Abstract
Mycoplasma species bovine group 7 bound plasminogen at the cell surface in a lysine-dependent manner. Cell-bound plasminogen was rapidly activated to plasmin by exogenous urokinase, and this activity was associated with plasminogen binding capacity. Binding assays using plasminogen modified with a trifunctional cross-linking agent revealed several binding proteins.
A small cell size and the total lack of a cell wall are phenotypic properties that distinguish mycoplasmas from other bacterial species. Many mycoplasmas lack genes involved in biosynthetic pathways (e.g., tricarboxylic acid cycle, oxidative phosphorylation, amino acid, and lipid synthesis pathways), which is reflected by their comparatively small genome sizes (28). Mycoplasmas are parasites of a wide variety of eukaryotes and prefer to scavenge macromolecules and essential nutrients from their host. Mycoplasmas exhibit a strict host range, typically displaying organ and tissue specificity, and are usually found in association with the surfaces of eukaryotic cells, although there is increasing evidence that some species can invade tissues (28, 34, 36).
Mycoplasmas play an important role in infectious diseases affecting mucosal surfaces, joints, and internal organs of humans, mammals, reptiles, arthropods, birds, and fish (see reference 28 and references therein). Several Mycoplasma species, including Mycoplasma bovis and Mycoplasma sp. bovine group 7, have been identified as etiological agents of mastitis and polyarthritis in cattle (9, 23). Mastitis is a chronic inflammatory disease in cattle and is the cause of substantial economic losses worldwide (19). Mechanisms of adherence, colonization, and invasion in mycoplasmas are comparatively poorly understood compared to those in bacterial pathogens that are more amenable to genetic manipulation. Bovine mycoplasmal pathogens are transferred between animals by direct contact, consumption of contaminated milk (mastitic milk), and/or inhalation of mucus droplets. Thus, these organisms require a mechanism(s) to travel to and invade the bovine mammary gland. Mycoplasma sp. bovine group 7 has long been implicated as a causative agent of polyarthritis and mastitis in Australian dairy herds (1, 9, 32). Recently, Mycoplasma sp. bovine group 7 was implicated as the cause of polyarthritis, mastitis, and abortion in a large, centrally managed dairy in southwest Sydney, Australia (9). Genetic fingerprinting studies of 24 epidemiologically related strains recovered from multiple tissue sites and body fluids of infected calves with polyarthritis, from mastitic milk, and from the stomach contents, lungs, and livers of aborted fetuses showed a clonal pattern (6). However, this clonal pattern was clearly distinguishable from the fingerprint patterns representative of group 7 strains recovered from unrelated outbreaks and sporadic episodes both within Australia and overseas (6). These data suggest that bovine group 7 can be a virulent, invasive organism able to cause systemic infection.
Plasminogen, a 92-kDa plasma glycoprotein, is activated to the proteolytic enzyme plasmin by eukaryotic and prokaryotic secreted activators (5, 24). Plasmin has a number of tightly regulated physiological functions, including the degradation of fibrin clots and extracellular matrix proteins during cell migration (17, 24). Plasminogen binds to exposed lysine residues on cell surfaces or in fibrin structures via lysine binding sites within the N-terminal binding domain of the molecule (4, 25). The C-terminal protease domain contains a classic serine protease triad (4, 25).
A number of bacterial pathogens are known to acquire host plasminogen on their cell surface to assist in the invasion and colonization of bovine mammary tissue (5, 16, 24). Despite the recognized importance attributed to mycoplasmal infections affecting cattle, the mechanisms used by mycoplasmas for tissue colonization and invasion remain elusive. Recently, studies by Yavlovich et al. (36) described the binding and activation of human plasminogen on the surface of Mycoplasma fermentans by a urokinase-type plasminogen activator (uPA). These events were shown to promote the invasion of HeLa cells by M. fermentans. The aims of our study were to (i) determine if Mycoplasma sp. bovine group 7 is capable of binding to and activating plasminogen and (ii) detect plasminogen binding proteins by using various ligand blotting procedures.
Bacterial strains and media.
