoxic lesion induced by bifunctional cisplatinum complexes (18).
Figure 1.
Structure of ACRAMTU giving atom numbering for non-exchangeable protons and sequences of the duplexes used in the NMR study.
The ultimate goal of the present study was to establish the base-step selectivity of intercalation of the acridine chromophore in ACRAMTU and the groove specificity of the 9-thiourea group. The preliminary DNA binding profile of ACRAMTU has been established previously using spectrophotometric drug–DNA titrations, ethidium–DNA fluorescence quenching and competitive drug displacement (17). The acridinium cation [pKa = 9.8 ± 0.1 (14)] showed strong binding to native DNA with Ki = 1.5 × 106 M–1 and an excluded site size (n) of ∼2 bp. In synthetic alternating copolymers, a slight preference for poly(dA-dT)2 over poly(dG-dC)2 was observed. The facile displacement of ACRAMTU by the minor groove binder distamycin A from calf thymus DNA was suggestive of non-intercalating thiourea lying in the minor groove of the polymer. Here, we have investigated the binding of the carrier ligand to model DNA duplexes by high-resolution NMR spectroscopy/molecular modeling and competition dialysis experiments. The data presented strongly suggest that the unique sequence and groove specificity of this adenine-affinic intercalator plays an important role in the molecular recognition between DNA and Pt-ACRAMTU.
MATERIALS AND METHODS
Drug and (bio)chemicals
The synthesis and characterization of the hydrochloride salt of ACRAMTU was described earlier (14). Biochemical grade chemicals (Fisher/Acros, DNase-free, where available) were used for the preparation of biological buffers. All buffers were made from doubly distilled 0.22 µm-filtered DNase/RNase-free water obtained from a Milli-Q A10 synthesis water purification system. All other chemicals and reagents were purchased from common vendors and used without further purification. Stock solutions of acridine were stored at 277 K in the dark.
DNA
The oligodeoxyribonucleotides were purchased from Integrated DNA Technologies Inc. (Coralville, IA). All DNA sequences were synthesized using phosphoramidite chemistry and desalted by the vendor. Oligomers used in the NMR experiments were further treated with Chelex-100 resin (styrene divinylbenzene copolymers containing paired iminodiacetate ions, Bio-Rad) for removal of transition metal ions and subsequently separated from minor failure sequences by size-exclusion chromatography (Sephadex G-25, millipore water, Bio-Rad Biologic HR FPLC workstation). All other oligomers were used as supplied. Quantification of the DNA was done spectrophotometrically in the appropriate buffers at 260 nm using the molar absorptivities provided by the vendor. Spectra were recorded on a Hewlett Packard 8453 UV-visible spectrophotometer equipped with a thermostatted circulating water bath.
Titrations and sample preparation for NMR spectroscopy
The oligonucleotides were dissolved in 500 µl of aqueous sodium phosphate buffer (10 mM phosphate, 100 mM NaCl, 0.1 mM EDTA, pH 6.8) and lyophilized. The palindromic sequences were annealed by slow cooling from 348 K to ambient temperature. The DNA–intercalator complexes were generated by titrating suitable aliquots of a saturated solution of ACRAMTU in D2O to the duplexes. Titrations were performed at 308 K in a 5-mm NMR tube, and a 1H NMR spectrum was recorded after each addition. Integration of the signals of the acridine H2/H7 and thymine methyl protons was used to establish the acridine/duplex ratio. All DNA and complex samples were lyophilized twice from 99.96% D2O and finally dissolved in 500 µl of 99.996% D2O to afford an approximate concentration in (modified) duplex of 3–4 mM. The pH* (uncorrected pH meter reading) of the solutions was 7.0–7.1. NMR samples were stored in the dark at 253 K between measurements.
Acquisition and processing of NMR data
NMR spectra were acquired at 500 MHz on a Bruker DRX-500 spectrometer equipped with an inverse 3-channel Z-gradient TBI probe and a variable temperature unit. The spectral width was 5000 Hz, and the carrier frequency was set to the frequency of the HDO signal in all experiments. Chemical shifts (p.p.m.) were referenced to the residual HDO signal, which was calibrated against external 3-(trimethylsilyl)-1-propanesulfonic acid sodium salt (DSS) for the various acquisition temperatures. One-dimensional (1-D) 1H NMR spectra were recorded with a total of 64 k data points, 32 transients and a recycle delay of 1 s. The time domain data were apodized with an exponential window function using a line broadening of 0.1–0.3 Hz and zero-filled to 128 k data points before Fourier transformation. Spectra were processed using the XWinNMR software (Bruker, Ettlingen, Germany). Two-dimensional (2-D) NMR data sets were acquired with 2048 complex points in t2, 512 points in t1, and 64 transients with a recycle delay of 2 s, except for the dqf-COSY (double quantum-filtered correlation spectroscopy) experiments, which were performed with 1024 complex points in t2, 256 points in t1, and 64 transients per t1 increment. A spin-lock time of 80 ms was used in all TOCSY (total correlation spectroscopy) experiments. 2-D NOESY (nuclear Overhauser enhancement spectroscopy) spectra were acquired with mixing times (τm) of 300 and 400 ms. All data were multiplied with optimized phase-shifted squared sinebell apodization functions and zero-filled to a final matrix of 2048 × 2048 data points before Fourier transformation. The WATERGATE technique (water suppression by gradient-tailored excitation) was used to suppress the residual HDO peak. 2-D data sets were processed with Felix 2000 (Molecular Simulations Inc., San Diego, CA) installed on a Silicon Graphics O2 workstation.
