Abstract
Candida albicans biofilms are formed through three distinct developmental phases and are associated with high fluconazole (FLU) resistance. In the present study, we used a set of isogenic Candida strains lacking one or more of the drug efflux pumps Cdr1p, Cdr2p, and Mdr1p to determine their role in FLU resistance of biofilms. Additionally, variation in sterol profile as a possible mechanism of drug resistance was investigated. Our results indicate that parent and mutant strains formed similar biofilms. However, biofilms formed by double and triple mutants were more susceptible to FLU at 6 h (MIC = 64 and 16 μg/ml, respectively) than the wild-type strain (MIC > 256 μg/ml). At later time points (12 and 48 h), all the strains became resistant to this azole (MIC ≥ 256 μg/ml), indicating lack of involvement of efflux pumps in resistance at late stages of biofilm formation. Northern blot analyses revealed that Candida biofilms expressed CDR and MDR1 genes in all the developmental phases, while planktonic cells expressed these genes only at the 12- and 48-h time points. Functionality of efflux pumps was assayed by rhodamine (Rh123) efflux assays, which revealed significant differences in Rh123 retention between biofilm and planktonic cells at the early phase (P = 0.0006) but not at later stages (12 and 48 h). Sterol analyses showed that ergosterol levels were significantly decreased (P < 0.001) at intermediate and mature phases, compared to those in early-phase biofilms. These studies suggest that multicomponent, phase-specific mechanisms are operative in antifungal resistance of fungal biofilms.
Microorganisms form biofilms on a variety of implanted medical devices, resulting in biofilm-associated infections that constitute a significant public health problem (13, 38). Both bacterial and fungal biofilms have been associated with significantly high antimicrobial resistance compared to their planktonically grown forms (4, 9, 51). Although bacterial biofilms have been studied in great detail (39, 40), the study of medically relevant fungal biofilms has only recently come to the forefront. Candida species, especially Candida albicans, are the most common fungi associated with biofilm-related infections (7, 38). Forty percent of patients with Candida isolated from intravenous catheters develop occult fungemia (2, 37), and the mortality rate for patients with catheter-related candidemia can be as high as 41% (37). Biofilm formation is also critical in the development of denture stomatitis, a superficial form of candidiasis that affects 65% of edentulous individuals (6-8).
We have recently developed denture and catheter models of fungal biofilms using physiological substrates and clinically relevant C. albicans strains (9). Using this model, we showed that C. albicans biofilms are highly resistant to the action of clinically important antifungal and antimicrobial agents including amphotericin B, chlorhexidine, nystatin, and fluconazole (9, 10, 29, 30). We also demonstrated that C. albicans biofilm formation proceeds in three developmental phases: (i) early phase (0 to 11 h), involving adhesion of fungal cells to the substrate, (ii) intermediate phase (∼12 to 30 h), during which the blastospores coaggregate and proliferate, forming communities while producing a carbohydrate-rich extracellular matrix (ECM), and (iii) maturation phase (∼31 to 72 h), in which the fungal cells are completely encased in a thick ECM (9). Additionally, our studies demonstrated that acquisition of antifungal resistance by C. albicans biofilms correlates with the developmental phases of these biofilms (9).
The multidrug resistance phenotype in planktonic C. albicans has previously been shown to be linked to proteins encoded by CDR1, CDR2, and MDR1 genes (41, 48). These proteins act as membrane-localized efflux pumps that pump drugs from the fungal cells. Rhodamine 123 (Rh123) is a known fluorescent substrate of efflux pumps responsible for multidrug resistance in mammalian cells, bacteria, and yeasts (11, 12, 17, 21). It has been suggested that altered membrane sterol composition, which affects membrane permeability, is a possible mechanism of azole resistance among C. albicans cells grown in suspension (23). Thus, even though the mechanisms involved in drug resistance associated with these fungal cells are well characterized, their roles in biofilm-associated resistance remain to be elucidated.
In this study, we investigated the mechanisms of C. albicans biofilm-associated fluconazole resistance at the genetic level as well as the functional level. A set of isogenic C. albicans strains lacking (i) the CDR1 or MDR1 gene (single-knockout mutants), (ii) both CDR1 and CDR2 genes (double-knockout mutant), and (iii) CDR1, CDR2, and MDR1 genes (triple-knockout mutant) was employed. The metabolic activities, dry weights, and viable cell counts of biofilms formed by these isogenic strains were compared to determine any possible effect of deletion of the CDR and/or MDR1 genes on biofilm formation. The contribution of efflux pumps to azole resistance of candidal biofilms was investigated by determining the fluconazole susceptibilities of biofilms formed by CDR and/or MDR1 deletion mutants. Expression of CDR and MDR1 genes at the early (6 h), intermediate (12 h), and mature (48 h) phases of biofilm development and in similarly grown planktonic cells was examined. Furthermore, we assessed the functional activity of efflux pumps by measuring the levels of Rh123 retained in C. albicans biofilms and planktonic cells at different developmental phases. Additionally, we investigated variation in sterol profile as a possible mechanism of azole resistance by comparing the sterol composition of C. albicans biofilms to that of planktonic cells. Our results suggest that (i) disruption of efflux pumps does not affect the biofilm formation abilities of the resulting mutants, (ii) efflux pumps contribute to azole resistance in early-phase C. albicans biofilms but not in later phases, and (iii) changes in sterol composition are involved in the resistance phenotype in the intermediate and mature phases of biofilm development.
MATERIALS AND METHODS
Strains.
