Abstract
Shiga toxin 2 (Stx2)-converting bacteriophages induced from 49 strains of Escherichia coli O157:H7 isolated during a recent outbreak of enterocolitis in Spain were examined in an attempt to identify the variability due to the stx2-converting phages. The bacterial isolates were divided into low-, medium-, and high-phage-production groups on the basis of the number of phages released after mitomycin C induction. Low- and medium-phage-production isolates harbored two kinds of phages but released only one of them, whereas high-phage-production isolates harbored only one of the two phages. One of the phages, φSC370, which was detected only in the isolates with two phages, showed similarities with phage 933W. The second phage, φLC159, differed from φSC370 in morphology and DNA structure. When both phages were present in the same bacterial chromosome, as occurred in most of the isolates, only φSC370 was detected in the supernatants of the induced cultures. If φLC159 was released, its presence was masked by φSC370. When φSC370 was absent, large amounts of φLC159 were released, suggesting that there was some regulation of phage expression between the two phages. To our knowledge, this is the first description of clonal variability due to phage loss. The higher level of phage production was reflected in the larger amounts of Stx2 toxin produced by the cultures. Some relationship between phage production and the severity of symptoms was observed, and consequently these observations suggest that the virulence of the isolates studied could be related to the variability of the induced stx2-converting phages.
Escherichia coli Shiga toxin-producing (STEC) strains are responsible for hemorrhagic diarrhea and hemolytic-uremic syndrome, which leads to acute renal failure in children (19, 22). E. coli O157:H7 is the most common virulent serogroup, but other serogroups have been described, such as O18, O26, O111, O128, and O138 (1). STEC is mainly transmitted by consumption of contaminated food and water and by personal contact (6, 17, 19). The number of outbreaks caused by STEC has increased drastically in all developed countries (19, 22).
Among other virulence factors, the pathogenicity of STEC is related to the production of Shiga toxin 1 (Stx1), Stx2, or variants of Stx2 (7, 22). The genes encoding Shiga toxins (stx) are located in the genomes of temperate lambdoid bacteriophages (21). Stx2-converting phages are heterogeneous because they have wide ranges of DNA structures, restriction patterns, host spectra, and morphologies (10, 26, 30, 31). Phages carry stx1 (20), stx2 (21), and stx2 variants (18, 29). They carry not only the Shiga toxin genes but also some regulatory genes; hence, they are responsible for the production of toxin and its regulation (31, 32). Intraintestinal phage transmission between different bacteria has been described for stx1 (2) and in vitro for stx2 (26). Additionally, treatment with inhibitors of DNA synthesis, such as trimethoprim-sulfamethoxazole and mitomycin C, results in phage induction and an increase in toxin production in various STEC strains (12).
The mobility of the stx2 characteristic mediated by phages suggests that a wide variety of E. coli strains (and other enterobacteria) are susceptible to infection by stx phages. Recent studies have indicated that strains of the most virulent enterohemorrhagic E. coli serotypes (O157:H7, O103:H2, O111:H−) belong to a few clone complexes (3, 11). However, the presence of variable DNA, which can be readily transferred by horizontal genetic exchange, has hampered our understanding of the phylogeny and pathogenesis of STEC. Moreover, genetic exchange has been demonstrated in lambdoid phages, and it has been proposed that these phages have a common gene pool from which DNA can be exchanged (9, 18). This exchange could influence the host strain, as has been described for E. coli O157:H7 (34), Shigella dysenteriae (15), and Shigella sonnei, (28), producing variations in toxin production and thus in the virulence of the host bacteria.
This study of stx2-converting phages was carried out with 49 strains of E. coli isolated from a recent outbreak of STEC infection reported in Spain, which occurred in the Barcelona area between 19 September and 5 November 2000 and affected three schools. Infections in two of the schools were caused by contaminated food supplied by the same caterers. In the third school the transmission was attributed to personal contact. The whole outbreak involved more than 200 patients, most of whom were children under the age of 10 years. Six children developed hemolytic-uremic syndrome. The outbreak was associated with ingestion of E. coli O157:H7 via contaminated sausages (4), although bacteria were not isolated from the suspected food.
Although all bacterial of the isolates were E. coli O157:H7 stx1− stx2+ and thus were assumed to be clonal, we were interested in examining the presence of stx2-converting phages in these bacteria. The presence and expression of stx2 varied from one isolate to another due to certain variability observed in the stx2-converting bacteriophages.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and media.
Forty-nine strains were isolated from children under the age of 10 years and from one adult from three schools in the Barcelona area in Spain in 2000. Fifty percent of the strains were isolated from patients showing symptoms of infection; most of the patients had primary infections, but nine patients had secondary infections (Table 1). The individuals with primary infections had direct contact with the contaminated food and exhibited symptoms before 2 October, whereas the individuals with secondary infections had no contact with the contaminated food but were in contact with individuals with primary infections and exhibited symptoms after 3 October. Analyses were also performed with asymptomatic subjects related to the outbreak, who could not be classified as individuals with primary or secondary infections. A total of 24 of the asymptomatic subjects were positive for E. coli O157:H7, and the strains were included in this study (Table 1). All of the isolates were E. coli O157:H7 stx1− stx2+.
TABLE 1.