Molecular modeling of the adduct [d(GGAGCTCC)2(ACRAMTU)2]
Calculations were performed using the program Discover interfaced with InsightII (version 2000, Molecular Simulations Inc., San Diego, CA) for structure visualization and analysis. The AMBER force field (19) was used in all molecular mechanics (MM) calculations. Three-dimensional starting structures of the B-form octamer d(GGAGCTCC)2 and the drug molecule were built using the Biopolymer module in InsightII. Fourteen sodium counter ions were placed at a distance of 3.0 Å from the phosphorous atoms to balance the negative charge of the DNA backbone. Solvent was simulated with a distance-dependent dielectric (ε = 4rij), and a cutoff of 20 Å was used for non-bonded interactions with a switching distance of 1.5 Å. Coulombic and 1–4 parameters were scaled by a factor of 0.5. New atom types were introduced for the sp2 hybridized thiourea carbon (CZ) and sulphur (ST) atoms. The assignment criteria for the atom type CA were slightly modified from the original AMBER type to describe the acridine ring carbon atoms. Ad-hoc force constants, bond lengths and bond angles were based on existing parameters and optimized to most accurately reproduce the solid state structure of 1-[2-(acridin-9-ylamino)ethyl]-1,3,3-trimethylthiourea (20). Atomic partial charges of the ligand were generated using a combination of the Gasteiger–Marsili and Hückel methods (21). The duplex and the drug were energy-minimized separately, followed by ‘manual’ insertion of the ligand into the GA/TC base steps from the minor groove.
Interproton NOE distances were calculated from cross peak volumes in NOESY spectra (τm = 400 ms) using Felix 2000 and exported to the Discover module. NOE intensities were calibrated against the H5–H6 (2.45 Å) cross peak of the C5 cytosine base and categorized as strong, medium and weak, with distance bounds of 1.5–2.5, 2.5–3.5 and 3.5–5.0 Å, respectively. For Discover restraints a flat-bottomed potential was used with an upper and lower energy penalty of 200 kcal mol–1 Å–2 and a maximum force of 5 kcal mol–1 Å–1. A total of 86 inter- and intranucleotide distance restraints and 11 intermolecular DNA–drug distance restraints for each ligand were included in the calculations. Additional restraints were introduced between the sodium ions and phosphorous atoms (10 kcal mol–1 Å–2) and Watson–Crick hydrogen bonds (100 kcal mol–1 Å–2) in the restricted molecular dynamics (rMD) simulations. The starting structure was subjected to steepest descent and conjugate gradient minimization until the convergence criteria of 0.01 kcal mol–1 Å–2 was satisfied. The minimized structure was subjected to an rMD–MM protocol consisting of a 10 ps heating phase from 0 to 300 K and 2 ns of dynamics at 300 K using a step size of 1 fs with restraints ‘on’. Along the trajectory, snapshots of the drug–DNA complex were saved every 10 ps. Each of the 200 high-energy structures was minimized according to the above protocol with restraints ‘on’ and then with all restraints ‘off’ to a final maximum derivative of 0.001 kcal mol–1 Å–2. The three lowest-energy structures identified in a cluster graph were superimposed, and the rms deviation from the mean of all atom coordinates was found to be <0.4 Å. Finally, an average structure was calculated using the program Analysis/InsightII. Non-bonded distances between the drug and the octamer were measured in InsightII, and plots were generated from car files using WebLab Viewer Pro (version 3.5; Molecular Simulations Inc., San Diego, CA).
Competition dialysis assay
All dialysis experiments were carried out in a buffer containing 6 mM Na2HPO4, 2 mM NaH2PO4, 1 mM Na2EDTA and 250 mM NaCl pH 7.0. Experiments were performed in a 28-well microdialyser (Gibco BRL) with a 1000 Da molecular weight cut-off membrane at 277 K and at room temperature. For each assay, a DNA concentration of 480 µM (base pairs) was used. Three 1 ml samples of each DNA sequence were pipetted into separate wells of the dialyser and equilibrated against 1000 ml of a 5 µM circulating dialysate solution of ACRAMTU for 60 h. Equal volumes of each sample were removed and treated with a 10% sodium dodecyl sulfate (SDS) solution to a final SDS concentration of 1%. The total concentration of the drug (Ct) present in each sample was determined spectrophotometrically using an extinction coefficient of ε413 = 9450 M–1 cm–1 (17). Absorbance correction was done for dilution effects resulting from the addition of SDS. The free ligand concentration (Cf) was determined using an aliquot of the dialysate solution after the 60 h equilibration period. The amount of drug bound to the oligomer duplexes was determined using the equation Cb = Ct – Cf, where Cb is the concentration of bound drug. The reported data are averages of three individual determinations.
RESULTS
Design of sequences
The self-complementary duplexes used in the NMR investigation are shown in Figure 1. Terminating GG was chosen in all cases to enhance the thermal stability (22) and reduce end fraying of the duplexes in NMR experiments performed at elevated acquisition temperatures. The thermal melting behavior of the unmodified sequences at NMR-relevant concentrations (10–3 M) was predicted from nearest-neighbor thermodynamic data. The melting points (Tm) determined for sequences I–III were 334 (±2), 333 (±2) and 311 (±2) K, respectively (23). The structural integrity of the duplexes was further confirmed by variable-temperature NMR spectroscopy. All sequences were designed to contain only one high-affinity intercalation site, which was hoped to facilitate the interpretation of NMR data. The central CG/CG step in sequence I has been shown to be the major intercalation site of various acridine-based chromophores, such as 9-amino-DACA (24). Simple intercalators usually show preferred intercalation into 5′-pyrimidine–purine-3′ steps (25). Early work on 9-aminoacridine, in fact, suggested that intercalation into the GC step is noticeably disfavored compared to the reverse CG sequence (26). The choice of sequence II containing a central 5′-purine–pyrimidine-3′ step was motivated by the outcome of the study of sequence I, which indicated multiple intercalation sites (vide infra). The central hypothesis investigated in this mechanistic work is that the acridine moiety of the platinum–acridine conjugate Pt-ACRAMTU dictates the sequence specificity of platinum binding. Sequence II contains the three dinucleotide steps, GG, AG and GC, which are the preferred sites of intra- and interstrand cross-link formation of classical platinum drugs (18). Thus, this duplex was useful in assessing if ACRAMTU intercalates into platinum-specific sites. The third potential intercalation site studied was TA/TA in sequence III based on our finding that ACRAMTU, unlike other 9-amino(anilino) derivatives (27), exhibits a high affinity to alternating AT (17).
