Abstract
Impairment of the omp25 gene in Brucella spp. leads to attenuated strains and confers protection to the host. Omp25 and Omp31, whose functions remain unknown, were the first characterized members of group 3 outer membrane proteins (Omps) (25 to 34 kDa). Recently, genomic and proteomic approaches identified five new putative members of this family, some of which are produced in B. melitensis or B. abortus. In the present study, using protein microsequencing, we identified new members of group 3 Omps proteins produced in B. suis. Since several monoclonal antibodies (MAbs) against Omp25 cross-reacted with other members of group 3 Omps, we also performed Western immunoblotting to compare wild-type B. suis with mutants systematically having B. suis omp25-related genes knocked out. We demonstrate the production of three paralogs of Omp31 and/or Omp25 in B. suis, and the existence of a common site of signal peptide cleavage (AXAAD), which is very similar to that present in the five homologous Omps of Bartonella quintana. The seven group 3 Omps were classified in four-subgroups on the basis of percentage amino acid sequence identities: Omp25 alone, the Omp25b-Omp25c-Omp25d cluster, the Omp31/31b subgroup, and the less related Omp22 protein (also called Omp3b). Together with previous data, our results demonstrate that all new members of group 3 Omps are produced in B. suis or in other Brucella species and we propose a nomenclature that integrates all of these proteins to facilitate the understanding of future Brucella interspecies study results.
Brucellae are small, nonmotile, gram-negative coccobacilli that are able to infect a broad range of wildlife and domestic mammals. They remain a major zoonotic disease source affecting humans worldwide and are also a focus of concern as potential biological warfare agents (20). Although BrucelIa spp. are not particularly host specific, three major (B. abortus, B. melitensis, and B. suis) and minor (B. canis, B. ovis, and B. neotomae) species each have distinct host preferences and pathogenicities for humans (29). Malta fever (also known as Mediterranean, Gibraltar, or undulant fever) and porcine brucellosis—caused by B. melitensis and B. suis infection of humans, respectively—are usually by far more clinically apparent than Bang's disease (B. abortus infection), whereas among the remaining species only B. canis causes anecdotal mild infections in humans (10).
The Brucella outer membrane was investigated to seek immunogenic and protective antigens for potential diagnostic and vaccine applications. The major outer membrane proteins (Omps) of Brucella spp. were thoroughly studied in this regard. They were classified according to their apparent molecular mass as 36- to 38-kDa Omps or group 2 porin proteins, and 25- to 27-kDa and 31- to 34-kDa Omps which belong to group 3 proteins (8). Genes encoding group 2 porin proteins consist of two genes, i.e., omp2a and omp2b, which are closely linked in the Brucella genome and share a great degree of identity (>85%). In the 1990s, two genes coding for group 3 proteins were identified and named omp25 and omp31. The predicted amino acid sequences of Omp25 and Omp31 share 34% identity (8). Studies using recombinant protein technology and monoclonal antibodies (MAbs) have suggested that the major Omps are not very relevant as antigens in infections with smooth (S) B. abortus or B. melitensis, i.e., low or no protective activity in the mouse model of infection, and low or no immunogenicity during host infection. However, group 3 proteins, in particular Omp31, appear as immunodominant antigen during the course of infection with rough (R) B. ovis in rams and as important protective antigen in the B. ovis mouse model of infection (4, 15). Interestingly, B. abortus is the only Brucella species in which omp31 (as part of a 25 kb DNA fragment) is deleted (30).
The other well known Omp25 protein has been shown to be involved in the virulence of B. melitensis, B. abortus, and B. ovis (12-14). Mutants lacking Omp25 are indeed attenuated in animal models of infection, and also provide levels of protection similar to or better than the currently used attenuated vaccine strain B. melitensis Rev.1. These mutant strains are thus interesting future vaccine candidates. The only putative function of Omp25 in relation with Brucella virulence described so far is its ability to prevent or inhibit TNF-α release from macrophages (19). Another clue of the possible involvement of Omp25 in virulence was recently obtained after an analysis of B. abortus mutants. It was found that insertion of the Tn5 transposon in either gene of the two component bvrR/bvrS system leads to attenuated strains. Analysis of the Omp25 and homologous protein contents in these mutants indicated that impairment of the bvrR/bvrS system hinders the production of Omp25 and Omp22 (also named Omp3b) (18). Hence, it was hypothesized that attenuation is a consequence of the absence of Omp25 or related proteins in B. abortus bvr/bvrS mutants.
Analysis of B. melitensis 16M and B. suis genomic sequences, revealed the presence of five other gene products homologous to Omp25 or Omp31, and indicated the existence of a multiple gene family for this type of Omps (11, 18). In the α-proteobacteria Bartonella quintana, whose growth requires an extraordinary high hemin concentration in the medium (5, 26) it was shown that the Omp, HbpA, acting as a hemin receptor possesses 34% amino acid sequence identity with Omp31. Both proteins are heat modifiable and exhibit eight β-sheets which appear to constitute a β-barrel structure in HbpA (23). Similarly to Brucella, four other HbpA-related proteins were documented (HbpBCDE) from the B. quintana genome and a recent study showed that these latter homologs are all produced in routine culture conditions (23).