Table 1 describes the C. albicans strains used in this study. Strains CAF2-1, DSY448, DSY465, DSY654, and DSY1050 were generous gifts from D. Sanglard (Lausanne, Switzerland), and strain GDH2346 was a gift from L. J. Douglas (Glasgow, United Kingdom). The wild-type strain CAF2-1 was used in Northern blotting, Rh123 efflux, and sterol analyses. Strains were grown overnight at 37°C in yeast nitrogen base (YNB) medium with amino acids (Difco Laboratories, Detroit, Mich.; catalog no. 0392-15-9) supplemented with 50 mM glucose.
TABLE 1.
Isogenic C. albicans strains used in this study
| Strain | Parent | Genotype |
|---|---|---|
| CAF2-1 | SC5314 | Δura3::imm434/URA3 |
| DSY448 | CAF2-1 | Δcdr1::hisG-URA3-hisG/Δcdr1::hisG |
| DSY465 | CAF2-1 | Δmdr1::hisG-URA3-hisG/Δmdr1::hisG |
| DSY654 | CAF2-1 | Δcdr1::hisG/Δcdr1::hisG Δcdr2::hisG-URA3-hisG/Δcdr2::hisG |
| DSY1050 | CAF2-1 | Δcdr1::hisG/Δcdr1::hisG Δcdr2::hisG/Δcdr2::hisG Δmdr1::hisG-URA3-hisG/Δmdr1::hisG |
Biofilm formation and quantitation.
Biofilms were formed on 1.5-cm2 denture acrylic (polymethylmethacrylate [PMA]) strips (Makki Dental Prosthetics, Inc., Middleburg Heights, Ohio) as described previously (9, 10). Briefly, a standard inoculum of 107 cells/ml from an overnight culture of the fungal strains was applied to the surface of PMA strips placed in a 12-well tissue culture plate. The cells were allowed to adhere for 90 min at 37°C. Nonadherent cells were removed from the strips by gentle washing with 5 ml of phosphate-buffered saline (PBS). Strips were then submerged in 4 ml of YNB medium supplemented with 50 mM glucose and incubated for various durations at 37°C on a rocker. Strips with no Candida cells served as negative controls. Control and experimental strips were then incubated at 37°C for various time periods. Planktonic cultures were grown in the same way as biofilms, in 12-well tissue culture plates, except that denture acrylic strips were not added to the wells. Biofilm and planktonic cultures were grown for 6, 12, and 48 h, corresponding to early, intermediate, and mature phases of development, respectively. Biofilms formed on PMA strips were quantified by (i) a tetrazolium XTT [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide] reduction assay and (ii) dry weight measurement, as described previously (9, 10). Assays were carried out in triplicate and were repeated on different days.
Cell viability assessment.
For assessing the viability of C. albicans cells, biofilms were scraped from denture strips and sonicated for 10 min in a water bath sonicator (Fisher Scientific Co., Pittsburgh, Pa.) to obtain a suspension of fungal cells. This suspension was incubated with 0.01% (vol/vol) trypan blue, and the viable candidal cells (not staining blue) were counted microscopically with a hemacytometer.
Antifungal susceptibility testing.
Antifungal susceptibilities of biofilm and planktonic C. albicans were determined as described previously (9). Briefly, biofilms were grown on PMA strips for 6, 12, or 48 h and the acrylic strips were transferred to wells containing different concentrations (0.5 to 256 μg/ml) of fluconazole (Pfizer Pharmaceuticals Group, New York, N.Y.). Strips were further incubated for 48 h, and metabolic activities of biofilms were measured by the XTT reduction assay as described previously (9, 10, 44). MIC was defined as the antifungal concentration which caused 50% reduction in metabolic activity of a C. albicans biofilm compared with control (incubated in the absence of drug). For each drug concentration, separate strips were used in triplicate and experiments were performed on different days. The antifungal susceptibility of planktonic cells was determined similarly (9).
PCR amplification of C. albicans CDR1 and -2 and MDR1 genes.
CDR1 and -2 and MDR1 genes were amplified by PCR using specific oligonucleotide primers. For CDR1 and CDR2 the forward primer was 5′-TATGTCAGATTCTAAGATGTC-3′ and the reverse primer was 5′-TCGATACCTTCACCTCTG-3′; for MDR1 the forward primer was 5′-AGGTGAACCCAATTCAAGTC-3′ and the reverse primer was 5′-ACAACTGGTTCCATAACGGT-3′. PCR conditions were as follows: denaturation (95°C, 2 min) followed by 33 cycles of denaturation (94°C, 1 min), annealing (50°C, 1 min), and extension (72°C, 3 min), ending with a 10-min extension at 72°C. PCR-amplified fragments were labeled with digoxigenin (DIG)-dUTP (Roche Molecular Biochemicals, Indianapolis, Ind.) according to the manufacturer's instructions and used as probes for Northern blot analyses.
Northern blot analysis.
To determine whether expression of CDR and MDR1 genes is altered during the developmental phases of biofilm formation, Northern analyses were performed on C. albicans (strain CAF2-1) biofilms and planktonic cells grown for 6, 12, or 48 h. Biofilm material was scraped from the surface of PMA strips, resuspended in PBS, and centrifuged (3,000 × g) to obtain a pellet. Planktonic cells were similarly collected. Total RNA was extracted from biofilm and planktonic cells and analyzed by Northern blotting as described previously (9, 25). UV-cross-linked RNA blots were prehybridized for 1 to 2 h at 50°C and then hybridized overnight with a DIG-labeled CDR or MDR1 probe (30 ng/ml) at 50°C. Hybridizing gene transcripts were detected with the DIG High Prime detection kit (Roche Molecular Biochemicals) according to manufacturer's instructions. 25S rRNA was used as a control for RNA loading.
Rh123 efflux assay.