Mycoplasma species bovine group 7 type strain PG50 (isolate 23) and field isolate 4 were obtained from a collection of group 7 mycoplasmas housed at the Elizabeth Macarthur Agricultural Institute, Camden, New South Wales, Australia, and their isolation has been described previously (9). M. bovis type strain NCTC 10131 (PG45) (a gift from R. Hirst) and field strain 20 (kindly supplied by J. Forbes-Faulkener) were also examined for their ability to bind plasminogen. Each of these strains was grown in a modified Friis broth or on Friis agar as described by Hum et al. (9). The mycoplasma cells were pelleted by centrifugation at 8,000 × g for 30 min in culture, followed by centrifugation at 11,000 × g for 30 min in phosphate-buffered saline (PBS) (pH 7.4). The number of viable cells was determined as CFU.
Human and bovine plasminogen sequence comparison.
Human and bovine glu-plasminogen (accession numbers P00747 and P06868, respectively) were shown to be 78% identical (82% conserved) at the amino acid level overall and were considered equivalent for experimental purposes (31).
Plasminogen isolation and modification.
Bovine and human glu-plasminogen were isolated from plasma by using lysine-Sepharose chromatography as described by Andronicos et al. (2). Both human and bovine glu-plasminogen were conjugated with fluorescein isothiocyanate (FITC) (isomer 1) (Sigma Chemical Co.) as described previously (7), and the differences in ratios of FITC conjugation to human and bovine plasminogen (factor of 2.45) were normalized after measuring the fluorescence of each preparation with a Biolumin 960 fluorimeter (Molecular Dynamics). Modification of plasminogen in this manner or with NHS-biotin (Pierce) has been shown to retain the lysine binding activity of the molecule (2, 27).
Human glu-plasminogen was conjugated to the trifunctional cross-linking agent sulfosuccinimidyl-2-[6-(biotinamido)-2-(p-azidobenzamido)-hexanoamido]-ethyl-1,3′-dithiopropionate(sulfo-SBED) (Pierce) according to the manufacturer's instructions. Plasminogen modified with this agent (referred to as S-plasminogen) retains its lysine binding capacity as assessed by lysine-Sepharose chromatography (data not shown).
FITC-plasminogen binding assay.
To perform binding and activation assays, mycoplasma cells were fixed in 1% paraformaldehyde dissolved in PBS at a stock concentration equivalent to 1013 cells/ml. For binding assays, a fixed stock solution of cells was pelleted by centrifugation at 14,000 × g for 15 min, washed three times with chilled PBSB (PBS containing 0.1% [wt/vol] bovine serum albumin), and resuspended in PBSB at a final concentration of 5 × 1012 cells/ml. The cells were then incubated with 100 μg of FITC-labeled plasminogen per ml in the absence or presence of the lysine analogue tranexamic acid (TA) (1 mM; Sigma Chemical Co.) for 45 min in the dark on ice. The cells were then washed twice as described above and finally resuspended in 300 μl of PBS for flow cytometry (FACSort; Becton Dickinson) as described previously (27). Autofluorescence levels for each mycoplasma strain were measured and subtracted from each assay result. All data were analyzed with CellQuest software (Becton Dickinson). Control experiments confirmed a direct correlation between the capacity to bind plasminogen and mycoplasma cell number (data not shown). Furthermore, binding of plasminogen to mycoplasma cells was concentration dependent (data not shown).
Plasminogen activation assay.
The generation of cell surface plasmin by uPA was detected by spectrophotometric determination with the chromogenic plasmin substrate Spectrozyme-PL (American Diagnostica Inc.). The optimal concentration of uPA was determined by construction of a kinetic concentration curve of plasmin activity resulting from the incubation of various concentrations of human uPA (Calbiochem) with plasminogen (100 μg/ml) and Spectrozyme-PL (250 μmol/liter). Fixed stock solutions of mycoplasmas were washed three times with cold PBSB, resuspended in PBSB at a final concentration of 5 × 1012 cells/ml, and incubated with various concentrations of plasminogen (0 to 100 μg/ml). The cells were then washed and resuspended in Tris-buffered saline (TBS) and incubated at 37°C with 30 IU of uPA per ml and 250 μmol of Spectrozyme-PL per liter in a final reaction volume of 100 μl. Color development was monitored at 415 nm over 45 min with a 96-well Spectra Max 250 microplate reader (Molecular Devices). The plasmin activity generated on the cell surface was extrapolated from a plasmin (American Diagnostica Inc.) standard curve.