The possible role of Omp25, which is just one member of an enlarging Omps family, in Brucella virulence prompted us to more carefully characterize putative proteins deduced from genes related to omp25 and omp31 in B. suis and in some other Brucella species. Indeed, other group 3 proteins identified in the genome whose functions might be similar to that of Omp25 would merit further investigation with respect to the potential development of new vaccines.
MATERIALS AND METHODS
Bacteria strains, plasmids, and growth conditions.
B. suis 1330 (ATCC 23444T), B. canis (23365T), B. melitensis 16 M (23448T), B. ovis Reo 198, and B. abortus 2308 strains were grown in tryptic soy broth medium (TS). For B. suis derived mutants, TS was supplemented with kanamycin at 50 μg ml−1. Brucella strains were grown for 16 to 20 h in TS at 37°C to the stationary phase (optical density at 600 nm [OD600] of 1.4 to 1.6) and collected by centrifugation. The pellets were resuspended in phosphate-buffered saline (PBS) to obtain an OD600 value of 0.5. From the various bacterial suspensions, 2-ml aliquots were sedimented for 2 min at 13,000 × g and lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Alternatively, pellets were resuspended in 0.5 ml of minimal medium at pH 4.5 or 7 and incubated for 6 h at 37°C as previously described (1, 24). Plasmid constructs were amplified in E. coli DH5α strain which was routinely grown in Luria-Bertani medium with appropriate antibiotics.
Omps N-terminal microsequencing by Edman degradation.
Brucella suis pellets from 2 liters of stationary culture were washed by centrifugation in 800 ml PBS and resuspended in the same volume of 25 mM ammonium acetate buffer (pH 4.0) containing 120 mM NaCl and incubated at 37°C for 2 h with shaking. Bacterial cells were sedimented by centrifugation and the supernatant was collected, sterilized by filtration through a 0.2-μm-pore-size membrane filter (Sartorius, Palaiseau, France). Supernatants were then concentrated 20-fold through a hydrophilic membrane (Amicon YM; Millipore, Bedford, Mass.) with a cutoff of 10 kDa before protein precipitation with chloroform/methanol. Brucella proteins were then separated on SDS-15% PAGE, blotted onto a polyvinylidene difluoride membrane (Immobilon-Pseq; Millipore) and stained with Coomassie blue. The four major spots between 25 and 30 kDa were subjected to Edman degradation using ABI 431A automated peptide sequencing, as previously described (1, 22).
Inactivation of the B. suis omp25 gene family by homologous recombination.
Knockout of genes homologous to omp25 was performed as previously described (1, 19). Briefly, complete DNA sequences encoding B. suis Omp25 and Omp31 paralogs were amplified by PCR with appropriate primers derived from B. melitensis 16 M or B. suis 1330 genomes. The primers used were: Omp25b (forward, ATGTTTGGAGGGAACCATGAAC; reverse, CGAAACGCCATCGCATTCCGTTGA), Omp25c (forward, TGTGGGACTATTTCCCGAGAG; reverse, CTATCAGAACTTGTAAGCGACACC), Omp25d (forward, CAACGCCATTCTTATTAGGA; reverse, GTCGGCAATCAGAACTTATAGG), Omp31b (forward, TATAGTGACAGACATTGGAGCC; reverse, CCGTTCCAGTTCTCGATTTA) and Omp22 (forward, TACAAAATGTTCAAGCGTTG; reverse, TTGGCCGGGAAAACCGGAGTAC). PCR fragments were introduced into pGEMT (Promega, Charbonnières, France) or pBluescript (Stratagene, Amsterdam, Netherlands). The kanamycin resistance gene cassette was inserted into each gene and these constructs were introduced into the suicide vector pCVD442 after digestion with SphI and SacI. Double recombination was verified by PCR amplification and Southern blotting on genomic DNA of B. suis Kanr Amps sucrose-resistant clones. Clones in which the kanamycin resistance gene cassette and the knocked out gene were in the opposite direction were selected for further studies.
DNA manipulation and Southern blotting.
Plasmid DNA was isolated from E. coli according to standard procedures (25). DNA treatments with restriction and modification enzymes were performed according to the manufacturer's instructions. Restriction fragments were purified after separating bands on low-melting-point agarose gels (Invitrogen, Cergy Pontoise, France) with a gel extraction kit (Qiagen, Courtaboeuf, France). B. suis chromosomal DNA was prepared as previously described (3). For Southern blotting, genomic DNA was purified and digested with HindIII enzyme. Southern blotting was performed using Biodyne B nylon membranes (Pall, Port Washington, N.Y.) with digoxigenin-labeled DNA probes using a random priming kit (Roche, Mannheim, Germany).
Western blot analysis.