To determine whether efflux pump activity varied with different developmental phases, the functional activity of these pumps was assayed as described previously (12). Briefly, C. albicans CAF2-1 was grown as a biofilm to 6, 12, or 48 h representing early, intermediate, and mature phases. At these time points, biofilms were scraped, resuspended in PBS, and sonicated for 10 min in a water bath sonicator (Fisher Scientific Co.). The number of fungal cells in the resulting suspension was determined with a hemacytometer. Planktonic cells were collected by aspiration, washed, and resuspended in PBS. Suspensions of C. albicans biofilms or planktonic cells (107 cells/ml for each) were incubated with 10 μM Rh123 at 37°C for 20 min and centrifuged at 12,000 × g in a microcentrifuge. The resulting pellet was washed twice, resuspended in 200 μl of PBS, and transferred to a 96-well plate. The fluorescence of the reaction mixture was recorded with a spectrofluorimeter (excitation and emission wavelengths of 485 and 538 nm, respectively). To determine whether cells assayed for Rh123 retention assay were metabolically active, we measured their metabolic activities using the Live/Dead kit, based on FUN-1 (2-chloro-4-[2,3-dihydro-3-methyl-{benzo-1,3-thiazol-2-yl}-methylidene]-1-phenylquinolinium iodide; Molecular Probes Inc., Eugene, Oreg.) by following the manufacturer's instructions. FUN-1 is a membrane-permeant nucleic acid-binding asymmetric halogenated cyanine dye that gives rise to cylindrical intravacuolar structures in metabolically active yeast cells (35). A biofilm or planktonic cell suspension (107 cells/ml) was incubated with FUN-1 for 45 min at 37°C, and fluorescence was estimated with a spectrofluorimeter (excitation and emission wavelengths, 485 and 585 nm, respectively). Rh123 retention by the cells was expressed as fluorescence accumulated per unit of metabolic activity.
Sterol extraction and analysis.
Sterols were extracted from C. albicans biofilms and planktonic cells and analyzed by gas-liquid chromatography (GLC) as described previously (26, 52). Briefly, Candida biofilms and planktonic cells grown to different time points were harvested and washed twice with PBS. The harvested cells were refluxed for 3 h with ethanolic KOH, filtered, and mixed with equal volumes of double-distilled water. Sterols were extracted from this mixture with 4 volumes of heptane. The extracted sterols were derivatized with hexamethyldisilazane and trimethlylchlorosilane and analyzed by GLC using an HP-6890 series gas-liquid chromatography system (Hewlett-Packard) with an OV-1 column (26, 52). Authentic sterol standards (Sigma) were used to identify various sterol intermediates based on their retention times relative to that of ergosterol. The level of each sterol was determined from the corresponding peak areas as a percentage of the total.
Statistical analyses.
All experiments were repeated at least three times on separate days. Comparison of multiple sets of data was performed by analysis of variance, and paired comparisons were performed by Student's t test using StatView software (version 4.5; Abacus Concepts).
RESULTS
Deletion of CDR1 and -2 and MDR1 genes in C. albicans mutants does not affect the abilities of these strains to form a biofilm.
To evaluate whether disruption of the CDR1, CDR2, or MDR1 gene leads to altered abilities to form biofilms, we determined the metabolic activities, dry weights, and viable-cell counts of biofilms formed by mutant strains with these genes disrupted. Biofilms were formed on denture acrylic strips as described previously (10). Our data showed that strains with the efflux pump(s) deleted (efflux pump-deleted strains) as well as the parent strain formed thick ECM-encased biofilms, showing similar morphologies when stained with Calcofluor white, a carbohydrate-binding dye (data not shown). Biofilms formed by all the strains predominantly contained blastospores, although some hyphae were also visible. These features were similar to those observed in biofilms formed by strain GDH2346, which was previously used to standardize our biofilm model (9). Moreover, biofilms formed by mutant and parent strains did not differ significantly in their metabolic activities (P > 0.05; data not shown), as determined by using the XTT tetrazolium dye-based assay, or dry weights (P > 0.05; data not shown). To examine the correlation between cell density and metabolic activity, we determined the numbers of viable cells in biofilms formed by the wild-type and mutant strains. Our data revealed no difference between the viable-cell counts in 48-h biofilms formed by mutant and parent strains (P > 0.05; data not shown). Moreover, there were no differences in viable-cell counts at the early and intermediate phases of biofilm development between wild-type and mutant strains (data not shown). Additionally, to ascertain that the disruption of genes does not lead to altered growth, we monitored the growth of these strains and found no differences in their growth curves (data not shown). Thus, our data demonstrate that biofilms formed by efflux pump-deleted strains were similar to those formed by the wild-type strain, indicating that any effects of these pumps on biofilm-associated drug resistance are not due to gross changes in biofilm structure or morphology.
Antifungal susceptibility of C. albicans biofilms is affected by drug efflux pumps in a developmental phase-specific manner.
In our previous studies, we have shown that C. albicans biofilms exhibit high resistance to fluconazole and that this resistance phenotype correlates with the corresponding developmental phase (early, intermediate, or maturation) of biofilms (9). Since efflux pumps have previously been implicated in azole resistance in planktonic C. albicans cells (48), we investigated whether the CDR1 and -2 and MDR1 genes play a role in biofilm-associated fluconazole resistance. The fluconazole susceptibilities of planktonic C. albicans and biofilms formed by efflux pump-deleted mutants were determined and compared to that of the parental strain. Antifungal susceptibility assays with planktonically grown isogenic C. albicans strains revealed drastically reduced fluconazole MICs compared to those for biofilms (Table 2). Our results showed that mature biofilms formed by mutant and parental strains were highly resistant to fluconazole (MIC > 256 μg/ml).
TABLE 2.