NMR studies
When ACRAMTU was titrated into buffered D2O solutions of the three oligonucleotides at room temperature, the proton resonances of the ligand and the oligomers in the NMR spectra broadened significantly. Characteristic changes in chemical shifts were observed for both the DNA and acridine protons. Signal shifts (mostly upfield) were most pronounced in the downfield region (6–8 p.p.m.). In variable-temperature spectra taken over a 278–338 K range (shown for [d(GGACGTCC)2ACRAMTU] and [d(GGAGCTCC)2(ACRAMTU)2] in Figs S1 and S2 of the Supplementary Material) a sharpening of all DNA and acridine signals was observed upon increasing the sample temperature, indicating that the oligomer–drug complexes are in an intermediate-to-fast exchange regime at, and below, room temperature (28). Quantitative titrations in the NMR tube and all data acquisitions for signal assignment and structure determination were carried out at elevated temperatures where signals could be resolved without compromising duplex stability. The proton signals in sequences I–III, the corresponding complexes, [d(GGACGTCC)2ACRAMTU], [d(GGAGCTCC)2(ACRAMTU)2], [d(GGTACC)2ACRAMTU] and free ACRAMTU were assigned with the help of 2-D NOESY, TOCSY and dqf-COSY spectra acquired at 308 (I and II) and 303 K (III) (Tables S1–S3 of the Supplementary Information).
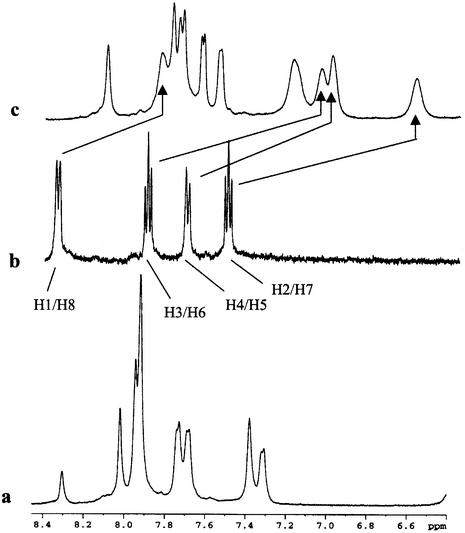
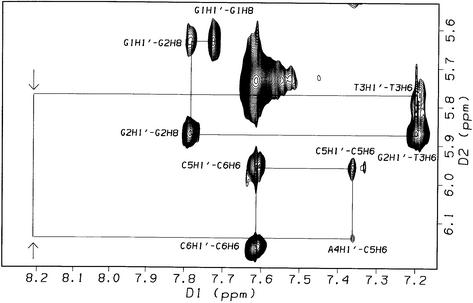
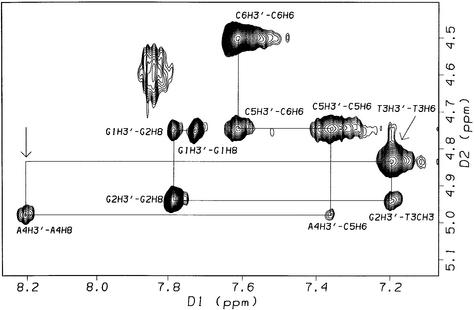
A common spectral feature in 1-D 1H NMR spectra of the three complexes proves to be the pronounced upfield shifts of –0.5 to –1 p.p.m. of the H1–H8 signals of the acridine moiety (see Fig. 1 for atom numbering) upon complex formation, shown for [d(GGACGTCC)2ACRAMTU] in Figure 2, indicating intercalative binding of the planar ring. The methylene H9 and H10 protons in the linker chain are affected in the same way but to a lesser extent. In the three complexes, the order of upfield shifts for these protons is H2/H7 > H3/H6 > H4/H5 >> H1/H8 >> H9 ≈ H10. In contrast, the methyl protons H11 and H12 are affected in the opposite way, resulting in a significant downfield shift of ∼0.3–0.35 p.p.m. The magnitude of chemical shift changes resulting for an intercalated chromophore depends on the relative positioning of the guest molecule with respect to the duplex (28). The observed Δδ values suggest that the H2–H7 containing portion of the planar base penetrates deeply into the base stack, while H1/H8 and the ethylene linker chain lie at the edge of a base pair and the thiourea methyl groups protrude into the DNA groove. The number of acridine and nucleobase proton signals (only one set of pairwise equivalent aromatic protons is observed for the intercalator) would be consistent with acridine intercalating into the unique central CG/CG, GC/GC and TA/TA base steps. A detailed 2-D NMR study, however, demonstrated that fast drug dissociation (‘on/off’) rates on the NMR timescale and exchange of ACRAMTU between multiple binding sites are responsible for the signal multiplicity and the temperature dependence of the line shapes.
Figure 2.
Downfield regions of 1H NMR spectra (500 MHz) showing (a) base proton signals of the sequence d(GGACGTCC)2, (b) aromatic proton signals H1–H8 of ACRAMTU and (c) characteristic chemical shift changes in the 1:1 DNA–drug complex. All spectra were taken at 308 K in 99.996% D2O, 10 mM phosphate, 100 mM NaCl, pH* 7.0.
2-D NOESY spectra (τm = 400 ms, 308 K) of the 1:1 complex, [d(GGACGTCC)2ACRAMTU], show critical intermolecular NOEs between the planar acridine chromophore and the minor-groove H1′ deoxyribose protons at the G2A3/T6C7 and C4G5/C4G5 base steps. Additional cross-peaks result from intermolecular contacts between C7H5 and aromatic acridine protons (Fig. S3). NOEs are observed for acridine H2/H7 and H3/H6, which are closest to the long axis of the chromophore (see Fig. 1), with H1′ sugar protons on both strands indicating that ACRAMTU penetrates the duplex parallel to the base-pair hydrogen bonding, the typical intercalation mode of acridine-type drugs (Nucleic Acid Data Base: http://ndbserver.rutgers.edu/NDB/). Several of the H1′ and H2′/H2′′ sugar protons at the above base steps are shifted upfield (Δδ ≤ –0.3 p.p.m.) with respect to the free oligomer. The most significant change in chemical shift of –0.5 p.p.m., however, is observed for minor-groove A3H2, while the base protons H5, H6 and H8, which are located in the major groove, are virtually unaffected by the presence of ACRAMTU. In summary, the above observations suggest that the single drug molecule in [d(GGACGTCC)2ACRAMTU] intercalates into, and is in fast dynamic exchange between, the central CG/CG and the two symmetry-related GA/TC sites. Rapid on/off rates also explain why internucleotide cross-peak intensities along the ‘NOE walk’ were not significantly reduced at these base steps. The chemical shift changes suggest that intercalation occurs most likely from the minor groove, but no NOEs were observed in this sequence between the 9-thiourea residue and the oligomer that would substantiate this notion.