Proteins of various Brucella strains were separated on SDS-15% PAGE before transfer onto Immobilon polyvinylidene difluoride membranes (Millipore) using a semidry transfer procedure. Omps immunodetection was performed with anti-Omp25 MAb, A59 05F01 C09 (6). Horseradish peroxidase-conjugated goat anti-mouse antibodies (Jackson Immunoresearch Laboratories Inc.) were used in combination with the ECL system (Amersham Bioscience, Orsay, France) for chemiluminescence and visualization on Kodak X-AR films (Sigma-Aldrich, Saint Quentin Falavier, France). The molecular markers were from Amersham Bioscience.
Computer-based sequence analysis.
The amino acid sequences of Omp25, Omp31 and their homologs shown in Fig. 2 were deduced from the Brucella suis genome available from The Institute for Genomic Research (http://www.tigr.org), and primers used for gene amplification were taken from B. melitensis genome available from the Institute of Molecular Biology and Medicine, University of Scranton, Scranton, Pa. Multiple sequence alignment of Omps amino acid sequences was carried out with ClustalW, 1.60 (28) and the dendrogram from MULTALIN (http://prodes.toulouse.inra.fr/multalin/multalin.html).
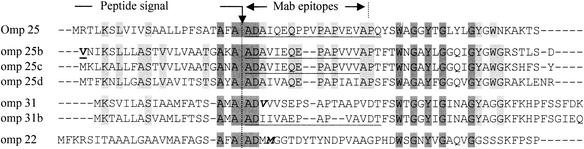
FIG. 2.
Alignment of N-terminal amino acid sequences deduced from genes present in the B. suis genome which are homologous to omp25/31. The underlined amino acid sequences were obtained by Edman degradation. The vertical arrow points to the peptide signal cleavage site. The first amino acid of the second row, depicted here V, was encoded by the alternative initiation codon GTG. V and M in boldface type in the fifth and seventh sequences, respectively, were initially considered as the first translated amino acid in B. melitensis omp22 and omp31 genes. The epitopes recognized by several MAbs against Omp25 such as A59 5F1 C9 are indicated between arrows above the Omp25 sequence (18). Dark shading indicates amino acids conserved in all N-terminal sequences of Omp25/31 family. Light gray shading points to additional amino acids conserved in the Omp25b-d subset some of which are also present in either Omp25 or Omp31/31b but not in Omp22.
RESULTS
Existence of four Omps subgroups based on the percentage amino acid sequence identity.
Analysis of the Brucella suis genome revealed the presence of five other paralogous of omp25/31 gene products which were almost identical to their B. melitensis homologs with the notable exception of one truncated form in B. melitensis (BME I 0402). The correspondence of the B. suis and B. melitensis gene designation in each strain is given in Fig. 1A (last two columns). The B. melitensis gene nomenclature was adapted in that each B. suis gene number is preceded by a roman numeral to define its location on the large (I) or small chromosome (II). Note that all genes were found to be located in chromosome I, except the omp31 gene. Further, the genes described in row 3 and row 4 are contiguous. For example in omp25c and omp25d genes are numbered I 119 and I 118 in B. suis, and I 1830 and I 1829 in B. melitensis.
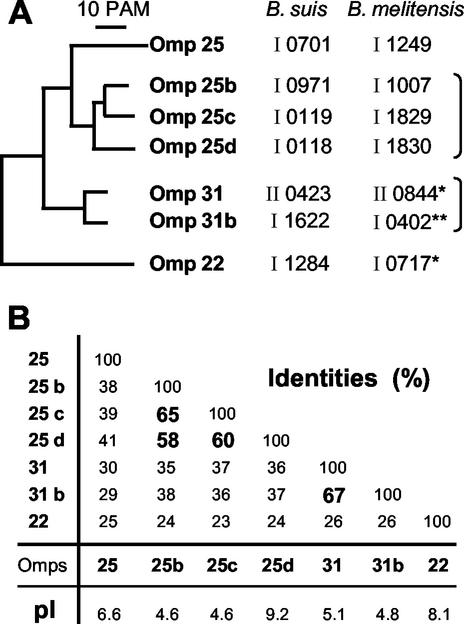
FIG. 1.
Global analysis of mature group 3 Omp proteins deduced from the B. suis genome sequenced by TIGR. (A) Neighbor-joining tree derived from distance matrix analysis of the group 3 Omps sequences. 10 PAM; 10 percent or point accepted mutations. It also indicates the correspondence between the proposed names and the gene numbers reported in the respective annotations of the B. suis and B. melitensis genomes. The roman numerals I and II before the gene number refer to large and small chromosomes, respectively. Note that the genes numbering in the large chromosome of the two species are in the opposite direction. (B) Percentage of identity between the proteins matured form compared two by two. Percentages around or above 60%, are shown in bold character, and allowed clustering of two multiple partner subgroups; the Omp25b-Omp25c-Omp25d on one side and the previous Omp31 together with Omp31b on the other. This panel also indicates the theoretical pI value of the various group 3 Omp proteins. Note that like Omp22, the theoretical 9.2 pI value of Omp25d is strongly basic compared to pI values of 4.6 of the two other members of the subgroup. * In contrast to the genome annotations, these two B. melitensis proteins are likely as long as those described for B. suis (Fig. 2). **, in B. melitensis, the Omp31b is truncated (see discussion).