MICs of fluconazole against different C. albicans strains grown to different time points as biofilms or planktonic cells
| Strain | Genotype | MIC (μg/ml) for strain grown as:
|
|||||
|---|---|---|---|---|---|---|---|
| Biofilm at:
|
Planktonic cells at:
|
||||||
| 6 h | 12 h | 48 h | 6 h | 12 h | 48 h | ||
| CAF2-1 | Wild type | >256 | >256 | >256 | 2 | 2 | 2 |
| DSY448 | Δcdr1 | 256 | >256 | >256 | 1 | 1 | 1 |
| DSY465 | Δmdr1 | 256 | >256 | >256 | 2 | 2 | 2 |
| DSY654 | Δcdr1 Δcdr2 | 64 | >256 | >256 | 1 | 1 | 1 |
| DSY1050 | Δcdr1 Δcdr2 Δmdr1 | 16 | 256 | >256 | 0.13 | 0.13 | 0.13 |
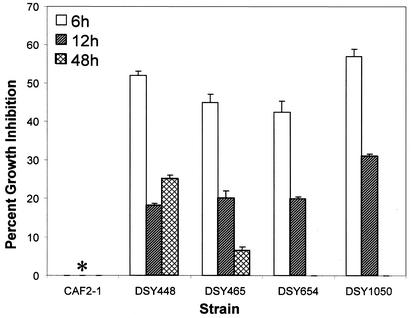
To examine whether the observed fluconazole resistance varies with developmental phases of biofilm formation, MICs of fluconazole for biofilms grown to early (6 h), intermediate (12 h), and mature (48 h) phases were determined. As shown in Table 2, early-phase biofilms formed by all mutants were more susceptible to fluconazole than intermediate- and mature-phase biofilms (Table 2). Additionally, at the 6-h time point, MICs of fluconazole against single (DSY448 and DSY465), double (DSY654), and triple (DSY1050) mutants were 256, 64, and 16 μg/ml, respectively. Moreover, biofilms formed by mutant strains exposed to a high concentration of fluconazole (256 μg/ml) exhibited a time-dependent decrease in growth inhibition, while this drug had no effect on a biofilm formed by the wild-type strain (Fig. 1). These results demonstrated that the azole resistance of biofilms formed by efflux pump-deleted mutant strains varies with the developmental phase and that efflux pumps play an additive role in contributing to antifungal resistance in early-phase biofilms.
FIG. 1.
Percent growth inhibition of C. albicans biofilms exposed to high concentration of fluconazole. Percentages of inhibition for biofilms grown to the early (6 h), intermediate (12 h), or late (48 h) phase of development and exposed to 256 μg of fluconazole/ml were determined. Strains used were: CAF2-1 (wild type), DSY448 (Δcdr1), DSY465 (Δmdr1), DSY654 (Δcdr1 Δcdr2), and DSY1050 (Δcdr1 Δcdr2 Δmdr1). For each strain, drug susceptibility decreased from the early to late phase of biofilm development. Additionally, deletion of two and three efflux pumps led to progressively decreasing susceptibility to fluconazole. Metabolic activity was normalized to the control without fluconazole, which was taken as 100%. Data (means ± standard deviations) are representative of three separate experiments. ∗, the wild-type (CAF2-1) strain showed 0% inhibition at all the time points.
Expression of genes encoding efflux pumps is temporally regulated in C. albicans biofilms.
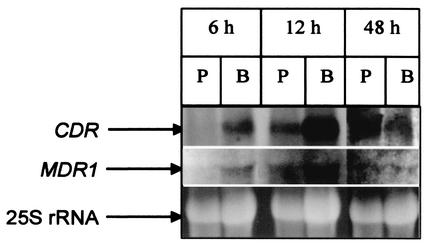
Since early-, intermediate-, and mature-phase biofilms showed differences in fluconazole resistance pattern, it is logical to assume that the CDR1 and -2 and MDR1 genes may be differentially expressed at the transcriptional level in these developmental phases. To determine whether developmental-phase-dependent biofilm-associated azole resistance is correlated with CDR1 and -2 and MDR1 expression at the mRNA level, we investigated the temporal expression of these genes in C. albicans biofilms at the transcriptional level by Northern blot analyses as described previously (9). The expression profile of the CDR1 and -2 and MDR1 genes in 6-, 12-, and 48-h biofilms formed by the wild-type strain was compared with that of planktonic C. albicans. As shown in Fig. 2, the CDR transcript was detected in the early-, intermediate-, and mature-phase biofilms of C. albicans. In contrast, expression of these genes in planktonic cells was detected only at 12 and 48 h (Fig. 2). The MDR1 transcript was detected in early-phase biofilms at 6 h but not in planktonic cells at the same time point (Fig. 2). Moreover, the levels of the MDR1 transcript for both planktonic cells and biofilms were highest at 12 h but minimal at 48 h (Fig. 2). The levels of MDR1 transcript at the 6-h time point did not differ from those at the 48-h time point. Therefore, our data showed that the expression of genes encoding efflux pumps in biofilm and planktonic cells is temporally regulated at different developmental phases.
FIG. 2.
Expression of CDR and MDR1 genes in (P) planktonic (P) and biofilm (B) forms of C. albicans. Total RNA was isolated from biofilms and planktonic cells grown for 6, 12, and 48 h. Sixty micrograms of total RNA was analyzed by Northern blotting using a CDR- or MDR1-specific probe as described in Materials and Methods. Top, CDR transcript; middle, MDR1 transcript; bottom, 25S rRNA (loading control). Results are representative of three separate experiments.
Functional analysis of drug efflux activity at the protein level.