Isolates used in this study
| Isolate(s) | Age (years) | Infection | Symptoms |
|---|---|---|---|
| 2 | 3 | Secondary | Diarrhea, fever |
| 7 | 6 | Primary | Bloody diarrhea, abdominal cramps |
| 11 | 9 | Primary | Diarrhea, abdominal cramps |
| 20 | 5 | Primary | No diarrhea, vomiting, fever |
| 22 | 4 | Primary | Diarrhea |
| 26 | 1 | Secondary | Bloody diarrhea, fever, vomiting |
| 39 | 5 | Primary | Diarrhea, abdominal cramps |
| 41 | 3 | Primary | Diarrhea |
| 65 | 7 | Secondary | No diarrhea, fever, vomiting |
| 81 | 8 | Primary | No diarrhea, abdominal cramps |
| 84 | 3 | Secondary | Bloody diarrhea |
| 104 | 3 | Secondary | No diarrhea, vomiting, fever |
| 118 | 3 | Primary | Diarrhea, fever |
| 135 | 4 | Primary | Diarrhea |
| 156 | 4 | Secondary | Diarrhea |
| 159 | 4 | Secondary | Diarrhea |
| 233 | 4 | Primary | Diarrhea, abdominal cramps |
| 360 | 2 | Secondary | Bloody diarrhea, fever, vomiting |
| 346 | 3 | Primary | Bloody diarrhea, HUSa |
| 360 | 2 | Primary | Bloody diarrhea, vomiting, HUS |
| 370 | 4 | Secondary | No diarrhea, fever, vomiting |
| 371 | 3 | Primary | No diarrhea, fever, vomiting |
| 451 | 6 | Primary | Diarrhea, abdominal cramps |
| 622 | 20 | Secondary | Diarrhea |
| 631 | 1 | Secondary | Diarrhea, vomiting |
| 61, 67, 71, 87, 90, 98, 122, 127, 128, 132, 143, 168, 237, 288, 301, 305, 487, 523, 624, 635, 636, 652, 659, 671 | 1-8 | None |
HUS, hemolytic-uremic syndrome.
E. coli laboratory strain DH5α was used as the host for propagation of bacteriophages induced from the isolates studied. E. coli O157:H7 strain ATCC 43889, which produces Stx2, and bacteriophage 933W (21) were used as positive controls for bacteria and phages, respectively, and for the bacteriophage immunity tests. E. coli O157:H7 strain ATCC 43888, which does not contain either stx1 or stx2, was used as a negative control in some experiments.
Luria-Bertani (LB) broth or LB agar was used for culture of bacteria.
PCR studies.
PCRs were performed with a GeneAmp 2400 PCR system (Perkin-Elmer, PE Applied Biosystems, Barcelona, Spain). A DNA template was prepared by suspending two colonies of each isolate in 50 μl of double-distilled water and heating the preparation at 96°C for 10 min prior to addition of the reaction mixture or by diluting isolated bacterial or phage DNA 1:20 in double-distilled water. A 5-μl portion of each PCR product was analyzed by agarose (1%) gel electrophoresis, and bands were visualized by ethidium bromide staining. Oligonucleotides used for PCR amplification and customized primers used for sequencing are shown in Table 2.
TABLE 2.
Oligonucleotides used for PCR analysis and stx2-flanking regions
| Primer | Nucleotide sequence | Target sequence | Reference |
|---|---|---|---|
| UP 378 | 5′-GCGTTTTGACCATCTTCGT-3′ | 378-bp fragment of stxA2 | 17 |
| LP 378 | 5′-ACAGGAGCAGTTTCAGACAG-3′ | ||
| S2Aup | 5′-ATGAAGTGTATATTATTTA-3′ | stxA2 subunit | This study |
| S2Alp | 5′-TTCTTCATGCTTAACTCCT-3′ | ||
| GK3 | 5′-ATGAAGAAGATGTTTATG-3′ | stxB2 subunit | 24 |
| GK4 | 5′-TCAGTCATTATTAAACTG-3′ | ||
| RevUP | 5′-ACGAAGATGGTCAAAACGC-3′ | Flanking region of φLC159, φSC370 | This study |
| WPrev1 | 5′-AATGAAGGTTAATTTATGG-3′ | Flanking region of φLC159 | This study |
| WPrev2 | 5′-ACTGCTTGCGTATGACCA-3′ | Flanking region of φLC159 | This study |
| WPrev3 | 5′-TCGACAGTCCAGCGATG-3′ | Flanking region of φLC159 | This study |
| WPB1 | 5′-ATTGAGTATTACGCCTGT-3′ | Flanking region of φLC159 | This study |
| WPB2 | 5′-GCCGCGTATTAAGCAACT-3′ | Flanking region of φLC159 | This study |
| Rev FB | 5′-CTGGCTGGATGAACTCCG-3′ | Flanking region of φSC370 | This study |
| For FB | 5′-AGGATTCATAAGGCTGCGC-3′ | Flanking region of φLC370 | This study |
| Rev FB2 | 5′-ACCATCCCTGTACTTTCAG-3′ | Flanking region of φSC370 | This study |
| Q-stx-f | 5′-CGGAGGGGATTGTTGAAGGC-3′ | Antiterminator Q | 30 |
| Q-up | 5′-ATACACTGGCGATAAAGAAG-3′ | Antiterminator Q | This study |
Preparation of DIG-labeled stxA2-specific gene probes.
A 378-bp DNA fragment of the stxA2 gene resulting from amplification with primers UP 378 and LP 378 (Table 2) was labeled with digoxigenin (DIG) and used as a probe. The probe was labeled by incorporating digoxigenin-11-deoxyuridine triphosphate (Roche Diagnostics, Barcelona, Spain) during PCR as described previously (18).
Standard DNA techniques.
DNA was digested with restriction endonucleases (Promega Co., Inc., Madison, Wis.), and restriction fragments were analyzed by separation on 0.7% agarose gels in 0.5× Tris-borate-EDTA buffer and stained with ethidium bromide. PCR products were purified with a PCR purification kit (Qiagen Inc., Valencia, Calif.). DNA fragments were purified from agarose gels by using a gel excision kit (Qiagen Inc.).
Isolation of temperate bacteriophages and preparation of phage lysates.