Distance matrix analysis carried out with B. suis group 3 Omps confirmed the existence of four distinct subgroups (Fig. 1A) as reported for B. abortus (18). Compared to Omp25, amino acid sequence identities of about 40% were found with a subgroup of three Omp25-like proteins named Omp25b, Omp25c, Omp25d, and about 30% with Omp31 and Omp31b, while the amino acid sequence identity between Omp25 and Omp22 was only 25% (Fig. 1B, first column). Omp22 name refers to its calculated mass of 22 kDa but was also called previously Omp3b (18). Within the three member subgroup (Omp25b, Omp25c, and Omp25d), the amino acid sequence identity was around 60%, while it reached 67% between Omp31 and its counterpart Omp31b. With these values taken into account, a dendrogram was designed to visualize the various subgroups (Fig. 1A). It also indicates the proposed nomenclature with corresponding gene numbers in the B. suis and B. melitensis genome. Besides the Omp25 alone, each gene of three-member subgroup related to Omp25 was named Omp25 plus one letter b, c or d. Omp31 and its related gene product called Omp31b also formed another subgroup. Finally, for the last gene product, the nomenclature Omp22 was chosen to indicate that this protein is somewhat different from the other Omp25/31 of the family. For the formerly characterized Omp25 and Omp31, the postfix letter “a” was omitted in order to keep their original names.
N-terminal Edman degradation analysis highlights the signal peptide cleavage site of all Omps.
In an attempt to analyze the proteome of proteins released in the acidic medium by B. suis, we isolated a fraction enriched with group 3 Omps (Boigegrain and Rouot, manuscript in preparation). N-terminal microsequence analysis of four proteins with an apparent molecular weight of 25 kDa and above provided three unambiguous N-terminal sequences (Fig. 2, see underlined sequences in rows 1, 2, or 3 and 6). As expected, one amino acid sequence corresponded to the Omp25 protein while the second one matched either Omp25b or Omp25c since the first 31 amino acids of the mature proteins were identical. The last sequence fitted with Omp31b but not with the well known Omp31. The fact that these three sequences started with the same two amino acids (AD) and possessed an identical PAPV stretch clearly indicated that B. suis produced Omp25/31 paralogs. Some of these potential group 3 Omps were already characterized by a proteomic approach. Hence, the one that we called Omp22 was described in B. abortus 2308 and also named Omp3b (18), while Omp25b and Omp25c homologs were identified in the B. melitensis proteome (31). Our detected N-terminal sequences which matched Omp25b and Omp25c, revealed the biosynthesis of at least one of them in B. suis. The third N-terminal microsequence was found in the deduced amino acid sequence of the omp31b paralog of omp31, demonstrating the production of Omp31b in Brucella spp for the first time.
Alignment of N-terminal amino acid sequences of the seven members of the Omp25/31 family revealed a consensus sequence (AXAAD) about 20 amino acids downstream from the N-terminal end. The presence in this conserved sequence of AD, i.e., the first two amino acids of our three microsequences, strongly suggested that a signal peptide of 19 to 24 amino acids was cleaved for all proteins within this stretch (Fig. 12 [see vertical arrow]). This finding indicated that translation of the B. melitensis Omp22 protein started, as for the B. suis homolog, 27 amino acids upstream from methionine, as initially predicted in the genome annotation (Fig. 2, last row). Sequence alignment also showed that among the 25 amino acids conserved in the 7 matured proteins, 13 were glycine residues (data not shown). Since they were located in domains predicted to adapt β sheet structures (8, 18), it could be hypothesized that these glycine residues play a key role in maintenance of the tertiary structure needed for the protein to function.
Immunological detection and characterization of Omp25 paralogs.
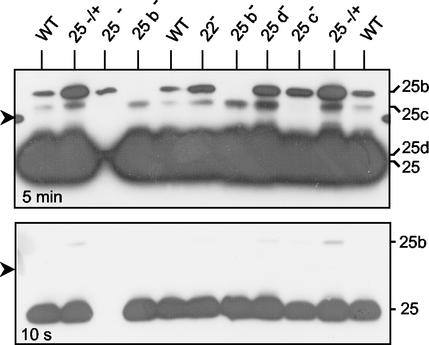
We then investigated whether the production of different members of Brucella group 3 proteins could be assessed immunologically. We reasoned that because of their relatively high degree of similarity some MAbs directed against Omp25 might also recognize other members of this Omps family (6, 7). A previous report (18) together with the above cleavage site results permitted us to localize, within the first 17 amino acids of the mature Omp25, the domain of epitopes recognized by three specific MAbs, including MAb A59 05F01 C09 (Fig. 2). In order to specify which proteins might be recognized by the antibodies, we individually knocked out the newly found group 3 Omps genes. Insertional inactivation of B. suis genes followed by homologous recombination (1) was successfully carried out for most genes, as verified by PCR on the genomic DNA (data not shown). However, in our hands, no clone could be obtained in which omp31b had properly inserted the kanamycin resistance gene cassette. Preliminary experiments indicated that MAb A59 05F01 C09 and to a lesser extent A19 12B10 F4 permitted Western blot detection of proteins of lower electrophoretic mobility than Omp25. Indeed, the films had to be overexposed to detect MAb cross-reactivity on membranes (Fig. 3). Examination of immunostaining obtained with the various B. suis omp mutants revealed that Omp25b exhibited the highest apparent molecular mass (about 34 kDa), followed by Omp25c (31 kDa). The inactivation of omp25 unmasked a faint labeling slightly above the bulk staining of Omp25. This reproducible labeling was also present when no proteins were loaded in adjacent tracks and corresponded to Omp25d (Fig. 3, lane 3).