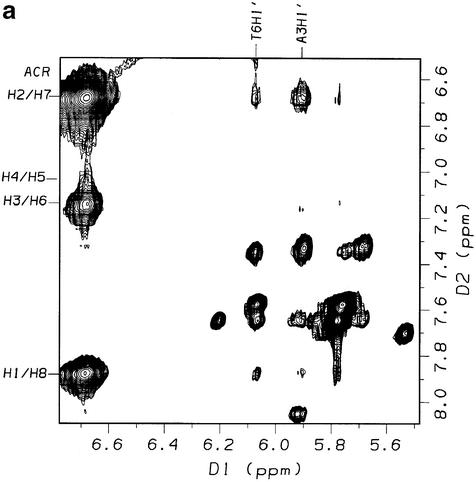
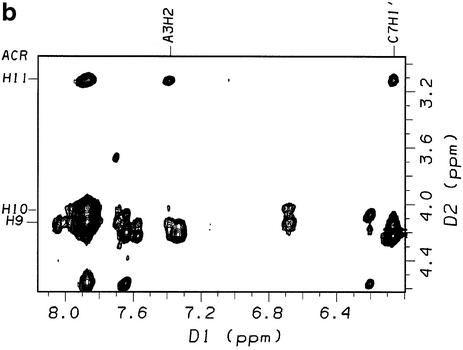
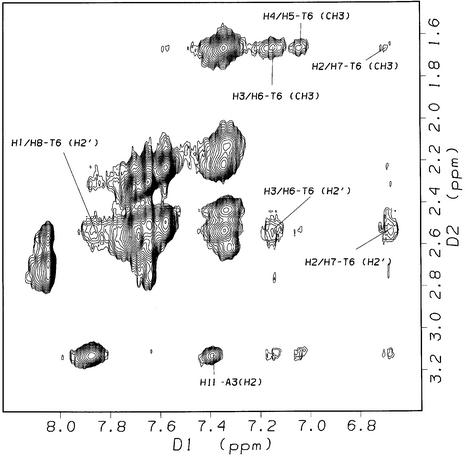
The most informative NMR data were obtained for the 1:2 complex of sequence II, which allowed for a detailed modeling study of the drug binding site using restrained dynamics simulations (vide infra). 2-D NOESY spectra (τm = 400 ms, 308 K) acquired for sequence II after addition of one equivalent of ACRAMTU showed no intermolecular NOEs involving the drug molecule and the GC/GC site, suggesting that reversing the CG dinucleotide sequence abolishes intercalation into the central base step. Instead, the spectra showed evidence for binding in the GA region of the sequence (data not shown). When a second equivalent (slight excess) of the ligand was titrated to the duplex to produce the 1:2 complex, [d(GGAGCTCC)2(ACRAMTU)2], the number of ligand–DNA NOEs increased. The new peaks revealed critical through-space connectivities between the thiourea side chain and the minor groove. The section of the NOESY spectrum presented in Figure 3a shows cross peaks resulting from NOEs between the aromatic acridine protons and T6H1′/A3H1′ at the intercalation site. The number of cross-peaks in this region of the spectrum is greatly reduced compared to the corresponding section for the complex [d(GGACGTCC)2ACRAMTU] (Fig. S3), suggesting that the GA/TC base step in sequence II is the only binding site. Critical intermolecular NOEs at this site are observed between the acridine H11 methyl protons and minor-groove A3H2, and between the aromatic acridine protons H3/H6, H4/H5 and T6CH3 in the major groove (Fig. 4). A very weak NOE also results for H2/H7–T6CH3, which was not included in the structure calculation (see Table 1). H9–H11 of the side chain in ACRAMTU give additional NOEs to A3H2 and C7H1′ (Fig. 3b). The combined 1-D and 2-D NMR data (characteristic Δδ for the 1:2 complex are listed in Table 2) are in agreement with ACRAMTU intercalating into the 5′-GA/TC base steps from the minor groove. The deep penetration of the duplex by the ligand results in major-groove contacts of the acridine chromophores and minor-groove contacts of the non-intercalating thiourea moieties, which are directed toward the ends of the duplex as suggested by the presence of H11–C7H1′ and H12-C8H1′ cross peaks. J-coupling analysis according to Rinkel and Altona (29) based on COSY spectra (Fig. S4) showed that the sums of vicinal couplings involving the H1′ deoxyribose protons (Σ1′) were larger than 14.5 Hz for the entire sequence, indicating that the duplex adopts a typical B-type form.
Figure 3.
Sections of the 2-D NOESY spectrum (500 MHz, τm = 400 ms, 308 K, 99.996% D2O, 10 mM phosphate buffer, 100 mM NaCl, pH* 7.0) of the 1:2 DNA–drug complex [d(GGAGCTCC)2(ACRAMTU)2] showing NOEs between H1′ sugar protons and the acridine chromophore (a) and between the thiourea side chain and minor groove protons of the duplex (b).
Figure 4.
Section of the 2-D NOESY spectrum (500 MHz, τm = 400 ms, 308 K, 99.996% D2O, 10 mM phosphate buffer, 100 mM NaCl, pH* 7.0) of the 1:2 DNA–drug complex [d(GGAGCTCC)2(ACRAMTU)2] showing NOEs between ACRAMTU and the protons both in the minor and major groove of the duplex.