Bacteria were grown from single colonies in Luria broth to the exponential growth phase. Mitomycin C was added to the cultures to a final concentration of 0.5 μg/ml. The cultures were then incubated overnight. Supernatants of the induced cultures were centrifuged at 10,000 × g for 10 min, filtered through low-protein-binding 0.22-μm-pore-size membrane filters (Millex-GP; Millipore, Bedford, Mass.), and treated with DNase (10 U/ml; Sigma-Aldrich, Madrid, Spain).
To measure the rate of phage production after induction of each isolate, the cultures were analyzed with a spectrophotometer at 600 nm every 30 min after addition of mitomycin C. The optical density was compared with the optical density of a control of the same culture without mitomycin C. Induction and optical density measurements were performed at least in triplicate for each isolate.
Screening for the presence of stx2-containing temperate bacteriophages.
A plate containing E. coli DH5α as the host strain was used to screen for the presence of temperate bacteriophages in the isolates. It was prepared with 500 μl of a log-phase culture of strain E. coli DH5α, 100 μl of 0.1 M CaCl2, and 3 ml of molten LB top agar; the mixture was then poured onto LB agar plates and allowed to solidify. Fifteen-microliter portions of a suspension of each phage lysate and phage 933W, used as a positive control, were dropped onto the plates. The plates were examined for the presence of lysis zones after incubation for 18 h at 37°C.
Additionally, phage lysates were diluted 10-fold. One hundred microliters of each dilution was then mixed with 100 μl of 0.1 M CaCl2 and 500 μl of a log-phase culture of E. coli DH5α and examined by the plaque assay by using a double-layer agar method (26). The plates were examined for the presence of plaques after incubation for 18 h at 37°C.
Plaque hybridization.
To determine the presence of the stx2 gene in the bacteriophages present in a lysis zone in which a drop of phage suspension was placed, the plaques were transferred to a nylon membrane (Hybond-N+; Amersham Pharmacia Biotech, Barcelona, Spain) by using a standard procedure (25) and hybridized at 65°C with a DIG-labeled stxA2 probe prepared as described above. Stringent hybridization was performed with a DIG DNA labeling and detection kit (Roche Diagnostics) used according to the manufacturer's instructions.
Isolation of phage DNA and detection of the stx2 gene.
Phage DNA was isolated from 200-ml cultures after induction with mitomycin C by the polyethylene glycol method and phenol-chloroform extraction as described previously (25). Purified DNA was digested with EcoRI, the fragments were separated by agarose (0.7%) gel electrophoresis, and bands were visualized by ethidium bromide staining. After electrophoresis, the DNA was transferred to nylon membranes (Hybond N+; Amersham Pharmacia Biotech) by capillary blotting (25). The membranes were hybridized with the DIG-labeled stxA2 fragment probe as described above. DNA was quantified with a spectrophotometer at 260 nm.
Isolation and detection of stx2 in bacterial DNA.
Chromosomal DNA was isolated from 40-ml cultures of each strain by lysozyme treatment and phenol-chloroform extraction as described previously (27). Purified DNA was diluted 1:3 in double-distilled water and digested with EcoRI. Electrophoresis, transfer to nylon membranes, and hybridization were carried out under the same conditions that were used for phage DNA.
Electron microscopy.
The stx2-converting bacteriophages obtained after induction of each isolate were purified by cesium chloride centrifugation (25). One drop of a phage suspension was deposited on a copper grid with carbon-coated Formvar film and stained with 2% KOH-phosphotungstic acid (pH 7.2) for 2.5 min. Samples were examined with a Hitachi E.M. 600 electron microscope operating at 80 kV.
Toxin production.
To compare the relative levels of production of Stx2 in the induced cultures, an enzyme immunoassay (Premier EHEC; Meridian Diagnostics Inc., Cincinnati, Ohio) was performed by using the manufacturer's instructions. Levels of expression by low-, medium-, and high-phage-production isolates with and without induction with mitomycin C were compared. E. coli DH5α was used as a negative control. Results were analyzed spectrophotometrically at two wavelengths (450 and 630 nm).
Characterization of the two types of phages observed.
Two phages were chosen as representatives for analysis of the two types of phage detected, those induced from isolate 159 and those induced from isolate 370. These phages are referred to below as φLC159 and φSC370. The characteristics of each phage are described in the Results.
Bacteriophage immunity test.
Phage immunity was tested on LB agar plates by spotting 10-μl portions of a suspension of phages φLC159, φSC370, and 933W onto four plates containing a top agar overlay; each overlay contained E. coli DH5α, E. coli C600 (933W), isolate 370, or isolate 159.
Cross-hybridization studies.
For cross-hybridization of digested DNAs of phages φSC370, φLC159, and 933W, SmaI-digested DNA of each phage was labeled with DIG by using a DIG labeling detection kit (Roche Diagnostics) and the manufacturer's instructions. For phage λ, the λ molecular weight marker restricted with EcoRI and HindIII and labeled with DIG was used. The labeled digested DNA of each phage was used as a probe in a Southern blot to detect homologies with the SmaI-digested DNAs of the other two phages and with phage λ DNA.
Sequencing of the stx2 gene and stx2-flanking regions encoded by temperate phages.
Since not all of the isolates produced the same number of phage after induction, two approaches were used to sequence the two types of phages observed, φLC159 and φSC370. When the rates of phage production after induction were high, resulting in highly concentrated phage DNA, as observed for the long-capsid phage φLC159, 3 μg of purified phage DNA was used for direct sequencing by standard genomic DNA methods (Perkin-Elmer, PE Applied Biosystems). The fragment was sequenced by chromosomal walking. The oligonucleotides used for sequencing are described in Table 2.