FIG. 3.
Immunological characterization of Omp25 family members by Western blotting. Knock out of individual B. suis omp25 paralogous genes was carried out as previously described in Materials and Methods. The different Brucella strains were grown at 37°C in TS broth rich medium to the stationary phase. Proteins were separated on SDS-PAGE (15% polyacrylamide), blotted and immunodetection was performed using enhanced chemiluminescence (ECL; Amersham Bioscience) with the monoclonal antibody A59 05F01 C09 directed against the N-terminal epitope of Omp25 (2). The same membrane was exposed for 5 min (upper panel) and 10 s (lower panel). Staining detected at about 26 kDa in the omp25 mutant was always observed and might be due to the presence of the Omp25d protein. The arrow on the left side refers to the 29-kDa marker.
The absence of some Omps could modify the production of others. Thus, compared to the B. suis parental strain (WT), inactivation of omp25c, omp25d and omp22 substantially increased Omp25b production (and Omp25c when present in the strain) (Fig. 3). On the contrary, the faint Omp25c labeling seen in the WT strain could not be detected in the omp25 mutant. Curiously, complementation of the omp25 (25+/−) gene led to an increase in the Omp25b protein level although the Omp25 production was similar to that of the parental strain. Because of the very peculiar N-terminal sequence of Omp22, immunodetection of this protein was not expected and the absence of labeling could not be interpreted as a lack of Omp22 production in B. suis.
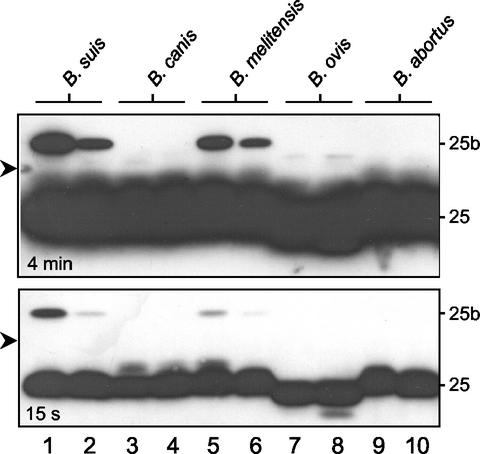
Further, we determined whether or not our A59 05F01 C09 MAb could also detect additional Omps in other Brucella species grown under routine culture conditions. Comparison of the staining of B. suis with other species indicated that only B. melitensis produced similar among of Omp25b (Fig. 4, odd-numbered lanes). On the contrary, B. canis, B. ovis, and B. abortus did not produce enough Omp25b to be detected under our experimental conditions. The reason for such absence (lack of gene or lack of protein production) remains to be determined.
FIG. 4.
Assessment of the presence of members of Omp25 family by Western blotting on various Brucella strains incubated in nutrient-deficient medium either at neutral or acidic pH. Wild-type B. suis, B. canis, B. melitensis, B. ovis and B. abortus A2308 bacteria were grown to the stationary phase in TS at 37°C. Aliquots of 2 ml with OD values adjusted to1.5 were washed in PBS by centrifugation. The resulting pellets were resuspended in either minimal medium at pH 7.0 (odd-numbered lanes) or pH 4.5 (even-numbered lanes), and further incubated for 6 h at 37°C. The presence of Omp25 paralogs was evaluated by Western blotting using the A59 05F01 C09 monoclonal antibody. The same membrane was exposed for 4 min (upper panel) and 15 s (lower panel). The data shown are from one representative experiment of the two performed. The arrow on the left side refers to the 29 kDa marker.
Omp25 paralogs production and Brucella virulence.
Since Omp25 production is suspected to play an important role in Brucella virulence (12-14, 18), we further investigated whether the production of group 3 Omps could be altered under various culture conditions. Indeed, it was shown that the production of VirB proteins composing the type IV secretion system were initiated (B. suis and B. canis) or increased (B. melitensis, B. ovis, or B. abortus) when grown in acidic nutrient-deprived medium (24). Figure 4 shows the Omp25 immunoreactivities obtained with the different strains incubated in deprived nutrient media at pH 7.0 (odd lanes) or pH 4.5, i.e., VirBs inducing conditions (even lanes). The lower panel of Fig. 4 clearly indicates that the Omp25 contents were roughly the same in all Brucella species tested, while Omp25b was present only in B. suis and B. melitensis. Interestingly, 6-h incubation of the various bacteria in acidic medium did not affect the Omp25 content, but strongly reduced the Omp25b content. It remains to be determined whether the difference in Omp25b production was due to a differential rate of turnover or transcription.