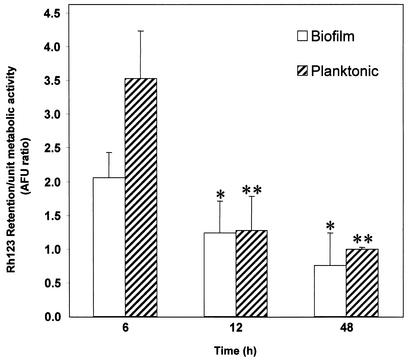
Functional activities of efflux pumps have traditionally been assayed with Rh123, a fluorescent substrate for these proteins which is retained in cells lacking functional efflux pumps (11, 12). We employed this Rh123-based assay to determine whether the levels of CDR and MDR1 mRNA are correlated with the efflux pump activity of biofilms and planktonic C. albicans at different developmental phases. Our results showed that, at 6 h, the level of Rh123 retained in biofilms was significantly lower than the corresponding levels in planktonic cells (P = 0.0006), indicating higher efflux pump activity in early-phase biofilms (Fig. 3). Moreover, compared to that of the early-phase biofilm, the levels of Rh123 at the intermediate and mature phases were significantly reduced (P < 0.05; Fig. 3). In contrast to results for the 6-h time point, at 12 and 48 h, no significant differences in Rh123 levels between biofilm and planktonic cells were found (P > 0.05). These results further confirmed our data obtained from Northern blot analyses and suggested that efflux pumps are important in biofilm-associated resistance only at the early phase of development.
FIG. 3.
Rh123 accumulation by early-, intermediate-, and mature-phase biofilms and planktonic cells of C. albicans. Data were analyzed by two-way analysis of variance, and a value of P <0.05 was considered significant. ∗, P < 0.05 versus 6-h biofilms; ∗∗, P < 0.0001 versus 6-h planktonic cells. AFU, arbitrary fluorescence units.
C. albicans biofilms have altered sterol composition at intermediate and mature phases compared to that at the early phase.
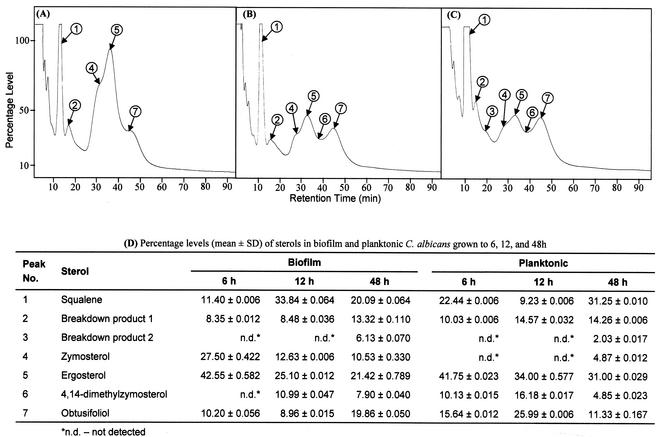
The cellular target of fluconazole in C. albicans is a cytochrome P-450 hemoprotein involved in the ergosterol biosynthetic pathway (19). Alterations in sterol composition have previously been linked to antifungal resistance in planktonic cells (18, 24, 47). Since our data showed no role of efflux pumps in the fluconazole resistance of intermediate- and mature-phase biofilms, we investigated whether this phenotype can be attributed to changes in sterol composition. Total membrane sterols were isolated from biofilms and planktonic cells and analyzed by GLC (26, 52). Representative chromatographs showing the patterns of sterols present in biofilms grown to different developmental phases are shown in Fig. 4A to C. The levels of individual sterols were calculated from their relative retention times compared to that of ergosterol and are tabulated in Fig. 4D. As can be seen in Fig. 4D, the level of ergosterol decreased by 41% between early- (6 h) and intermediate (12 h)-phase biofilms (Fig. 4D; P < 0.001). Moreover, ergosterol level was reduced by 50% at mature phase, compared to that for early-phase biofilms (Fig. 4D; P < 0.001). In contrast, planktonically grown cells showed only 18% reduction in ergosterol level between the 6- and 12-h time points (Fig. 4D; P = 0.0021). Furthermore, the levels of other ergosterol intermediates fluctuated with no apparently consistent pattern (Fig. 4D). These results show that the sterol composition of C. albicans biofilms is modulated during different developmental phases, which likely contributes to candidal biofilm resistance at intermediate and mature phases.
FIG. 4.
Variations in sterol profiles of C. albicans biofilms at different developmental phases. Sterol patterns for biofilms grown to the early (A), intermediate (B), or mature (C) phase were determined by GLC. (D) Percentages of sterols identified in C. albicans biofilms and planktonic cells (chromatograph not shown), determined from the corresponding peak areas and retention times relative to ergosterol. Peaks 1 to 7 (A to C) represent sterols described in panel D. SD, standard deviation.
DISCUSSION
Bacterial as well as fungal biofilms are characterized by significantly enhanced resistance to antimicrobial agents (4, 10, 13, 15, 45). Multiple mechanisms including drug efflux pumps, ECM, metabolic quiescence, and unique architecture have been proposed to explain bacterial biofilm-associated drug resistance (3, 14, 16, 31, 33, 50). Efflux pumps are critically involved in the antimicrobial resistance of planktonically grown bacteria, but their role in biofilm-associated resistance varies. Thus, while efflux pumps have been implicated in the resistance of Pseudomonas aeruginosa biofilms to low doses of ofloxacin, these proteins are not involved in resistance to other antimicrobials including ciprofloxacin, chloramphenicol, and tobramycin (5, 14). Similarly, biofilms formed by Escherichia coli demonstrate the involvement of an efflux pump in resistance to a low concentration of ciprofloxacin (0.004 mg/liter) but not to a higher concentration (0.1 mg/liter) (34). These studies indicate a dose- and drug-dependent role for efflux pumps in antimicrobial resistance of bacterial biofilms.