Plasminogen ligand blotting analysis.
Plasminogen ligand blotting analyses were conducted as previously described (2) with minor modifications. Whole mycoplasma cell lysates were prepared in nonreducing sample buffer, and the proteins were fractionated on sodium dodecyl sulfate (SDS)-10% polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were washed once with TNCM (50mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM CaCl2, 3 mM MgCl2) and blocked with TNCM-2% polyvinyl pyrrolidine 40 (TNCM-PVP-40) overnight at room temperature. The membranes were then probed with 5 nM biotinylated glu-plasminogen in the absence or presence of 100 mM ɛ-amino caproic acid in TNCM-PVP-40 containing 0.05% (vol/vol) Tween 20 (TNCMT) for 45 min and washed for 1 h with three changes of TNCMT. Membranes were probed with a 1:4,000 dilution of horseradish peroxidase (HRP)-conjugated neutravidin (Calbiochem) in TNCMT-PVP-40 for 1 h, followed by three washes with TNCMT and one wash with TNCM. The blots were then developed by enhanced chemiluminescence (SuperSignal; Pierce).
Cell surface sulfo-SBED-plasminogen binding analysis.
A novel approach was developed for the detection of cell surface plasminogen binding proteins by using S-plasminogen. Sulfo-SBED is built on a biocytin backbone and contains two chemically reactive groups, a sulfo-NHS arm for ligand coupling with a cleavable disulfide bridge and a photosensitive phenyl azide that is activated by long-wave UV light (8). Since the sulfo-NHS arm and phenyl azide arm are only 22.8 Å apart, S-plasminogen can be cross-linked to cell surface receptor proteins via the phenyl azide group (8). Lysis of the cells in a buffer containing a disulfide reducing agent allows biotin label transfer to the plasminogen receptor(s). Similar methods have been used successfully to detect cell surface receptors. For example, Rabin et al. (26) modified lipopolysaccharide with a radiolabeled sulfo-NHS-based photoactive cross-linker to identify its major binding site on human monocytes.
The S-plasminogen binding assay was performed with live mycoplasma cells under the same conditions as the FITC-plasminogen binding assays with the following modifications. First, to maintain cell viability (>90%), mycoplasma cells were centrifuged at 11,000 × g. Cell viability was monitored by propidium iodide (Sigma Chemical Co.) staining of samples and measurement with a Biolumin 960 fluorimeter; loss of viability corresponds to increased fluorescence due to nuclear staining (27). Following incubation, unbound S-plasminogen was removed by washing with PBS, and the cells were exposed to long-wavelength UV light (365 nm) for 20 min at a distance of 5 cm on ice by using the UV Stratalinker 1800 (Stratagene). These conditions gave maximal cross-linking activity (data not shown). The cells were then pelleted as described above and resuspended in reducing SDS-polyacrylamide gel electrophoresis (SDS-PAGE) buffer at an approximate final concentration of 2.5 × 1014 cells/ml. The whole-cell lysates were separated by SDS-12% PAGE, and proteins were transferred to PVDF membranes. For detection of biotin-labeled proteins, the membranes were washed in TBST (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% [vol/vol] Tween 20), blocked in TBST-10% milk powder at room temperature for 1 to 2 h, rinsed in TBST, and incubated with a 1:8,000 dilution of horseradish peroxidase-conjugated neutravidin dissolved in 2% milk-TBST for 1 h at room temperature. After three washes with TBST and one wash with TBS, the complexes were detected by enhanced chemiluminescence.