When the production of phage did not result in a high concentration of DNA, as observed for phage φSC370, phage DNA was purified from 500 ml of induced bacterial cultures and digested with various restriction enzymes, including BamHI, ClaI, EcoRI, EcoRV, KpnI, PstI, SalI, and SmaI (Promega Co.). The digested DNA was transferred to a nylon membrane, and the presence of the stx2 gene was determined by Southern blot hybridization as described above. A 4.8-kb EcoRI restriction fragment containing the stx2 gene was excised from the agarose gel and purified as described above. Five microliters of the purified fragment was used as a template for PCRs performed with stx2 reverse primers and forward primers for Q and stxB2-flanking regions as described in Table 2. PCR products were used directly for sequencing, and 4,189 bp between primers Q-up and Rev FB (Table 2) of the 4.8-kb fragment was sequenced.
Sequencing was performed with an ABI PRISM Big Dye II terminator cycle sequencing Ready Reaction kit (Perkin-Elmer, PE Applied Biosystems) by using an ABI PRISM 3700 DNA analyzer (Perkin-Elmer, PE Applied Biosystems) according to the manufacturer's instructions. All sequences were determined in duplicate.
Nucleotide sequence analysis, searches for open reading frames (ORFs), and searches for homologous DNA sequences in the EMBL and GenBank database libraries were performed with the Wisconsin Package, version 10.2 (Genetics Computer Group, Madison, Wis.). BLAST analyses were done with tools available on the web (http://www.ncbi.nlm.nih.gov).
Nucleotide sequence accession numbers.
The nucleotide sequence of the 5,812-bp fragment of phage φLC159 DNA and the nucleotide sequence of the 4,189-bp fragment of phage φSC370 containing the stx2 gene and stx2-flanking regions have been deposited in the EMBL database library under accession numbers AF548456 and AF548457, respectively.
RESULTS
Presence of stx2-converting bacteriophages.
stx2-converting bacteriophages were detected in all 49 isolates on the basis of the formation of a lysis zone, which gave a positive signal when the phages were hybridized with the stxA2-DIG probe. However, the rate of phage production after induction differed from one isolate to another (Table 3). Phage induced from high- and medium-phage-production isolates produced a confluent lysis zone in the spot area, whereas low-phage-production isolates produced only isolated plaques, which sometimes were not very visible. The negative control E. coli O157:H7 strain ATCC 43888 showed no lysis. Enumeration of plaques was difficult because they were barely discernible on the agar plates. Plaques were observed only after plaque blot hybridization. However, although positive signals were obtained after hybridization, the plaques were still too small to count accurately (data not shown).
TABLE 3.
Induction of stx2-converting phages and detection of the stx2 gene in phage and chromosomal DNAs
| Phage production after induction | Isolates | Optical density after inductiona | Lysisb | Amt of isolated phage DNA (μg) | Size of fragment containing stx2 detected in phage DNA by Southern bloting (kb)c | Size(s) of fragments containing stx2 detected in chromosomal DNA by Southern bloting (kb)c |
|---|---|---|---|---|---|---|
| Low | 22, 39, 41, 61, 67, 71, 81, 87, 90, 98, 122, 127, 128, 132, 135, 143, 168, 237, 288, 301, 305, 371, 487, 523, 635, 636, 652, 659, 671 | >0.6 | + | <0.5 | 4.8 | 4.8, 5.8 |
| Medium | 2, 7, 11, 20, 26, 65, 84, 104, 118, 156, 233, 346, 369, 370, 451, 622, 624, 631 | 0.3-0.6 | ++ | 0.5-3 | 4.8 | 4.8, 5.8 |
| High | 159, 360 | <0.3 | ++ | >3 | 5.8 | 5.8 |
Measured after 24 h.
Detected directly on the agar plate and by hybridization. +, isolated plaques that were not very visible; ++, confluent lysis zone in the spot area.
Fragment(s) obtained after hybridization with an stxA2-DIG probe of the EcoRI-digested DNA.
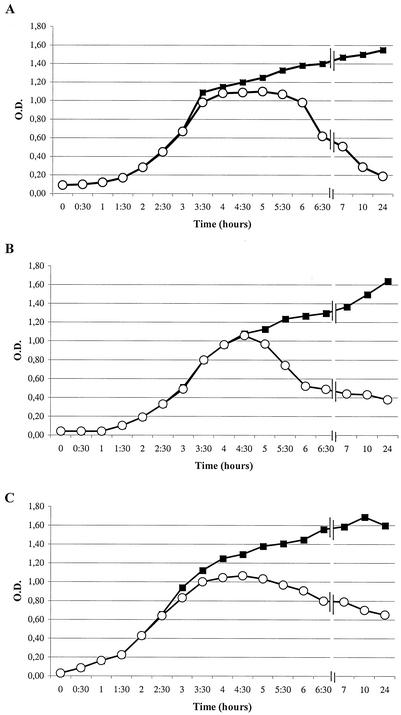
On the basis of the optical densities of the cultures after lysogenic induction, isolates were classified into three groups: isolates producing large, medium, and small amounts of stx2-converting phages, corresponding to optical densities of <0.3, between 0.3 and 0.6, and > 0.6, respectively (Fig. 1). Results described below (phage DNA size and restriction analysis and electron microscopy results) indicated that no phages other than stx2-converting phages were detected in the isolates studied.
FIG. 1.
Comparison of bacteriophage production after induction with mitomycin C of high-phage-production isolates (A), medium-phage-production isolates (B) and low-phage-production isolates (C). Symbols: ▪, without mitomycin C; ○, with mitomycin C. O.D., optical density.
Isolation of phage DNA and detection of the stx2 gene.