Antifungal resistance of planktonically grown C. albicans has been linked to the expression of efflux pumps such as Cdr1p, Cdr2p, and Mdr1p (1, 32, 42, 48, 49, 53). Therefore, in this study, we investigated the role of efflux pumps in the antifungal resistance of C. albicans biofilms. We determined the antifungal susceptibilities of biofilms formed by mutants carrying single, double, or triple deletion mutations of the CDR and MDR1 genes. Our results showed that, at the early phase of development, biofilms formed by these mutants were more susceptible to fluconazole than those formed by the wild-type strain. However, among the mutants, the triple-knockout strain was the most susceptible (MIC = 16 μg/ml), indicating the involvement of efflux pumps in the azole resistance of early-phase biofilms. Interestingly, at later developmental phases (12 and 48 h), biofilms formed by the mutants displayed complete resistance to fluconazole (MIC ≥ 256 μg/ml), similar to those formed by the wild-type parent strain. These results indicate that efflux pumps contribute to azole resistance in the early phase of biofilm formation but not in the later phases. To further investigate the role of efflux pumps in azole resistance, we determined the levels of CDR and MDR1 gene mRNA in biofilms formed by the wild-type strain. Our results clearly showed that expression of CDR genes is temporally regulated during C. albicans biofilm formation, with higher levels of gene transcripts detected in early- and intermediate-phase biofilms than in planktonic cells. Interestingly, the CDR and MDR1 genes were expressed at all developmental phases in C. albicans biofilms but only after 12 h in planktonic cells.
Using a different model of biofilm formation, Ramage et al. (43) recently reported that efflux pumps including Cdr1p, Cdr2p, and Mdr1p are not involved in C. albicans biofilm-associated drug resistance. Our results are in agreement with those of Ramage et al. (43) regarding the role of efflux pumps in the drug resistance of mature C. albicans biofilms. However, these investigators examined the role of efflux pumps only at 24 and 48 h, at which times the biofilms were already completely formed. We have previously shown that C. albicans biofilms pass through three distinct developmental phases: early (6 h), intermediate (12 h), and mature (48 h) (9). In the present study, we investigated the phase-dependent expression of the CDR and MDR1 genes during biofilm formation and demonstrated that efflux pumps contribute to candidal resistance only at the early phase. Our data demonstrate that it is prudent to examine biofilm-related processes, including resistance, at all three phases of biofilm development. Another difference between our study and that reported previously (43) is that we employed triple-knockout mutants in addition to single and double mutants, which were more susceptible to fluconazole than the wild type at the 6-h growth phase. These studies further confirmed the role of efflux pumps in azole resistance during the early phase of biofilm development.
Antifungal susceptibility assays revealed that biofilms formed by mutants lacking the CDR and MDR1 genes were resistant to fluconazole (MIC ≥ 256 μg/ml) at intermediate and mature phases. However, Northern blot analyses showed that, during these phases, C. albicans biofilms expressed the CDR and MDR1 genes at the mRNA level. One possible reason for this discordance could be that mRNA expression at these phases is not translated into corresponding functional proteins. In this regard, previous investigations have shown that gene expression at the mRNA level is not always correlated at the functional protein level and that it is not possible to deduce protein levels from transcript analyses (20). Therefore, we decided to perform functional analyses of efflux pump proteins using the previously described Rh123-based method (12). In this assay, lower retention of Rh123 by cells indicates higher pump activity, since Rh123 retention is a measure of drug resistance mediated by efflux pumps. Our results showed that, at 6 h, biofilms have less Rh123 retention than planktonic cells, indicating that the former is more resistant to the drug at this time point. This correlated well with our Northern blot analyses, which showed expression of CDR and MDR1 genes at 6 h in biofilms but not in planktonic C. albicans cells. Subsequent reductions in Rh123 retention (indicating increased pump activity) at 12 and 48 h also correlated with the Northern blot analysis results. Therefore, these studies revealed that efflux pump proteins are functional in intermediate- and mature-phase biofilms, although they do not seem to play a role in azole resistance at these developmental phases.
Since efflux pumps did not appear to contribute to the azole resistance of C. albicans biofilms at intermediate and mature phases, we decided to investigate whether the observed resistance is due to changes in membrane integrity mediated by variations in sterol composition. The effect of sterols on the fluidity and asymmetry of the membrane has a significant effect on the sensitivity and resistance of C. albicans cells to antifungals (19, 28). Hitchcock et al. (22) earlier showed that a C. albicans strain which is resistant to both polyene and azole groups of antifungal antibiotics had a larger lipid content and lower polar lipid-to-neutral lipid ratio than other strains. The main distinctive feature of the lipid composition of this C. albicans strain was the absence of ergosterol, which was replaced by methylated sterols, mainly lanosterol, 24-methylene-24,25-dihydrolanosterol, and 4-methylergostadiene-3-ol. These investigators suggested that the altered membrane sterol pattern provided a common basis for antifungal resistance by preventing the binding of a drug and/or reducing its permeability (22). Recent reports also suggest that changes in the status of the membrane lipid phase and asymmetry could contribute to azole resistance in C. albicans (27, 36). Moreover, microarray analyses revealed that the ERG2 (encoding the ergosterol biosynthesis pathway enzyme C-8 sterol isomerase) and CDR1 genes are upregulated in a planktonically grown fluconazole-resistant strain of C. albicans (46). Therefore, we reasoned that changes in sterol patterns may also play a role in biofilm-associated drug resistance.