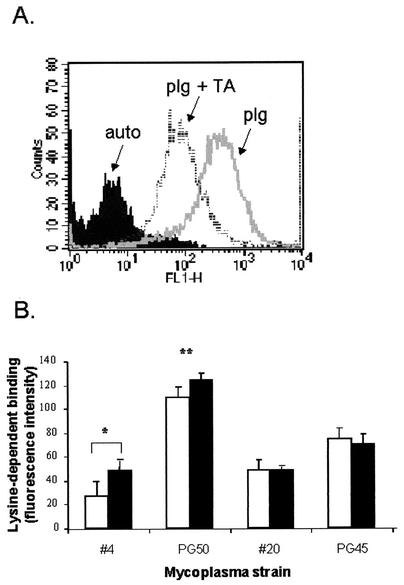
The ability of human and bovine plasminogen to bind in a lysine-dependent manner to cell surfaces of paraformaldehyde-fixed mycoplasmal strains was assessed semiquantitatively by flow cytometry (Fig. 1). A comparison of normalized human and bovine FITC-plasminogen lysine-dependent binding to two strains of Mycoplasma sp. bovine group 7 was made under optimal conditions (Fig. 1B). For both human and bovine plasminogen, the binding capacity was fourfold higher in strain PG50 than in strain 4 (P < 0.01). While there was no significant difference between human and bovine plasminogen binding to PG50, there was a significant difference with strain 4 (0.01 > P < 0.05). Control experiments using two strains of M. bovis (PG45 and field strain 20) also showed no significant difference between human and bovine plasminogen binding capacities, both of which were similar to that of strain 4 (Fig. 1B). This result suggests that in most cases there is no species difference in the utilization of plasminogen. McCoy et al. (18), who studied human, equine, and porcine plasminogen binding to human equine and porcine strains of streptococci, also found that there was no species preference for the host plasminogen.
FIG. 1.
Cell surface FITC-plasminogen binding to mycoplasma strains. (A) Representative histogram plot of FITC-labeled human plasminogen binding to strain PG50, showing relative FITC fluorescence intensity (FLH-1) verses cell count. Autofluorescence relates to background levels of cellular fluorescence measured in the absence of FITC-plasminogen. The amount of lysine-dependent binding was measured as the difference between the geometric means of FITC-plasminogen (plg) and FITC-plasminogen plus TA (plg+TA). (B) Comparison of human (open bars) and bovine (filled bars) lysine-dependent FITC-plasminogen binding to four mycoplasmal strains. The concentrations of human FITC-plasminogen (50 μg/ml), TA (1 mM), and cells (5 × 1012 cells/ml) remained constant for each assay. Bars indicate means ± standard errors of the means (n = 3). **, statistically significant difference between strain PG50 and strain 4 for both human and bovine plasminogen binding (P < 0.05). *, significant difference between human and bovine plasminogen binding in strain 4 only (P < 0.01).
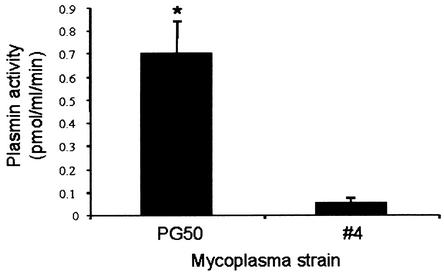
Activation of bound plasminogen by uPA was concentration dependent for both strains of Mycoplasma sp. bovine group 7 (data not shown). At 100 μg of plasminogen per ml, PG50 generated approximately 10-fold more plasmin than strain 4 (Fig. 2), reflecting its high human/bovine plasminogen binding capacity (Fig. 1). We were unable to show any plasmin activity on cells in the absence of added plasminogen or uPA. This observation indicates that these mycoplasmas do not produce their own plasminogen activators. Yavlovich et al. (36) also found that plasminogen bound to M. fermentans could not be activated in the absence of added activators. The level of plasminogen activation on the cell surface of both strains under these conditions may not reflect their activation potential in vivo, as other activators such as tissue type plasminogen activator (33) may more efficiently activate plasminogen on these strains.
FIG. 2.
Cell surface plasmin activity. Fixed mycoplasma strains PG50 and 4 were incubated with 100 μg of human glu-plasminogen per ml. Plasminogen activation by uPA (30 IU) was then determined by incubation with the Spectrozyme-PL plasmin substrate for 30 min. Bars indicate means ± standard errors of the means (n = 3). *, statistically significant difference between strain PG50 and strain 4 (P < 0.05).