Isolation of phage DNA after induction from all the isolates studied indicated that some isolates had a higher rate of phage production than other isolates. After three independent inductions of each culture, the isolates exhibiting optical densities higher than 0.6 produced lower concentrations of phage DNA than the isolates exhibiting lower optical densities, although the concentration of DNA obtained varied slightly from one experiment to another. From a 200-ml culture no more than 0.5 μg of phage DNA was obtained from the low-phage-production isolates, whereas 3 μg of phage DNA was obtained from the high-phage-production isolates. The medium-phage-production isolates yielded between 0.5 and 3 μg of DNA.
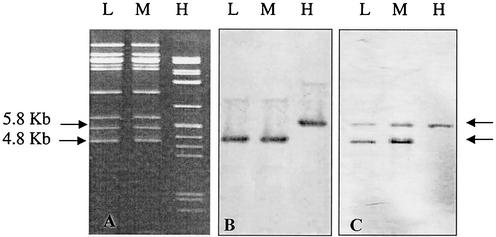
The stx2 gene was located in a fragment of the DNA that was the same size (4.8 kb) in phages induced from most of the isolates (Fig. 2); however, in phages induced from two of the isolates, isolates 159 and 360 (Table 3), the stx2 gene was located in a 5.8-kb DNA fragment. DNAs of phages isolated from all medium- and low-phage-production isolates showed the same restriction patterns when the phage DNAs were analyzed with a range of enzymes (data not shown), and the patterns were always different from those produced by the phages induced from isolates 159 and 360. The genome sizes of the phages released from all the isolates were calculated to be 60 ± 0.2 kb based on the sum of the sizes of the EcoRI or SmaI restriction fragments of the phage DNA.
FIG. 2.
Detection of the stx2 gene by Southern blot analysis of the EcoRI restriction patterns of phage and chromosomal DNAs of the low-phage-production isolates (lanes L), medium-phage-production isolates (lanes M), and high-phage-production isolates (lanes H). (A) EcoRI restrictions patterns of phage DNAs. (B) Southern blot hybridization with stxA2-DIG probe of the phage DNAs shown in panel A. (C) Southern blot hybridization with stxA2-DIG probe of the chromosomal DNAs extracted from the same isolates.
The phage DNAs obtained with isolates 159 and 360 had the same restriction patterns, and no differences were observed between them when they were tested with several enzymes. The genome sizes of these phages, calculated from the sum of the sizes of the EcoRI or SmaI restriction fragments of phage DNA, were 48 ± 0.3 kb.
Isolation and detection of stx2 in bacterial DNA.
Restriction analysis and Southern blot analysis revealed two bands of different sizes (4.8 and 5.8 kb) for low- and medium-phage-production isolates but only one band at 5.8 kb for the two high-phage-production isolates (isolates 159 and 360) (Fig. 2 and Table 3), indicating that there were two copies of the stx2 gene in all of the isolates except isolates 159 and 360.
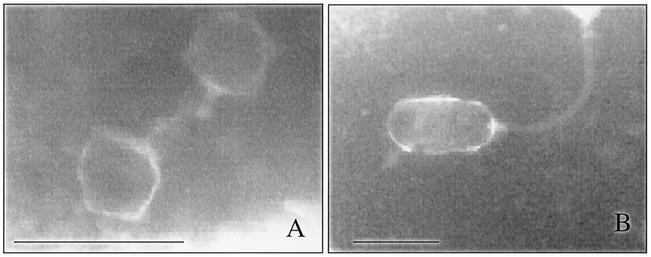
Electron microscopy.
Two different kinds of phages were also observed with the electron microscope. The medium- and low-phage-production isolates released a phage with an isometric capsid that was approximately 60 nm diameter and a short tail (Fig. 3A). Isolates 159 and 360 produced a phage with a capsid that was 45 by 90 nm and a long tail. (Fig. 3B).
FIG. 3.
Electron micrographs of the two main types of bacteriophages. (A) Phage φSC370; (B) phage φLC159. Bars = 100 nm.
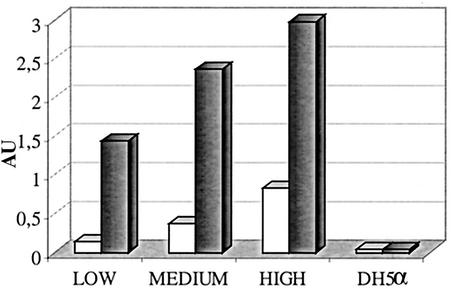
Toxin production.
Results of the toxin production analysis are shown in Fig. 4, in which the amounts of toxin produced are expressed in absorbance units. Isolate 975, which was used as a representative of the low-phage-production group, produced less toxin than isolate 370, which was used as a representative of the medium-phage-production group. Isolate 159, a representative of the high-phage-production group, produced the most toxin. Toxin production increased after addition of mitomycin C, and the number of absorbance units was proportionally higher in each group. As expected, DH5α did not produce toxin (Fig. 4.)
FIG. 4.
Toxin production evaluated with low-, medium-, and high-phage-production isolates with and without induction with mitomycin C. Values on the y axis correspond to absorbance units (AU) at two wavelengths (450 and 630 nm). The negative control was strain DH5α. Open bars, without mitomycin C; shaded bars, with mitomycin C.
Characterization of phages.
Two phages were chosen as representatives for analysis of the two types of phages detected in the set of isolates studied. Phage φSC370, obtained after induction of isolate 370, was chosen as a representative of the low- and medium-phage-production isolates, since these isolates released different amounts of the same kind of phage. Phage φLC159, induced from isolate 159, was selected as a representative of the two high-phage-production isolates.
(i) Cross-hybridization studies.