Table 1. Calculated and measured distances (Å) for the energy-minimized structure of the 1:2 DNA–drug complex [d(GGAGCTCC)2(ACRAMTU)2].
| Atoms involved | Calculated rangea | Site Ib | Site IIb | Mean |
|---|---|---|---|---|
| ACRH1/T6H1′ | 3.5–5.0 (w) | 5.26 | 5.34 | 5.30 |
| ACRH2/T6H1′ | 3.5–5.0 (w) | 3.62 | 3.68 | 3.65 |
| ACRH3/T6CH3 | 2.5–3.5 (m) | 3.41 | 3.36 | 3.39 |
| ACRH4/T6CH3 | 2.5–3.5 (m) | 3.57 | 3.61 | 3.59 |
| ACRH11/C7H1′ | 2.5–3.5 (m) | 2.33 | 2.33 | 2.33 |
| ACRH12/C8H1′ | 3.5–5.0 (w) | 3.27 | 3.38 | 3.33 |
| ACRH6/A3H8 | 3.5–5.0 (w) | 4.00 | 4.03 | 4.02 |
| ACRH7/A3H8 | 3.5–5.0 (w) | 3.23 | 3.25 | 3.24 |
| ACRH9/A3H2 | 3.5–5.0 (w) | 4.09 | 4.01 | 4.05 |
| ACRH10/A3H2 | 3.5–5.0 (w) | 3.65 | 3.59 | 3.62 |
| ACRH11/A3H2 | 2.5–3.5 (m) | 3.98 | 3.93 | 3.96 |
aBased on NOE cross-peak volumes in NOESY spectra (τm = 400 ms, 308 K) calculated with Felix 2000. Letters in parentheses refer to the intensity of the NOE: m, medium; w, weak.
bAverage distances for the acridine–DNA complex based on NMR data acquired in a fast exchange regime. Values are given for the two intercalation sites in the freely minimized (‘constraints off’) 1:2 adduct.
Table 2. Changes in NMR chemical shift (Δδ) for minor groove and major groove protons of the oligonucleotide and ACRAMTU protons upon formation of the 1:2 DNA–drug complex [d(GGAGCTCC)2(ACRAMTU)2]a.
| Oligonucleotide | H6/H8 | H5/CH3 | H2 | H1′ | H2′ | H2′′ | ||
|---|---|---|---|---|---|---|---|---|
| G2 | –0.18 | –0.12 | –0.13 | –0.21 | ||||
| A3 | –0.09 | –0.42 | –0.21 | –0.15 | –0.18 | |||
| T6 | –0.17 | +0.04 | –0.05 | –0.04 | –0.03 | |||
| C7 |
+0.12 |
–0.05 |
|
–0.07 |
0.00 |
–0.01 |
|
|
| ACRAMTU | H1/H8 | H2/H7 | H3/H6 | H4/H5 | H9 | H10 | H11 | H12 |
| –0.52 | –0.87 | –0.80 | –0.72 | –0.30 | –0.34 | +0.25 | +0.24 |
a500 MHz, D2O, 308 K, pH* 7.0; shifts are relative to DSS. Δδ = δadduct–δfree DNA/free ACRAMTU (p.p.m.).
In the 1:1 complex [d(GGTACC)2ACRAMTU], intercalation of the drug into the central TA/TA step could be established only indirectly. The absence of both the T3H1′–A4H8 and T3H3′–A4H8 cross peaks along the NOE walk in 2-D NOESY spectra (τm = 300 ms, 303 K) can be explained with selective intercalation of ACRAMTU into this base step (Fig. 5). The absence of the intranucleotide A4H1′–A4H8 cross peak indicates an altered glycosidic bond angle due to perturbation of the duplex structure at the intercalation site. Curiously, no intermolecular NOEs were detected in NOESY spectra taken at various mixing times and acquisition temperatures, indicating that the binding mode in this case may significantly differ from that identified in sequences I and II.
Figure 5.
Sections of the 2-D NOESY spectrum (500 MHz, 303 K, 99.996% D2O, 10 mM phosphate buffer, 100 mM NaCl, pH* 7.1) of the 1:1 DNA–ligand complex, [d(GGTACC)2ACRAMTU]. The arrows indicate disruption of the A4H8–T3H1′/A4H8–A4H1′ (top) and A4H8–T3H3′ (bottom) NOE walk at the proposed TA/TA intercalation site.
Equilibrium binding experiments
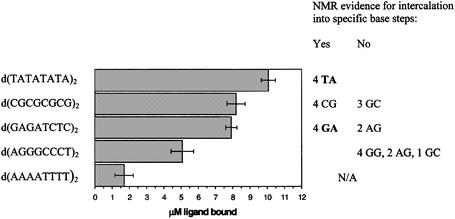
To further elucidate the thermodynamic basis of the sequence selectivity of ACRAMTU, its binding affinity was studied in five palindromic octanucleotides using a customized version of the competition dialysis assay of Ren and Chaires (30). The sequences d(TATATATA)2, d(CGCGCGCG)2 and d(GAGATCTC)2 contained the intercalation sites identified by NMR spectroscopy. To allow for direct comparison, duplexes of identical length containing an equal number of intercalation sites were chosen. In addition, the sequence d(AGGGCCCT)2 was used in this study, which contained the base steps that had proven to be disfavored recipient sites for ACRAMTU. The sequence d(AAAATTTT)2, a known low-affinity sequence for acridines (27), was included for comparison. In experiments performed at 277 K (Fig. 6), the alternating TA sequence showed the highest affinity followed by CG and GA. At room temperature, the association of ACRAMTU with d(TATATATA)2 decreased dramatically due to melting of the duplex (Fig. S5), corroborating the fact that ACRAMTU is a double strand-specific DNA binder. The observed preference for TA over CG confirms the trend in affinities established previously in the corresponding alternating copolymers, poly(dA-dT)2 and poly(dG-dC)2 (17). The amount of drug bound to the GA and CG sequences was virtually identical, while the binding levels in the other purine–purine sequences, AG, GG and AA, were significantly reduced.