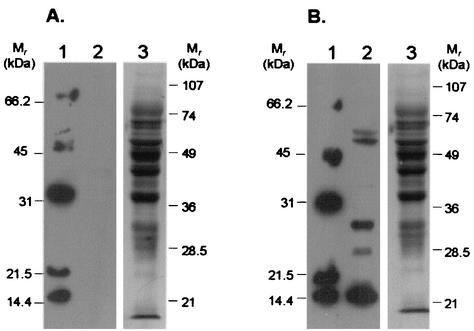
Since maximal plasminogen binding and activation occurred with the group 7 type strain PG50, the following experiments focused on this strain. Ligand blots of whole-cell lysates of PG50 with biotinylated plasminogen showed the presence of five easily distinguishable lysine-dependent binding proteins ranging in molecular mass from 14 to 55 kDa (Fig. 3). Overexposed blots showed the presence of two other weakly binding plasminogen receptor proteins with approximate molecular masses of 18 and 40 kDa (data not shown). The presence of multiple plasminogen binding proteins in other Mycoplasma strains (36) and various other prokaryotes (3, 15, 22, 30, 35) has been reported previously.
FIG. 3.
Biotinylated plasminogen ligand blot with whole-cell lysates of strain PG50. Whole-cell lysates were fractionated by SDS-12% PAGE (approximately 30 μg/lane), and the proteins were transferred to PVDF and subjected to ligand blotting with 5 nM biotinylated human glu-plasminogen (lanes 2) in the presence (A) or absence (B) of 100 mM ɛ-amino caproic acid. Lanes 1, biotinylated low-molecular-mass markers (Bio-Rad). After ligand analysis, the blots were washed and briefly stained with amido black to show the amount of protein transferred (lanes 3). All blots shown were derived from gels run, transferred, and probed in parallel and were exposed onto the same piece of autoradiograph film so that a direct comparison could be made between them. Note the absence of lysine-dependent plasminogen binding proteins in panel A (lane 2). Molecular masses of prestained molecular mass markers are as indicated.
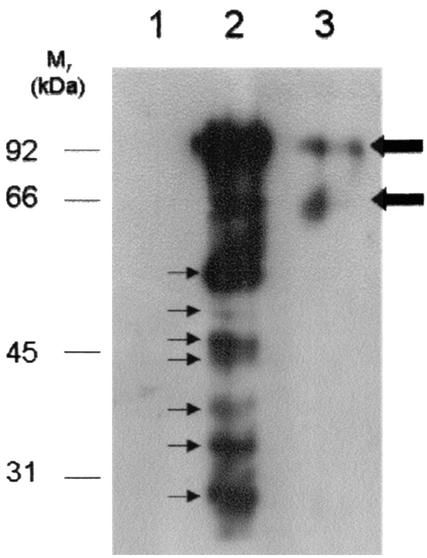
Ligand blots of whole-cell lysates do not definitively distinguish bona fide cell surface plasminogen binding proteins from proteins that reside intracellularly and fortuitously bind plasminogen (27). A second assay was developed to identify cell surface plasminogen binding proteins by using S-plasminogen. By this method, it is clear that there are multiple (at least six) plasminogen binding proteins of relatively equal abundances on the cell surface of strain PG50, ranging in size from approximately 20 to 55 kDa (Fig. 4, lane 2). Lower blot exposure times (data not shown) confirmed the presence of two distinguishable plasminogen binding proteins at approximately 45 kDa. Furthermore, a weak plasminogen binding protein band with an approximate molecular mass of 50 kDa was also evident (Fig. 4, lane 2). Control assays performed in the absence of S-plasminogen (Fig. 4, lane 1), or in the presence of S-plasminogen and TA (Fig. 4, lane 3) indicated that there were no detectable naturally biotinylated proteins present in the whole-cell lysate of strain PG50 and that none of the S-plasminogen binding occurred in a non-lysine-dependent manner, as biotin had not been transferred to any of the proteins. It was possible that trace levels of S-plasminogen (92 kDa) or light (25 kDa) and heavy chains (65 kDa) of S-plasmin in the preparation contributed to the biotinylated proteins detected in Fig. 4. To investigate this, the blots were also probed with a rabbit antiplasminogen polyclonal antibody followed by a goat anti-rabbit-HRP polyclonal antibody (data not shown). Proteins with apparent molecular masses of 185, 92, and 66 kDa were detected, corresponding to S-plasminogen dimer, S-plasminogen monomer, and S-plasmin heavy chain, respectively (data not shown). These results indicate that all other bands shown in Fig. 4 are likely to represent plasminogen binding proteins of mycoplasmal origin.