We then evaluated whether the bvrS/bvrR two component system required for the production of B. abortus Omp25 (18) was also needed for B. suis Omp25 production. However, many attempts to knock out this two component system failed for unknown reasons. It should be noted that the B. abortus strains inactivated on bvrS or bvrR used in all previous studies were obtained by transposon mutagenesis and not by an appropriate resistance gene cassette insertion and homologous recombination. In agreement with this observation with B. abortus, Köhler et al. recently isolated in a macrophage survival screening test, several viable mutants in which the miniTn5 had been inserted in B. suis bvr genes (21). Interestingly, when using these bvrS/bvrR transposon mutagenized B. suis mutants, no significant differences in Omp25 or Omp25b and Omp25c content compared to the WT B. suis were observed by immunoblotting (data not shown). This finding indicated that a functional bvrS/bvrR system was not directly involved in Omp25 production in B. suis.
DISCUSSION
On the basis of the present study and previous reports, it appears that all proteins deduced from new group 3 Omps genes found in B. melitensis and/or B. suis genomes were produced in some Brucella species under routine culture conditions. Omp25 and Omp25b were thus identified in B. suis and B. melitensis. The absence of Omp25b cross-immunoreactivity in the other Brucella species examined indicates that this protein is poorly or not at all produced in vitro. Omp25d, with a very basic pI value, has not yet been clearly identified in B. suis and B. melitensis. However, it might correspond to spot 9 (pI >8.5) detected immunologically in B. abortus by Guzman-Verri et al. (18). Omp31b identified in B. suis in the present study was not detected by the proteomic approach carried out by the Delvecchio group (31). This is not surprising since there is a 232-nucleotide deletion at the 5′ end of the B. melitensis omp31b gene. This deletion, further confirmed by N. Vizcaino (personal communication), leads to a protein devoid of any signal peptide and thus is nonfunctional. Several Omp22 isoforms have been characterized by mass spectrometry in B. abortus species (18) but not yet in other species. All putative Brucella Omps can thus be produced and we hypothesize that they are located in the outer membrane of the bacteria, based upon (i) their high similarities with the well known outer membrane protein Omp25/31, (ii) possession of a C-terminal phenylalanine (27), and (iii) the confirmation or prediction of a secretory signal peptide. With regards to this latter point, the cleavage site within the AXAAD Brucella sequence is very similar to that of B. quintana Omps (XQAADV) leading to proteins which started with an AD dipeptide in both bacteria.
To facilitate and clarify future studies carried out with various Brucella species, a nomenclature including all known members of the groups 3 Omps family was elaborated (Fig. 2, upper panel). It has two main advantages. First, the names of the originally identified Omp25 and Omp31 proteins remain unchanged, thus allowing unambiguous bibliographic searches for past articles. Secondly, the nomenclature takes into account the existence of four subgroups in the whole family based on the percentage amino acid sequence identities between members. The gene or protein names immediately indicate the subgroup to which the protein belongs: Omp25, Omp25 + one letter (three members), Omp31 (± a letter) and Omp22. Moreover, this amino acid sequence identity based classification might also correspond to different functions. Interestingly, when identities between Brucella and Bartonella Omps were determined, we found that Hbps exhibited a higher percentage identity with the three-member Omp25 b-Omp25c-Omp25d subgroup (up to 37%) than with Omp31 (33%), Omp25 (31%), or Omp22 (25%). This finding indicates that all Bartonella Hbps are not distributed in the four distinguishable categories of Brucella but are closely related to the three-member Omp25b-Omp25c-Omp25d subgroup. In this context, it is worth noting that the hemin binding property first demonstrated with HbpA, is also shared by other members of the Bartonella Hbp family (23).
This study also provided evidence of the differential production of Omp25 in various Brucella species. Hence, besides the presence of Omp25 with or without (B. abortus) Omp31, each Brucella species possesses a typical pattern of different group3 Omps. Methods used to identify Omps (two-dimensional matrix-associated laser desorption ionization-time-of-flight, microsequencing, cross-immunological reactivity) were qualitative and did not enable us to quantitatively determine the relative distribution of Omps in each species. Although the first identified proteins (Omp25 and Omp31) appear to be the major ones of group 3, the relative contents of all group 3 Omp members might have to be reassessed with antibodies directed to epitopes identical among the whole Omps family in order to validate this assumption. Similarly, the numerous immunologically identified Omp25 isoforms might have to be reconsidered in the light of the cross-reactivity of MAbs against Omp25 subgroups. In this regard, Brucella mutants with each omp gene knocked out clearly established that Omps of group 3 exhibiting an apparent molecular weight higher than 30 kDa on SDS-PAGE did not derive from omp25 but rather from omp25b and omp25c genes. The very slow electrophoretic mobilities of these proteins compared to their theoretical molecular weights might result from posttranslational modifications in these new Omps. It was suggested that Omp25 is covalently linked to peptidoglycan (9) or LPS molecules (16, 17), and proteomic studies should confirm whether or not this applies for Omp25b and Omp25c.