Figure 6.
Results of the competition dialysis assay for ACRAMTU. Base steps relevant to the ‘NMR sequences’ are shown on the right. The bars give averages of 3–4 individual experiments.
Molecular modeling of the 1:2 complex [d(GGAGCTCC)2(ACRAMTU)2]
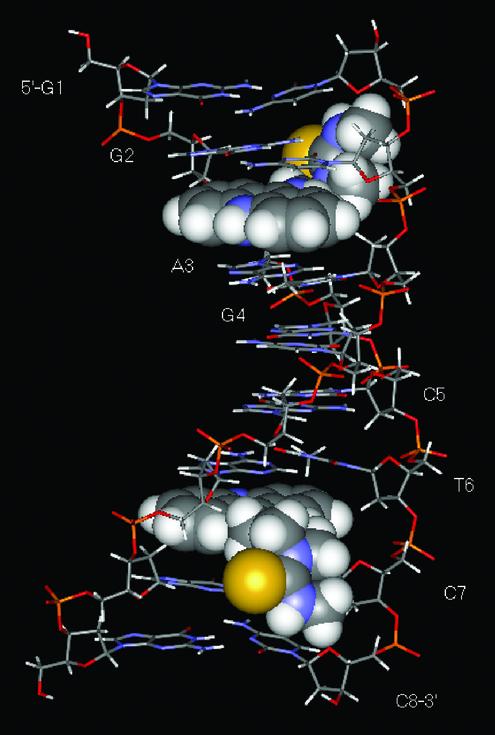
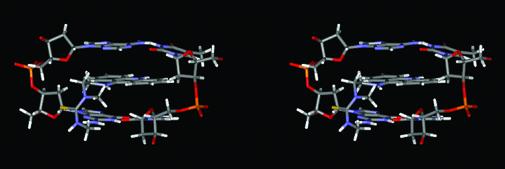
A number of NOEs were indentified in the 2-D NOESY spectra of the 1:2 complex [d(GGAGCTCC)2(ACRAMTU)2] between ACRAMTU and the double-stranded oligomer. Insertion of the planar chromophore into the duplex occurs in a manner that produces NOEs between the drug molecule and both DNA strands at, and beyond, the intercalation site. NOE-derived distance information in conjunction with MD and MM calculations was used to generate an energy-minimized structure of this complex. The NMR restraints were based on a total of 108 selected NOEs that gave unambiguous and well-resolved cross peaks, including 22 drug–DNA through-space connectivities. The minimized structure of the complex shows pseudo 2-fold symmetry with the two ACRAMTU molecules intercalating into the GA/TC base steps from the minor groove of a B-form duplex (Fig. 7). The thiourea side chains are oriented away from each other. Calculated and measured distances between ligand and DNA non-exchangeable protons are summarized in Table 1. A stereoview of the intercalation site (Fig. 8) shows that the planar chromophore is deeply buried in the helical stack to maximize π-stacking interactions with the sandwiching base pairs while the thiourea moiety is oriented almost perpendicular to the acridine ring and makes contact with the walls of the minor groove.
Figure 7.
Energy-minimized averaged AMBER structure of the 1:2 complex [d(GGAGCTCC)2(ACRAMTU)2] resulting from a 2 ns quenched molecular dynamics simulation.
Figure 8.
Stereoview of the drug binding site in [d(GGAGCTCC)2(ACRAMTU)2] from the minor groove.
DISCUSSION
In this study the preferential intercalation of ACRAMTU into the 5′-CG/CG, 5′-GA/TC and 5′-TA/TA base steps was demonstrated. In two of the guanine-rich model duplexes studied by NMR spectroscopy, the ligand has been unequivocally shown to penetrate the base stack from the minor groove. Previous physico-chemical studies indicated that the binding of ACRAMTU to native DNA was an order of magnitude stronger than that of 9-methylaminoacridine (17). The combined NMR/modeling data suggest that hydrophobic interactions between the alkylated thiourea group and the minor groove may be responsible for the increased binding affinity of ACRAMTU compared to the simple intercalator. The distinct minor-groove affinity of ACRAMTU is in contrast to acridine-4-carboxamides (24,31), whose protonated side chains in the 4-position protrude into the major groove where they make guanine-specific hydrogen bond contacts with nucleobase nitrogen. The exact intercalation modes of other 9-substituted acridines, such as amsacrine, are still unknown. Recently, polycyclic acridine derivatives have been described that show little groove specificity leading to multiple orientations of the drug molecule at the intercalation site (32). The relative positioning of side chains in intercalator molecules may be an important factor in the recognition of the DNA adducts by DNA processing enzymes.
ACRAMTU displays a truly unique DNA binding profile. Both the TA and CG steps appear to be high-affinity intercalation sites. The thermodynamically rigorous equilibrium dialysis experiments demonstrate that the TA sequence is favored over the CG sequence only by a factor of 1.2. Previous physical data acquired in copolymers suggested a 1.6-fold preference (17). In addition to this dual selectivity, ACRAMTU shows an unprecedented tolerance of the purine–purine GA/TC base step. 2-D NMR and equilibrium binding data demonstrate that the affinities for this site and for the classical pyrimidine–purine sequence CG/CG are virtually the same. Very few intercalators have shown purine–purine (or pyrimidine–pyrimidine) selectivity (33). In sequences containing multiple adjacent purines, a distinct preference of ACRAMTU for the 5′-GA sequence over AG, GG and AA was observed.