FIG. 4.
Protein blot analysis of mycoplasma cell surface S-plasminogen binding proteins from strain PG50. Cells were incubated either in the absence of S-plasminogen (lane 1), in the presence of S-plasminogen alone (lane 2), or in the presence of S-plasminogen and 1 mM TA (lane 3) prior to UV exposure. After UV exposure (which is required to transfer biotin to plasminogen binding proteins), whole-cell lysates were prepared and fractionated by reducing SDS-12% PAGE (approximately 15 μg/lane), and the proteins were transferred to PVDF membranes. The blot shows biotin-labeled lysine-dependent plasminogen binding proteins (thin arrows) detected with neutravidin-HRP followed by chemiluminescence. Thick arrows indicate traces of S-plasminogen detected with neutravidin-HRP. Molecular masses of molecular mass markers are as indicated.
Many studies conducted on streptococcal pathogens have shown that there are variations in plasminogen binding capacity between species and strains of species (10, 11, 12, 13, 18). A direct comparison between plasminogen binding to the four different strains of mycoplasmas further supported this observation, as the different species displayed variations in plasminogen binding capacity.
Numerous diverse plasminogen receptors on mammalian cells (14, 20, 27, 29) and bacterial pathogens (3, 15, 21, 22, 30, 35), including mycoplasmas (36), have been reported. Yavlovich et al. (36), using an 125I-labeled plasminogen ligand blot and autoradiography detection system with M. fermentans plasma membranes, detected two putative plasminogen receptors of approximately 32 and 55 kDa. We detected several potential receptors in Mycoplasma sp. bovine group 7 by using either ligand blots of whole-cell extracts or the novel S-plasminogen label-transfer assay with intact live cells, which was designed to ensure the detection of cell surface binding moieties. While the proteins detected by both methods fall into the same molecular mass range (i.e., ∼15 to 55 kDa), at this stage it is not possible to say whether the repertoires of proteins detected by either method are identical. Future experiments will be aimed at identifying these proteins and confirming them as bona fide plasminogen receptors not only by determining the ability of recombinant proteins to bind to and activate plasminogen but also by providing evidence of cell surface localization. Nevertheless, it is clear that a number of different plasminogen binding proteins were detected on the group 7 type strain PG50 cell surface, suggesting that there is no one protein responsible for localizing plasminogen on these mycoplasmas.
This is the first study to report plasminogen binding and activation at the cell surface of bovine mycoplasmas associated with mastitis. Currently, the molecular basis by which mycoplasmal mastitis pathogens invade and stably infect the host bovine mammary gland remains largely elusive. A number of other nonmycoplasmal mastitis pathogens are known to become pathogenic by using the host plasminogen system (5, 10, 12). It is therefore plausible that plasminogen binding and activation are a virulence determinant of mycoplasmas and provides a means of invading the mammary gland, resulting in bovine mastitis pathology.