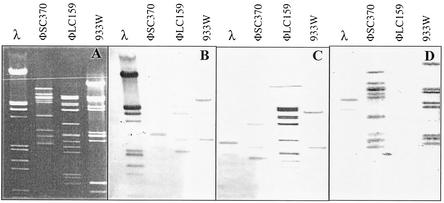
Cross-hybridization of EcoRI-digested DNAs of phages φSC370, φLC159, and 933W revealed some DNA homology between φSC370 and 933W (Fig. 5). However, phage φLC159 was different. All phages exhibited low levels of similarity with bacteriophage λ.
FIG. 5.
Agarose gel electrophoresis and Southern blot hybridization of phage DNAs. (A) Agarose gel electrophoresis of SmaI-digested DNAs of phages φSC370, φLC159, and 933W. The molecular weight marker was bacteriophage lambda DNA digested with EcoRI and HindIII. (B) Hybridization with lambda marker as the gene probe. (C) Hybridization with SmaI-digested DNA of phage φLC159 as the gene probe. (D) Hybridization with SmaI-digested DNA of phage φSC370 as the gene probe.
(ii) Bacteriophage immunity test.
E. coli DH5α was sensitive to phages φSC370 and φLC159. Strain 370 was resistant to φSC370 and φLC159, whereas strain 159 was resistant to φLC159 but sensitive to φSC370. These results indicated that phage φLC159 was present in isolate 370 and that phage φSC370 was not present in wild-type strain 159. These differences in immunity also indicated that phages φSC370 and φLC159 are in different bacteriophage immunity groups.
(iii) Sequencing of the stx2 gene and stx2-flanking regions encoded by temperate phages φLC159 and φSC370.
A 5,812-bp fragment of φLC159 DNA and a 4,189-bp fragment of φSC370 DNA containing the stx2 gene and stx2-flanking regions were sequenced to find out whether they had the structure described previously for other stx-converting phages (23, 30).
The total nucleotide sequence of the stx2 gene present in both phages exhibited 100% homology with the stx2 sequences carried by phages described previously (13, 23).
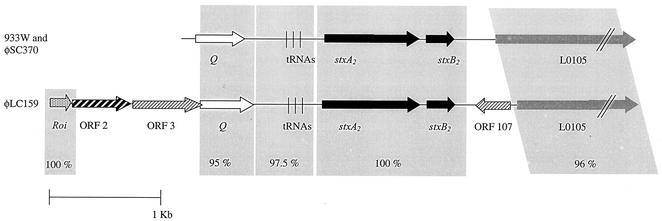
The 4,189-bp fragment of φSC370 DNA including the stx2 gene and flanking regions exhibited 100% homology with the corresponding fragment of phage 933W (23). Although the restriction patterns of this phage indicated that the total DNA of φSC370 is not identical to the total DNA of 933W, the fragment studied, which included the stx2 gene A and B subunits, the Q gene, and a fragment of the L0105 region (Fig. 6), showed that the stx2-flanking regions of φSC370 did not differ from those studied by other workers (13, 23).
FIG. 6.
Comparison of φSC370 and φLC159 or 933W stx2-flanking regions. The arrows indicate the relative lengths and directions of ORFs. The solid arrows indicate the stx2 genes. The percentages of sequence identity for the two phages and for phages φLC159 and 933W are indicated by shaded boxes. Bar = 1 kb. Gene designations are described in the text.
The first attempt to sequence phage φLC159 with the same set of primers used to sequence phage φSC370 was unsuccessful since the primers did not amplify the target DNA. The sequence of phage φLC159 was different from the sequence of phage φSC370 (Table 4 and Fig. 6). The stx2 gene sequence was again identical to the sequences of phage φSC370 and phages described previously. However, the sequence of the stx2-flanking regions was significantly different from the sequences in other phages. The sequence most closely related to the fragment studied was a 5,606-bp sequence describing the stx2 vhdA and stx2 vhdB genes for the type 2 variant subunit (accession number AB071845; submitted to the EMBL gene bank by T. Hamabata [unpublished data]), which exhibited 99% homology with the fragment of phage φLC159 sequenced in this study. However, we did not identify the ORFs present in this fragment except for the stx2 gene.
TABLE 4.
Description of ORFs in the sequence of the 5,812-bp fragment of phage φLC159
| ORF | Gene | DNA homology
|
Protein homology
|
Position | Length (bp) | ||
|---|---|---|---|---|---|---|---|
| Gene (accession no.) | % Homology | Protein | % Homology | ||||
| ORF 1 | roi | roi of 933W (AF125520) | 100 | Roi | 51-272 | 222 | |
| ORF 2 | ninG | L23 of phage 1639 (AJ304858) | 99 | NinG ORF 204 of phage 933W | 80 | 272-844 | 573 |
| ninG-ORF 204 of H-19 (AF034975) | 98 | ||||||
| ORF 3 | ser/thr | EDL933W serine/threonine phosphatase (AE005492) | 98 | Serine/threonine phosphatase | 99 | 875-1546 | 672 |
| nin 221 of phage λ (J02450) | 95 | ||||||
| ORF 4 | Q | Q of bacteriophage 21 (M58702) | 97 | Q protein of bacteriophage 21 | 95 | 1642-2022 | 381 |
| ileZ | ileZ of 933W (AF125520) | 100 | 2337-2411 | 75 | |||
| argN | argN of 933W (AF125520) | 93 | 2419-2495 | 77 | |||
| rho | rho of 933W (AF125520) | 100 | 2523-2546 | 24 | |||
| ORF 5 | stxA2 | stxA2 of 933W (AF125520) | 100 | StxA2 | 2601-3560 | 960 | |
| ORF 6 | stxB2 | stxB2 of 933W (AF125520) | 100 | StxB2 | 3572-3841 | 270 | |
| ORF 7 | ORF 107 | ORF 107 of E. coli stx2 gene phages (AJ251452) | 100 | Unknown protein | 4138-4461 | 324 | |
| ORF 8 | ORF 645 | ORF 645 fragment of stx2 (AJ251452) | 99 | 4684-5812 | >1,128 | ||
| ORF 645 fragment of stxB2 (AJ251520) | 99 | ||||||
Eight ORFs were identified (Table 4). The stx2 gene in phage φLC159 (ORFs 5 and 6) was found downstream of the homologues of the ileZ and argN genes. A sequence homologue of the Q antiterminator gene was found upstream of the ileZ gene, which exhibited 97% homology with the Q gene of bacteriophage 21 (accession number M58702); the protein exhibited 95% identity with the encoded Q protein of bacteriophage 21.