Sterol analyses of C. albicans biofilms and planktonic cells grown to different time points revealed that, at 6 h, biofilm and planktonic C. albicans had similar ergosterol levels. However, at later stages, the ergosterol level of the biofilm was significantly less than that of planktonic cells. During biofilm development, the ergosterol level was reduced by 41% at intermediate phase and by 50% at the mature phase, compared to that for early-phase (6-h) biofilms. Moreover, levels of other intermediate sterols, such as zymosterol, 4,14-dimethylzymosterol, obtusifoliol, in the biofilm and planktonic C. albicans were also significantly altered. Notably, differences in sterol profiles were more pronounced at 12 and 48 h, the same growth phases when efflux pumps do not seem to have any role on drug resistance. These results suggest that membrane sterol composition is a critical component of biofilm-associated azole resistance at the intermediate and mature phases. It is possible that there exists a threshold level of ergosterol that contributes to resistance. Changes in sterol profile may lead to altered membrane permeability and hence prevent or retard the entry of antifungal agents into candidal cells. Altered sterol levels can also influence fluconazole resistance indirectly i.e., mediated by a cell wall protein or lipid.
In conclusion, we present evidence that, while efflux pumps play a critical role in azole resistance in early-phase biofilms, alteration in sterol composition is an important mechanism of antifungal resistance at the intermediate and mature phases of biofilm formation. To the best of our knowledge, and based on the current literature available, these results demonstrate for the first time that antifungal resistance in C. albicans biofilms is a developmental-phase-specific multifactorial phenomenon.
Acknowledgments
We thank Ryan Munyon, Eric Bugyris, Katie Krepkowich, and Maher Balkis for technical assistance.
This work was supported by the Center for AIDS Research at Case Western Reserve University/University Hospitals of Cleveland (AI-36219), and the National Institutes of Health (RO1-DE13992 and 1RO1DE13932-01A1) to M.A.G.
Editor: T. R. Kozel
REFERENCES
- 1.Albertson, G. D., M. Niimi, R. D. Cannon, and H. F. Jenkinson. 1996. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob. Agents Chemother. 40:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anaissie, E. J., J. H. Rex, O. Uzun, and S. Vartivarian. 1998. Predictors of adverse outcome in cancer patients with candidemia. Am. J. Med. 104:238-245. [DOI] [PubMed] [Google Scholar]
- 3.Anderl, J. N., M. J. Franklin, and P. S. Stewart. 2000. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44:1818-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baillie, G. S., and L. J. Douglas. 1999. Candida biofilms and their susceptibility to antifungal agents. Methods Enzymol. 310:644-656. [DOI] [PubMed] [Google Scholar]
- 5.Brooun, A., S. Liu, and K. Lewis. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budtz-Jorgensen, E. 1990. Etiology, pathogenesis, therapy, and prophylaxis of oral yeast infections. Acta Odontol. Scand. 48:61-69. [DOI] [PubMed] [Google Scholar]
- 7.Budtz-Jorgensen, E. 1990. Histopathology, immunology, and serology of oral yeast infections. Diagnosis of oral candidosis. Acta Odontol. Scand. 48:37-43. [DOI] [PubMed] [Google Scholar]
- 8.Budtz-Jorgensen, E., and T. Lombardi. 1996. Antifungal therapy in the oral cavity. Periodontol. 2000 10:89-106. [DOI] [PubMed] [Google Scholar]
- 9.Chandra, J., D. M. Kuhn, P. K. Mukherjee, L. L. Hoyer, T. McCormick, and M. A. Ghannoum. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandra, J., P. K. Mukherjee, S. D. Leidich, F. F. Faddoul, L. L. Hoyer, L. J. Douglas, and M. A. Ghannoum. 2001. Antifungal resistance of candidal biofilms formed on denture acrylic in vitro. J. Dent. Res. 80:903-908. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhary, P. M., and I. B. Roninson. 1991. Expression and activity of P-glycoprotein, a multidrug efflux pump, in human hematopoietic stem cells. Cell 66:85-94. [DOI] [PubMed] [Google Scholar]
- 12.Clark, F. S., T. Parkinson, C. A. Hitchcock, and N. A. Gow. 1996. Correlation between rhodamine 123 accumulation and azole sensitivity in Candida species: possible role for drug efflux in drug resistance. Antimicrob. Agents Chemother. 40:419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 14.De Kievit, T. R., M. D. Parkins, R. J. Gillis, R. Srikumar, H. Ceri, K. Poole, B. H. Iglewski, and D. G. Storey. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 45:1761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donlan, R. M. 2000. Role of biofilms in antimicrobial resistance. ASAIO J. 46:S47-S52. [DOI] [PubMed] [Google Scholar]
- 16.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egner, R., F. E. Rosenthal, A. Kralli, D. Sanglard, and K. Kuchler. 1998. Genetic separation of FK506 susceptibility and drug transport in the yeast Pdr5 ATP-binding cassette multidrug resistance transporter. Mol. Biol. Cell 9:523-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghannoum, M. A., N. M. Moussa, P. Whittaker, I. Swairjo, and K. H. Abu-Elteen. 1992. Subinhibitory concentration of octenidine and pirtenidine: influence on the lipid and sterol contents of Candida albicans. Chemotherapy 38:46-56. [DOI] [PubMed] [Google Scholar]
- 19.Ghannoum, M. A., and L. B. Rice. 1999. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 12:501-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gygi, S. P., Y. Rochon, B. R. Franza, and R. Aebersold. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19:1720-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerle, S. P., B. Rothen-Rutishauser, S. D. Kramer, M. Gunthert, and H. Wunderli-Allenspach. 2000. P-glycoprotein in cell cultures: a combined approach to study expression, localisation, and functionality in the confocal microscope. Eur. J. Pharm. Sci. 12:69-77. [DOI] [PubMed] [Google Scholar]
- 22.Hitchcock, C. A., K. J. Barrett-Bee, and N. J. Russell. 1987. The lipid composition and permeability to azole of an azole- and polyene-resistant mutant of Candida albicans. J. Med. Vet. Mycol. 25:29-37. [DOI] [PubMed] [Google Scholar]
- 23.Hitchcock, C. A., K. J. Barrett-Bee, and N. J. Russell. 1989. The lipid composition and permeability to the triazole antifungal antibiotic ICI 153066 of serum-grown mycelial cultures of Candida albicans. J. Gen. Microbiol. 135:1949-1955. [DOI] [PubMed] [Google Scholar]
- 24.Hitchcock, C. A., N. J. Russell, and K. J. Barrett-Bee. 1987. Sterols in Candida albicans mutants resistant to polyene or azole antifungals, and of a double mutant C. albicans 6.4. Crit. Rev. Microbiol. 15:111-115. [DOI] [PubMed] [Google Scholar]
- 25.Hoyer, L. L., S. Scherer, A. R. Shatzman, and G. P. Livi. 1995. Candida albicans ALS1: domains related to a Saccharomyces cerevisiae sexual agglutinin separated by a repeating motif. Mol. Microbiol. 15:39-54. [DOI] [PubMed] [Google Scholar]
- 26.Ibrahim, A. S., and M. A. Ghannoum. 1996. Chromatographic analysis of lipids, p. 52-79. In R. Prasad (ed.), Manual on membrane lipids. Springer-Verlag, New York, N.Y.