ACRAMTU itself showed a cytotoxic effect in cancer cell lines (14) that is possibly mediated by topoisomerase enzymes (34). This novel sulphur-modified 9-aminoacridine derivative has been introduced as a substitution-inert (non-leaving) carrier group in Pt-ACRAMTU, the prototype of a new class of cytotoxic platinum–acridine conjugates. In a recent study (35) we have identified the covalent adducts formed by Pt-ACRAMTU in calf thymus DNA by in-line liquid chromatography/mass spectrometry (LC/MS) and NMR spectroscopy. In a substantial fraction of adducts, platinum was found to bind to adenine at GA and TA dinucleotide sites. These findings indicate that platinum has been ‘hijacked’ away from its natural target, guanine-N7, resulting in the platination of sites in DNA usually not favored by this metal. In the light of the current findings it appears that the covalent platinum adducts are formed at the preferred intercalation sites of the carrier ligand. Based on the groove specificity of ACRAMTU, established by NMR spectroscopy in this study, it is proposed that thiourea-linked platinum in the conjugate is directed into the minor groove where it binds to adenine, possibly at the N3 position. While adenine-N3 is the major target of various alkylating agents (36), including nitrogen mustards linked to 9-anilinoacridine derivatives (37), platination of this site in double-stranded DNA is unknown. In summary, we have explored the molecular recognition between ACRAMTU and DNA in model duplexes using combined NMR spectroscopy/molecular modeling and equilibrium binding experiments. We hypothesize that in a substantial fraction of covalent DNA adducts of Pt-ACRAMTU the intercalator dictates the sites of platination, leading to covalent lesions hitherto unknown for platinum-based drugs. These findings open new avenues in the design of DNA-targeted organic–inorganic hybrid pharmacophores.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Marcus W. Wright for his assistance with solving NMR-related problems. This work was supported by the American Cancer Society (Grant IRG 93-035-6) and the North Carolina Biotechnology Center (Grant 2001-ARG-0010).
REFERENCES
- 1.Denny W.A. (2002) Acridine derivatives as chemotherapeutic agents. Curr. Med. Chem., 9, 1655–1665. [DOI] [PubMed] [Google Scholar]
- 2.Baguley B.C. (1991) DNA intercalating antitumor agents. Anti-Cancer Drug Des., 6, 1–35. [PubMed] [Google Scholar]
- 3.Topcu Z. (2001) DNA topoisomerases as targets for anticancer drugs. J. Clin. Pharm. Ther., 26, 405–416. [DOI] [PubMed] [Google Scholar]
- 4.Geminéz-Arnau E., Missailidis,S. and Stevens,M.F.G. (1998) Antitumour polycyclic acridines. Part 4. Physico-chemical studies on the interactions between DNA and novel tetracyclic acridine derivatives. Anti-Cancer Drug Des., 13, 431–451. [PubMed] [Google Scholar]
- 5.McCrystal M.R., Evans,B.D., Harvey,V.J., Thompson,P.I., Porter,D.J. and Baguley,B.C. (1999) Phase I study of the cytotoxic agent N-[2-(dimethylamino)ethyl]acridine-4-carboxamide. Cancer Chemother. Pharmacol., 44, 39–44. [DOI] [PubMed] [Google Scholar]
- 6.Bridewell D.J.A., Finlay,G.J. and Baguley,B.C. (1999) Mechanism of cytotoxicity of N-[2-(dimethylamino)ethyl]acridine-4-carboxamide and its 7-chloro derivative: the roles of topoisomerases I and II. Cancer Chemother. Pharmacol., 43, 302–308. [DOI] [PubMed] [Google Scholar]
- 7.Ferrante K., Winograd,B. and Canetta,R. (1999) Promising new developments in cancer chemotherapy. Cancer Chemother. Pharmacol., 43, S61–S68. [DOI] [PubMed] [Google Scholar]
- 8.Temple M.D., McFadyen,W.D., Holmes,R.J., Denny,W.A. and Murray,V. (2000) Interaction of cisplatin and DNA-targeted 9-aminoacridine platinum complexes with DNA. Biochemistry, 39, 5593–5599. [DOI] [PubMed] [Google Scholar]
- 9.Perrin L.C., Prenzler,P.D., Cullinane,C., Phillips,D.R., Denny,W.A. and McFadyen,W.D. (2000) DNA targeted platinum complexes: synthesis cytotoxicity and DNA interactions of cis-dichloroplatinum(II) complexes tethered to phenazine-1-carboxamides. J. Inorg. Biochem., 81, 111–117. [DOI] [PubMed] [Google Scholar]
- 10.Whittaker J., McFadyen,W.D., Baguley,B.C. and Murray,V. (2001) The interaction of DNA-targeted platinum phenanthridinium complexes with DNA in human cells. Anti-Cancer Drug Des., 16, 81–89. [PubMed] [Google Scholar]
- 11.Kohn K.W., Orr,A., O’Connor,M., Guziec,L.M. and Guziec,F.S.,Jr (1994) Synthesis and DNA-sequence selectivity of a series of mono- and difunctional 9-aminoacridine nitrogen mustards. J. Med. Chem., 37, 67–72. [DOI] [PubMed] [Google Scholar]
- 12.Valu K.K., Gourdie,T.A., Boritzki,T.J., Gravatt,G.L., Baguley,B.C., Wilson,W.R., Wakelin,L.P.G., Woodgate,P.D. and Denny,W.A. (1990) DNA-directed alkylating agents. 3. Structure-activity relationships for acridine-linked aniline mustards: consequences of varying the length of the linker chain. J. Med. Chem., 33, 3014–3019. [DOI] [PubMed] [Google Scholar]
- 13.McConnaughie A.W. and Jenkins,T.C. (1995) Novel acridine-triazenes as prototype combilexins: synthesis, DNA binding, and biological activity. J. Med. Chem., 38, 3488–3501. [DOI] [PubMed] [Google Scholar]
- 14.Martins E.T., Baruah,H., Kramarczyk,J., Saluta,G., Day,C.S., Kucera,G.L. and Bierbach,U. (2001) Design, synthesis, and biological activity of a novel non-cisplatin-type platinum–acridine pharmacophore. J. Med. Chem., 44, 4492–4496. [DOI] [PubMed] [Google Scholar]