Acknowledgments
We are grateful for the excellent technical assistance of Wendy Forbes. We thank Robert Hirst and Judy Forbes-Faulkner for kindly supplying M. bovis type strain PG45 and field strain 20, respectively.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Alexander, P. G., K. J. Slee, S. McOrist, L. Ireland, and P. J. Coloe. 1985. Mastitis in cows and polyarthritis and pneumonia in calves caused by Mycoplasma species bovine group 7. Aust. Vet. J. 62:135-136. [DOI] [PubMed] [Google Scholar]
- 2.Andronicos, N. M., M. Ranson, J. Bognacki, and M. S. Baker. 1997. The human ENO1 gene product (recombinant human α-enolase) displays the characteristics required for a plasminogen binding protein. Biochem. Biophys. Acta 1337:27-39. [DOI] [PubMed] [Google Scholar]
- 3.Broeseker, J. A., M. D. Boyle, and R. Lottenberg. 1988. Characterisation of the interactions of human plasmin with its specific receptor on a group A streptococcus. Microb. Pathog. 5:19-27. [DOI] [PubMed] [Google Scholar]
- 4.Castellino, F. J. 1995. Plasminogen, p. 495-510. In K. A. High and H. R. Roberts (ed.), Molecular basis of thrombosis and hemostasis. Marcel Dekker, Inc., New York, N.Y.
- 5.Coleman, J. L., and J. L. Benach. 1999. Use of the plasminogen activation system by microorganisms. J. Lab. Clin. Med. 134:567-576. [DOI] [PubMed] [Google Scholar]
- 6.Djordjevic, S. P., W. A. Forbes, J. Forbes-Faulkner, P. Kuhnert, S. Hum, M. A. Hornitzky, E. M. Vilei, and J. Frey. 2001. Genetic diversity among Mycoplasma species bovine group 7: clonal isolates from an outbreak of polyarthritis, mastitis, and abortion in dairy cattle. Electrophoresis 22:3551-3561. [DOI] [PubMed] [Google Scholar]
- 7.Goding, J. W. 1976. Conjugation of antibodies with fluorochromes: modification to the standard methods. J. Immunol. Methods 13:215-226. [DOI] [PubMed] [Google Scholar]
- 8.Hermanson, G. T. 1996. Bioconjugate techniques, p. 289-291. Academic Press, Inc., New York, N.Y.
- 9.Hum, S., A. Kessell, S. P. Djordjevic, R. Rheinberger, M. Hornitzky, W. Forbes, and J. Gonsalves. 2000. Mastitis, polyarthritis and abortion caused by Mycoplasma species bovine group 7 in dairy cattle. Aust. Vet. J. 78:744-750. [DOI] [PubMed] [Google Scholar]
- 10.Leigh, J. A. 1993. Activation of bovine plasminogen by Streptococcus uberis. FEMS Microbiol. Lett. 114:67-72. [DOI] [PubMed] [Google Scholar]
- 11.Leigh, J. A., S. M. Hodgkinson, and R. A. Lincoln. 1998. The interaction of Streptococcus dysgalactiae with plasmin and plasminogen. Vet. Microbiol. 61:121-135. [DOI] [PubMed] [Google Scholar]
- 12.Leigh, J. A., and R. A. Lincoln. 1997. Streptococcus uberis acquires plasmin activity following growth in the presence of bovine plasminogen through the action of its specific plasminogen activator. FEMS Microbiol. Lett. 154:123-129. [DOI] [PubMed] [Google Scholar]
- 13.Lincoln, R. A., and J. A. Leigh. 1998. Characterisation of the interaction of bovine plasmin with Streptococcus uberis. J. Appl. Microbiol. 84:1104-1110. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Alemany, R., P. Correc, L. Camoin, and P. Burtin. 1994. Purification of the plasmin receptor from human carcinoma cells and comparison to alpha enolase. Thromb. Res. 75:371-381. [DOI] [PubMed] [Google Scholar]
- 15.Lottenberg, R., L. E. DesJardin, H. Wang, and M. D. Boyle. 1992. Strepokinase-producing streptococci grown in human plasma acquire unregulated cell-associated plasmin activity. J. Infect. Dis. 166:436-440. [DOI] [PubMed] [Google Scholar]
- 16.Lottenberg, R., D. Minning-Wenz, and M. D. P. Boyle. 1994. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends Microbiol. 2:20-24. [DOI] [PubMed] [Google Scholar]