Upstream of the Q gene, ORFs 1, 2, and 3 were detected (Table 4 and Fig. 6). ORF 1 exhibited 100% homology with the roi gene of phage 933W (accession number AF125520). ORF 2 exhibited high levels of homology with sequences of phages 1639, H-19, and 933W coding for a protein similar to the NinG protein (ORF 204) of bacteriophages lambda and P22, and the protein exhibited 80% amino acid identity with the NinG protein of phage 933W. ORF 3 exhibited similarity to the gene encoding a serine/threonine protein phosphatase of E. coli O157:H7 strain EDL 933 and to the nin 221 gene of phage λ. The protein homology data confirmed the similarity of the protein to a serine/threonine protein phosphatase.
Downstream of the stx2 gene two ORFs were detected, including ORF 7, which appeared to be identical to ORF 107 previously described downstream of the stx2 region in stx2c phage and other stx2+ strains (30) (accession numbers AJ251452, AJ250954, AJ251483, and AJ251520). Finally, the fragment of ORF 8 sequenced exhibited a high level of homology with the corresponding fragment ORF 645 described for stx2c and other stx2+ strains (accession numbers AJ251452 and AJ251520).
DISCUSSION
In this study the variability of the E. coli O157:H7 strains isolated from a single outbreak due to stx2-converting bacteriophages was analyzed. The field epidemiology and molecular epidemiology of the isolates (data not shown) indicate that they had a common origin. After the studies described here, we concluded that stx2-converting phages conferred certain variability to the isolates. Not all the isolates produced the same amount of phage after lysogenic induction, and the variability allowed us to classify them in three main groups. Although the criteria used for this classification are subjective, we intended only to clarify the expression results. The coherence of the results was demonstrated after several replications of induction for each isolate, which showed that the differences were reproducible. The three categories selected could also be demonstrated by the absorbance data for the cultures after phage induction and by the amounts of phage DNA obtained. For a given culture, a high level of phage production corresponded to low absorbance due to the bacterial cell lysis after phage release, and consequently more phage DNA was obtained from the culture.
Variability was also observed because low- and medium-phage-production isolates harbored two phages, although they released only one phage, phage φSC370. If φLC159 was released, its presence in the supernatants of the induced cultures was masked by phage φSC370. We do not know whether the second phage, φLC159, was also released from the low- and medium-phage-production isolates at low concentrations that could not be detected. In contrast, high-phage-production isolates harbored only one phage, φLC159, and had only one copy of the toxin gene, and phage φSC370 was not present in their chromosomes.
There are differences in the morphological and genetic structures of the two phages. Phage φSC370 has many similarities with phage 933W (23), although it is not identical. The sequence of the 4,189-bp fragment, the length of the DNA, the morphology, and the bacteriophage immunity test results indicated that phage φSC370 is related to bacteriophage 933W. The second phage, φLC159, has different characteristics. Its morphology is similar to that of phage H-19 (20), but it harbors the stx2 gene instead of stx1. It has shorter DNA than phage φSC370 and differs in its genetic patterns.
The sequence of the phage φLC159 DNA fragment indicated that the sequence of the stx2 gene is highly conserved, and the analysis of the flanking regions indicated that the structure is similar to that of other stx2-converting phages (30). A homologue of the antiterminator gene Q is located upstream the stx2genes and probably has an equivalent function in the regulation of stx gene expression. With the Q gene homologue, stx genes could be transcribed from the pR′ promoter as a part of late gene expression under the influence of the Q-like gene product synthesized after inactivation of the phage repressor (20, 32). Although a homologue of the pR′ promoter has not been identified in the sequence studied, some other sequence may have its function, since phage production and toxin production are directly related to Q-activated pR′-promoted transcription (32). Upstream of the Q region the presence of the roi gene (ORF 1) suggests similarities with phage 933W, but ORFs 2 and 3 appeared to be different. Genes located downstream of the stx2 genes seemed to be quite similar to phages described previously (30), although the regions were only partially sequenced. Phage φLC159 flanking regions appeared to be like those in a typical stx2 phage but had a mosaic structure that makes the phage slightly different from other phages, in accordance with the findings of other workers, who suggested that there is a common mosaic structure in stx2 and in general lambdoid, double-stranded DNA phages (8, 30).
The variability of the phage-converted strains was reflected in toxin production, in accordance with the findings of other workers (12, 14, 16). Greater phage release was associated with higher levels of toxin production, and it could produce a more severe infection. In our study to some extent this effect could be observed by looking at the symptoms. Although not all the low-phage-production isolates were from asymptomatic patients, all the asymptomatic patients were infected by one isolate belonging to the low-phage-production group. In contrast, all the medium- and high-production isolates caused symptoms.