The fact that the impairment of the B. suis two component bvrR/bvrS system did not hinder the production of the various Omp25/31 proteins indicates that these Omps were not involved in the virulence attenuation of the bvrR/bvrS mutants of B. suis. This contrasts with observations on the corresponding bvrR/bvrS B. abortus mutants which have lost their ability to produce Omp25 and Omp22 (18). Indeed, B. abortus differs from B. suis in that it is the only species which cannot produce Omp31 because of the 25 kb DNA deleted fragment including omp31 (30). Thus, the absence of Omp25 in the B. abortus bvrR/bvrS mutants would possibly result from the concomitant lack of the bvrR/bvrS system and Omp31 (or another protein encoded by the missing DNA fragment). To validate or refute this assertion, it would be interesting to investigate whether or not blockade of the bvrS/bvrR system hinders Omp25 production in the other Omp31 producing Brucella species. As far as the interrelation of group 3 Omps production is concerned, the enhanced production of Omp25b, after other omp genes had been knocked out, suggests the existence of compensatory regulation between expressed genes, as reported for Bartonella homologous proteins (23). However, omp gene expression exhibits some degree of autonomous regulation since, under in vitro VirB inducing conditions, the level of Omp25b is decreased independently of that of Omp25.
Finally, it appears that conserved amino acids in the group 3 Omps family are located in the predicted eight transmembrane β-strands and in the intracellular loops joining them, thus defining the bulk structure of proteins of this family. Since inter-Omp variations mainly concern the length and composition of the extracellular loops, these variations might confer differences in protein functions such as binding capacities. Further research is clearly needed to achieve progress on characterization of the function of each member of group 3 Omps, and determination of their respective involvements in strain attenuation or virulence. The knock out mutants described here will be very helpful in this quest.
Acknowledgments
We thank Véronique Maurin for providing us with B. suis 1330 mutants impaired in omp31 and omp25 genes and also the complemented omp25. We also thank S. Kohler and S. Ouahrani-Bettache for the bvrS/bvrR mutants of B. suis. We also thank the Institute of Molecular Biology and Medicine (University of Scranton, Scranton, Pennsylvania) and TIGR for providing access to preliminary B. melitensis and B. suis sequence data, respectively.
This work was supported by INSERM and the French Cancer Research Association (ARC #5566).
Editor: D. L. Burns
REFERENCES
- 1.Alvarez-Martinez, M.-T., J. Machold, C. Weise, H. Schmidt-Eisenlohr, C. Baron, and B. Rouot. 2001. The Brucella suis homologue of the Agrobacterium tumefaciens chromosomal virulence operon chvE is essential for sugar utilization but not for survival in macrophages. J. Bacteriol. 183:5343-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aragon, V., R. Diaz, E. Moreno, and I. Moriyon. 1996. Characterization of Brucella abortus and Brucella melitensis native haptens as outer membrane O-type polysaccharides independent from the smooth lipopolysaccharide. J. Bacteriol. 178:1070-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, S. F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seichman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. John Wiley & Sons., New York, N.Y.
- 4.Bowden, R. A., S. M. Estein, M. S. Zygmunt, G. Dubray, and A. Cloeckaert. 2000. Identification of protective outer membrane antigens of Brucella ovis by passive immunization of mice with monoclonal antibodies. Microbes Infect. 2:481-488. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, J. A., S. A. Coleman, L. S. Smitherman, and M. F. Minnick. 2000. Hemin-binding surface protein from Bartonella quintana. Infect. Immun. 68:6750-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cloeckaert, A., P. de Wergifosse, G. Dubray, and J. N. Limet. 1990. Identification of seven surface-exposed Brucella outer membrane proteins by use of monoclonal antibodies: immunogold labeling for electron microscopy and enzyme-linked immunosorbent assay. Infect. Immun. 58:3980-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cloeckaert, A., J. M. Verger, M. Grayon, M. S. Zygmunt, and O. Grepinet. 1996. Nucleotide sequence and expression of the gene encoding the major 25-kilodalton outer membrane protein of Brucella ovis: evidence for antigenic shift, compared with other Brucella species, due to a deletion in the gene. Infect. Immun. 64:2047-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cloeckaert, A., N. Vizcaino, J. Y. Paquet, R. A. Bowden, and P. H. Elzer. 2002. Major outer membrane proteins of Brucella spp.: past, present and future. Vet. Microbiol. 90:229-247. [DOI] [PubMed] [Google Scholar]