- 17.Markus, G. 1996. Conformational changes in plasminogen, their effect on activation, and the agents that modulate activation rates: a review. Fibrinolysis 10:75-85. [Google Scholar]
- 18.McCoy, H. E., C. C. Broder, and R. Lottenberg. 1991. Streptokinases produced by pathogenic group C streptococci demonstrate species-specific plasminogen activation. J. Infect. Dis. 164:515-521. [DOI] [PubMed] [Google Scholar]
- 19.Miles, H., W. Lesser, and P. Sears. 1992. The economic implications of bioengineered mastitis control. J. Dairy Sci. 75:596-605. [DOI] [PubMed] [Google Scholar]
- 20.Miles, L. A., C. M. Dahlberg, J. Plescia, J. Felez, K. Kato, and E. F. Plow. 1991. Role of cell-surface lysines in plasminogen binding to cells: identification of alpha enolase as a candidate plasminogen receptor. Biochemistry 30:1682-1691. [DOI] [PubMed] [Google Scholar]
- 21.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pancholi, V., and V. A. Fischetti. 1998. Alpha enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 23.Pfutzner, H., and K. Sachse. 1996. Mycoplasma bovis as an agent of mastitis, pneumonia, arthritis and genital disorders in cattle. Rev. Sci. Technol. 15:1477-1494. [DOI] [PubMed] [Google Scholar]
- 24.Pollanen, J., R., W. Stephens, and A. Vaheri. 1991. Directed plasminogen activation at the surface of normal and malignant cells. Adv. Cancer Res. 57:273-328. [DOI] [PubMed] [Google Scholar]
- 25.Ponting, C. P., J. M. Marshall, and S. A. Cederholm-Williams. 1992. Plasminogen: a structural review. Blood Coag. Fibrinol. 3:605-614. [PubMed] [Google Scholar]
- 26.Rabin, R. L., M. M. Bieber, and N. N. H. Teng. 1993. Lipopolysaccharide and peptidoglycan share binding sites on human peripheral monocytes. J. Infect. Dis. 168:135-142. [DOI] [PubMed] [Google Scholar]
- 27.Ranson, M., N. M. Andronicos, M. J. O'Mullane, and M. S. Baker. 1998. Increased plasminogen binding is associated with metastatic breast cancer cells: differential expression of plasminogen binding proteins. Br. J. Cancer 77:1586-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redlitz, A., B. J. Fowler, E. F. Plow, and L. A. Miles. 1995. The role of an enolase-related molecule in plasminogen binding to cells. Eur. J. Biochem. 227:407-415. [DOI] [PubMed] [Google Scholar]
- 30.Rosey, E. L., R. A. Lincoln, P. N. Ward, R. J. Yaney, Jr., and J. A. Leigh. 1999. PauA: a novel plasminogen activator from Streptococcus uberis. FEMS Microbiol. Lett. 178:27-33. [DOI] [PubMed] [Google Scholar]
- 31.Schaller, J., P. W. Moser, M. G. K. Dannegger, S. J. Rosselet, U. Kamper, and E. E. Rickli. 1985. Complete amino-acid sequence of bovine plasminogen comparison with human plasminogen. Eur. J. Biochem. 142:267-278. [DOI] [PubMed] [Google Scholar]
- 32.Simmons, G. C., and L. A. Y. Johnston. 1963. Arthritis in calves caused by Mycoplasma sp. Aust. Vet. J. 39:11-14. [Google Scholar]
- 33.Tarshis, M., B. Morag, and M. Mayer. 1993. Mycoplasma cells stimulate in vitro activation of plasminogen by purified tissue-type plasminogen activator. FEMS Microbiol. Lett. 106:201-204. [DOI] [PubMed] [Google Scholar]
- 34.Taylor-Robinson, D., H. A. Davies, P. Saratheandra, and P. M. Furr. 1991. Intracellular location of mycoplasmas in cultured cells demonstrated by immunocytochemistry and electron microscopy. Int. J. Exp. Pathol. 73:705-714. [PMC free article] [PubMed] [Google Scholar]
- 35.Ullberg, M., G. Kronvall, I. Karlsson, and B. Wiman. 1990. Receptors for human plasminogen on gram-negative bacteria. Infect. Immun. 58:21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yavlovich, A., A. A. Higazi, and S. Rottem. 2001. Plasminogen binding and activation by Mycoplasma fermentans. Infect. Immun. 69:1977-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]