- 27.Kohli, A., Smriti, K. Mukhopadhyay, A. Rattan, and R. Prasad. 2002. In vitro low-level resistance to azoles in Candida albicans is associated with changes in membrane lipid fluidity and asymmetry. Antimicrob. Agents Chemother. 46:1046-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kontoyiannis, D. P. 2000. Efflux-mediated resistance to fluconazole could be modulated by sterol homeostasis in Saccharomyces cerevisiae. J. Antimicrob. Chemother. 46:199-203. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn, D. M., J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2002. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect. Immun. 70:878-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn, D. M., T. George, J. Chandra, P. K. Mukherjee, and M. A. Ghannoum. 2002. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob. Agents Chemother. 46:1773-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen, T., and N. E. Fiehn. 1996. Resistance of Streptococcus sanguis biofilms to antimicrobial agents. APMIS 104:280-284. [PubMed] [Google Scholar]
- 32.Lyons, C. N., and T. C. White. 2000. Transcriptional analyses of antifungal drug resistance in Candida albicans. Antimicrob. Agents Chemother. 44:2296-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 34.Maira-Litran, T., D. G. Allison, and P. Gilbert. 2000. An evaluation of the potential of the multiple antibiotic resistance operon (mar) and the multidrug efflux pump acrAB to moderate resistance towards ciprofloxacin in Escherichia coli biofilms. J. Antimicrob. Chemother. 45:789-795. [DOI] [PubMed] [Google Scholar]
- 35.Millard, P. J., B. L. Roth, H. P. Thi, S. T. Yue, and R. P. Haugland. 1997. Development of the FUN-1 family of fluorescent probes for vacuole labeling and viability testing of yeasts. Appl. Environ. Microbiol. 63:2897-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukhopadhyay, K., A. Kohli, and R. Prasad. 2002. Drug susceptibilities of yeast cells are affected by membrane lipid composition. Antimicrob. Agents Chemother. 46:3695-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen, M. H., J. E. Peacock, D. C. Tanner, A. J. Morris, M. L. Nguyen, D. R. Snydman, M. M. Wagener, and V. L. Yu. 1995. Therapeutic approaches in patients with candidemia. Evaluation in a multicenter, prospective, observational study. Arch. Intern. Med. 155:2429-2435. [PubMed] [Google Scholar]
- 38.Nicastri, E., N. Petrosiillo, P. Viale, and G. Ippolito. 2001. Catheter-related bloodstream infections in HIV-infected patients. Ann. N. Y. Acad. Sci. 946:274-290. [DOI] [PubMed] [Google Scholar]
- 39.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 40.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 41.Prasad, R., P. De Wergifosse, A. Goffeau, and E. Balzi. 1995. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr. Genet. 27:320-329. [DOI] [PubMed] [Google Scholar]
- 42.Prasad, R., S. K. Murthy, V. Gupta, and R. Prasad. 1995. Multiple drug resistance in Candida albicans. Acta Biochim. Pol. 42:497-504. [PubMed] [Google Scholar]
- 43.Ramage, G., S. Bachmann, T. F. Patterson, B. L. Wickes, and J. L. Lopez-Ribot. 2002. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J. Antimicrob. Chemother. 49:973-980. [DOI] [PubMed] [Google Scholar]
- 44.Ramage, G., W. K. Vande, B. L. Wickes, and J. L. Lopez-Ribot. 2001. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob. Agents Chemother. 45:2475-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramage, G., B. L. Wickes, and J. L. Lopez-Ribot. 2001. Biofilms of Candida albicans and their associated resistance to antifungal agents. Am. Clin. Lab 20:42-44. [PubMed] [Google Scholar]
- 46.Rogers, P. D., and K. S. Barker. 2002. Evaluation of differential gene expression in fluconazole-susceptible and -resistant isolates of Candida albicans by cDNA microarray analysis. Antimicrob. Agents Chemother. 46:3412-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanati, H., P. Belanger, R. Fratti, and M. Ghannoum. 1997. A new triazole, voriconazole (UK-109,496), blocks sterol biosynthesis in Candida albicans and Candida krusei. Antimicrob. Agents Chemother. 41:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart, P. S. 2002. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 292:107-113. [DOI] [PubMed] [Google Scholar]
- 51.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135-138. [DOI] [PubMed] [Google Scholar]
- 52.Vandenheuvel, F. A., and A. S. Court. 1968. Reference high-efficiency nonpolar packed columns for the gas-liquid chromatography of nanogram amounts of steroids. I. Retention time data. J. Chromatogr. 38:439-459. [DOI] [PubMed] [Google Scholar]
- 53.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]