- 9.Cloeckaert, A., M. S. Zygmunt, P. de Wergifosse, G. Dubray, and J. N. Limet. 1992. Demonstration of peptidoglycan-associated Brucella outer-membrane proteins by use of monoclonal antibodies. J. Gen. Microbiol. 138:1543-1550. [DOI] [PubMed] [Google Scholar]
- 10.Corbel, M. 1997. Brucellosis: an overview. Emerg. Infect. Dis. 3:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DelVecchio, V., V. Kapatral, R. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. Elzer, S. Hagius, D. O'Callaghan, J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmonds, M. D., A. Cloeckaert, N. Booth, W. T. Fulton, S. Hagius, J. V. Walker, and P. H. Elzer. 2001. Attenuation of a Brucella abortus mutant lacking a major 25 kDa outer membrane protein in cattle. Am. J. Vet. Res. 62:1461-1466. [DOI] [PubMed] [Google Scholar]
- 13.Edmonds, M. D., A. Cloeckaert, S. D. Hagius, L. E. Samartino, W. T. Fulton, J. V. Walker, F. M. Enright, N. J. Booth, and P. H. Elzer. 2002. Pathogenicity and protective activity in pregnant goats of a Brucella melitensis omp25 deletion mutant. Res. Vet. Sci. 72:235-239. [DOI] [PubMed] [Google Scholar]
- 14.Edmonds, M. D., A. Cloeckaert, and P. H. Elzer. 2002. Brucella species lacking the major outer membrane protein Omp25 are attenuated in mice and protect against Brucella melitensis and Brucella ovis. Vet. Microbiol. 2373:1-17. [DOI] [PubMed] [Google Scholar]
- 15.Estein, S. M., J. Cassataro, N. Vizcaíno, M. Zygmunt, A. Cloeckaert, and R. A. Bowden. 2003. The recombinant Omp31 from Brucella melitensis alone or associated with rough lipopolysaccharide induces protection against Brucella ovis infection in BALB/c mice. Microbes Infect. 5:85-93. [DOI] [PubMed] [Google Scholar]
- 16.Gamazo, C., and I. Moriyon. 1987. Release of outer membrane fragments by exponentially growing Brucella melitensis cells. Infect. Immun. 55:609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamazo, C., A. I. Vitas, I. Moriyon, I. Lopez-Goni, and R. Diaz. 1993. Brucella group 3 outer membrane proteins contain a heat-modifiable protein. FEMS Microbiol. Lett. 112:141-146. [DOI] [PubMed] [Google Scholar]
- 18.Guzman-Verri, C., L. Manterola, A. Sola-Landa, A. Parra, A. Cloeckaert, J. Garin, J. Gorvel, I. Moriyon, E. Moreno, and I. Lopez-Goni. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. USA 99:12375-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jubier-Maurin, V., R.-A. Boigegrain, A. Cloeckaert, A. Gross, M.-T. Alvarez-Martinez, A. Terraza, J. Liautard, S. Kohler, B. Rouot, J. Dornand, and J.-P. Liautard. 2001. Major outer membrane protein Omp25 of Brucella suis is involved in inhibition of tumor necrosis factor alpha production during infection of human macrophages. Infect. Immun. 69:4823-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufmann, A. F., M. I. Meltzer, and G. P. Schmid. 1997. The economic impact of a bioterrorist attack: are prevention and postattack intervention programs justifiable? Emerg. Infect. Dis. 3:83-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köhler, S., V. Foulongne, S. Ouahrani-Bettache, G. Bourg, J. Teyssier, M. Ramuz, and J.-P. Liautard. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. USA 99:15711-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machold, J., C. Weise, Y. Utkin, V. Tsetlin, and F. Hucho. 1995. The handedness of the subunit arrangement of the nicotinic acetylcholine receptor from Torpedo californica. Eur. J. Biochem. 234:427-430. [DOI] [PubMed] [Google Scholar]
- 23.Minnick, M. F., K. N. Sappington, L. S. Smitherman, S. G. Andersson, O. Karlberg, and J. A. Carroll. 2003. Five-member gene family of Bartonella quintana. Infect. Immun. 71:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouot, B., M.-T. Alvarez-Martinez, C. Marius, P. Menanteau, L. Guilloteau, R. A. Boigegrain, R. Zumbihl, D. O'Callaghan, N. Domke, and C. Baron. 2003. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect. Immun. 71:1075-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Sander, A., S. Kretzer, W. Bredt, K. Oberle, and S. Bereswill. 2000. Hemin-dependent growth and hemin binding of Bartonella henselae. FEMS Microbiol. Lett. 189:55-59. [DOI] [PubMed] [Google Scholar]
- 27.Struyve, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vizcaino, N., A. Cloeckaert, J. Verger, M. Grayon, and L. Fernandez-Lago. 2000. DNA polymorphism in the genus Brucella. Microbes Infect. 2:1089-1100. [DOI] [PubMed] [Google Scholar]
- 30.Vizcaino, N., A. Cloeckaert, M. S. Zygmunt, and L. Fernandez-Lago. 2001. Characterization of a Brucella species 25-kilobase DNA fragment deleted from Brucella abortus reveals a large gene cluster related to the synthesis of a polysaccharide. Infect. Immun. 69:6738-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner, M. A., M. Eschenbrenner, T. A. Horn, J. A. Kraycer, C. V. Mujer, S. Hagius, P. Elzer, and V. G. DelVecchio. 2002. Global analysis of the Brucella melitensis proteome: identification of proteins expressed in laboratory-grown culture. Proteomics 2:1047-1060. [DOI] [PubMed] [Google Scholar]