- 15.Zákovská A., Nováková,O., Balcarová,Z., Bierbach,U., Farrell,N. and Brabec,V. (1998) DNA interactions of antitumor trans-[PtCl2(NH3)(quinoline)]. Eur. J. Biochem., 254, 547–557. [DOI] [PubMed] [Google Scholar]
- 16.Keck M.V. and Lippard,S.J. (1992) Unwinding of supercoiled DNA by platinum ethidium and related complexes. J. Am. Chem. Soc., 114, 3386–3390. [Google Scholar]
- 17.Baruah H., Rector,C.L., Monnier,S.M. and Bierbach,U. (2002) Mechanism of action of non-cisplatin type DNA-targeted platinum anticancer agents: DNA interactions of novel acridinylthioureas and their platinum conjugates. Biochem. Pharmacol., 64, 191–200. [DOI] [PubMed] [Google Scholar]
- 18.Jamieson E.R. and Lippard,S.J. (1999) Structure, recognition, and processing of cisplatin-DNA adducts. Chem. Rev., 99, 2467–2498. [DOI] [PubMed] [Google Scholar]
- 19.Pearlman D.A., Case,D.A., Caldwell,J.W., Ross,W.S., Cheatham,T.E., Debolt,S., Ferguson,D., Seibel,G. and Kollman,P. (1995) AMBER, a package of computer programs for applying molecular mechanics, normal-mode analysis, molecular dynamics and free-energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun., 91, 1–41. [Google Scholar]
- 20.Barry C.G., Turney,E.C., Day,C.S., Saluta,G., Kucera,G.L. and Bierbach,U. (2002) Thermally inert metal ammines as light-inducible DNA-targeted agents. Synthesis, photochemistry, and photobiology of a prototypical rhodium(III)–intercalator conjugate. Inorg. Chem., 41, 7159–7169. [DOI] [PubMed] [Google Scholar]
- 21.Gasteiger J. and Marsili,M. (1978) A new model for calculating atomic charges in molecules. Tetrahedron Lett., 3181–3184. [Google Scholar]
- 22.SantaLucia J. Jr, Allawi,H.T. and Seneviratne,P.A. (1996) Improved nearest-neighbor parameters for predicting DNA duplex stability. Biochemistry, 35, 3555–3562. [DOI] [PubMed] [Google Scholar]
- 23.SantaLucia J. Jr, (1998) A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA, 95, 1460–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams A., Guss,J.M., Collyer,C.A., Denny,W.A. and Wakelin,L.P.G. (1999) Crystal structure of the topoisomerase II poison 9-amino-[N-(2-dimethylamino)ethyl]acridine-4-carboxamide bound to the DNA hexanucleotide d(CGTACG)2. Biochemistry, 38, 9221–9233. [DOI] [PubMed] [Google Scholar]
- 25.Neidle,S. (2002) Nucleic Acid Structure and Recognition. Oxford University Press, Oxford, UK, pp. 89–138. [Google Scholar]
- 26.Young P.R. and Kallenbach,N.R. (1981) Binding of 9-aminoacridine to deoxydinucleoside phosphates of defined sequences: preferences and stereochemistry. J. Biol. Chem., 145, 785–813. [DOI] [PubMed] [Google Scholar]
- 27.Wilson W.R., Baguley,B.C., Wakelin,L.P.G. and Waring,M.J. (1981) Interaction of the antitumour drug m-AMSA (4′-(9-acridinylamino)methanesulphon-m-anisidide) and related acridines with nucleic acids. Mol. Pharmacol., 20, 404–414. [PubMed] [Google Scholar]
- 28.Wilson,W.D., Li,Y. and Veal,J.M. (1992) NMR analysis of reversible nucleic acid-small molecule complexes. In Hurley,L.H. (ed.), Advances in DNA Sequence Specific Agents. JAI Press, New York, NY, pp. 89–165.
- 29.Rinkel L.J. and Altona,C. (1987) Conformational analysis of the deoxyribofuranose ring in DNA by means of sums of proton-proton coupling constants–a graphical method. J. Biomol. Struct. Dyn., 4, 621–649. [DOI] [PubMed] [Google Scholar]
- 30.Ren J. and Chaires,J.B. (1999) Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry, 38, 16067–16075. [DOI] [PubMed] [Google Scholar]
- 31.Adams A., Guss,J.M., Denny,W.A. and Wakelin,L.P.G. (2002) Crystal structure of 9-amino-N-[2-(4-morpholinyl)ethyl]-4-acridinecarboxamide bound to d(CGTACG)2: implications for structure-activity relationships of acridinecarboxamide topoisomerase poisons. Nucleic Acids Res., 30, 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bostock-Smith C.E., Giménez-Arnau,E., Missailidis,S., Laughton,C.A., Stevens,M.F.G. and Searle,M.S. (1999) Molecular recognition between a new pentacyclic acridinium salt and DNA sequences investigated by optical spectroscopic techniques, proton nuclear magnetic resonance spectroscopy, and molecular modeling. Biochemistry, 38, 6723–6731. [DOI] [PubMed] [Google Scholar]
- 33.Lisgarten J.N., Coll,M., Portugal,J., Wright,C.W. and Aymami,J. (2002) The antimalarial and cytotoxic drug cryptolepine intercalates into DNA at cytosine–cytosine sites. Nature Struct. Biol., 9, 57–60. [DOI] [PubMed] [Google Scholar]
- 34.Brow J.M., Pleatman,C.R. and Bierbach,U. (2002) Cytotoxic acridinylthiourea and its platinum conjugate produce enzyme-mediated DNA strand breaks. Bioorg. Med. Chem. Lett., 12, 2953–2955. [DOI] [PubMed] [Google Scholar]
- 35.Barry,C.G. and Bierbach,U. (2002) Detection of DNA damage sites of a novel platinum–acridine anti-tumor agent by liquid chromatography–mass spectrometry. Presented at the 54th Southeast Regional Meeting of the American Chemical Society, Charleston, SC, November 2002; Paper 252.
- 36.Denny W.A. (2001) DNA minor groove alkylating agents. Curr. Med. Chem., 8, 533–544. [DOI] [PubMed] [Google Scholar]
- 37.Fan J.-Y., Ohms,S.J., Boyd,M. and Denny,W.A. (1999) DNA adducts of 9-anilinoacridine mustards: characterization by NMR. Chem. Res. Toxicol., 12, 1166–1172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.