Both high-phage-production isolates (isolates 159 and 360) were isolated from patients with secondary infections, who were siblings. These isolates lacked one of the phages, although we do not know whether the phage was lost before or after infection of the two secondary carriers. Nevertheless, assuming that the wild-type strain had lost one of the two phages, this should have had a clear effect on phage release and hence toxin production. The presence of the two temperate phages in the same bacterial genome seemed to affect the induction of both phages. It had a stronger effect on the long-capsid phage, which was not detected after induction when the isometric-capsid phage was present. Once the isometric-capsid phage disappeared, the long-capsid phage could be induced, and very large amounts were induced. This appears to be a mechanism of phage repression that acts when both phages are present. Watarai et al. (33) suggested some mechanisms of regulation in temperate phages in studies of the two converting phages isolated from the outbreak in Japan in 1996. In that case, the workers showed that when one of the phages was transduced to a C600 strain, the amounts of the phage produced were less than the amounts produced in the wild-type strain harboring two phages, which is the opposite of the effect observed here.
Clonal variability produced by changes either in the phage or in the flanking regions has been described by other workers (5, 11). However, to our knowledge, this is the first description of clonal variability due to loss of a phage.
The biological significance of clonal variability in either phage or toxin production remains unknown, but it surely results in variability, producing a mixed population that allows the bacteria to adapt to the most favorable conditions. In this case isolates showing higher phage induction produced more toxin and hence were more virulent. However, these strains had higher mortality rates due to the higher phage induction and consequent lysis. In contrast, this effect was reduced with the fraction of low-phage-production strains, which showed lower virulence but had higher survival rates.
Also, for phages implication in clonal variability is advantageous since being integrated as a prophage that is more or less inducible increases the chances of persistence. Clonal variability is advantageous either as a prophage, in which case the conditions for bacterial survival are good, or as a virion when conditions for bacterial survival are bad.
Clonal variability may also influence epidemiological studies by making it difficult to show linkage among epidemiologically related O157 isolates or by undervaluing Shiga toxin-producing E. coli as the causal agent of infectious enteritis.
Our results suggest that the variability induced by stx2-converting phages in strains is related to virulence. This variability could be observed even in closely related strains, like the strains studied here, which were isolated from the same outbreak. Since phages are an important element of gene exchange, more studies on stx2-converting phages are needed to increase our knowledge of the epidemiology and pathogenicity of STEC strains.
Acknowledgments
We thank Cristina Valdivieso for excellent technical assistance.
This work was supported by the project BMC2000-0549 of the Ministerio de Ciencia y Tecnologia, by the Generalitat de Catalunya (2001SGR00099), and by the Centre de Referència en Biotecnologia (CeRBa).
Editor: A. D. O'Brien
REFERENCES
- 1.Acheson, D. W. K., and G. T. Keusch. 1996. Which Shiga toxin-producing types of Escherichia coli are important? ASM News 62:302-306. [Google Scholar]
- 2.Acheson, D. W. K., J. Reidl, X. Zhang, G. T. Keusch, J. J. Mekalanos, and M. K. Waldor. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiba, M., T. Sameshima, and M. Nakazawa. 2000. Clonal turnover of enterohemorrhagic Escherichia coli O157:H7 in experimentally infected cattle. FEMS Microbiol. Lett. 194:79-83. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. 2001. Brot de gastroenteritis per E. coli O157:H7 en diferents escoles de Catalunya. Bol. Epidemiolog. Cataluna 22:59-64.
- 5.Datz, M., C. Janetzki-Mittmann, S. Franke, F. Gunzer, H. Schmidt, and H. Karch. 1996. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lamboid phage gene p and Shiga-like toxin structural genes. Appl Environ Microbiol. 62:791-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyle, M. P. 1997. Escherichia coli O157, p. 171-191 In M. P. Doyle, L. R. Beuchat, and T. J. Montville, (ed.), Food microbiology: fundamentals and frontiers. ASM Press, Washington, D.C.
- 7.Friedrich, A. W., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 8.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The association of IS1203 and stx and p genes in E. coli O157:H7, p. 178. In G. Duffy, P. Garvey, J. Coia, Y. Waterson, and D. A. McDowell (ed.), Pathogenicity and virulence of verotoxigenic E. coli. Proceedings of Concerted Action CT98-3935 Meeting 3. The National Food Center, Dublin, Ireland.
- 10.Johansen, B. K., Y. Wasteson, P. E. Granum, and S. Brynestas. 2001. Mosaic structure of Shiga-toxin-2 encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology 147:1929-1936. [DOI] [PubMed] [Google Scholar]
- 11.Karch, H., H. Russman, H. Schmidt, A. Schwarzkopf, and J. Hessemann. 1995. Long-term shedding and clonal turnover of enterohemorrhagic Escherichia coli O157 in diarrheal diseases. J. Clin. Microbiol. 33:1602-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohler, B., H. Karch, and H. Schmidt. 2000. Antibacterials that are used as growth promoters in animal husbandry can affect the production of Shiga-toxin-2-converting bacteriophages and Shiga toxin 2 from Escherichia coli strains. Microbiology 146:1085-1090. [DOI] [PubMed] [Google Scholar]
- 13.Makino, K., K. Yokohama, Y. Kubota, et al. 1999. Complete nucleotide sequence of the prophage VT2.Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet. Syst. 74:227-239. [DOI] [PubMed] [Google Scholar]
- 14.Matsushiro, A., K. Sato, H. Miyamoto, T. Yamamura, and T. Honda. 1999. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J. Bacteriol. 181:2257-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonough, M. A., and J. R. Butterton. 1999. Spontaneous tandem amplification and deletion of the Shiga toxin operon in Shigella dysenteriae 1. Mol. Microbiol. 34:1058-1069. [DOI] [PubMed] [Google Scholar]
- 16.Mühldorfer, I., J. Hacker, G. T., Keusch, D. W. Acheson, H. Tschäpe, A. V. Kane, A. Ritter, T. Ölschläger, and A. Donohue-Rolfe. 1996. Regulation of the Shiga-like toxin II operon in Escherichiacoli. Infect. Immun. 64:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muniesa, M., and J. Jofre. 1998. Abundance in sewage of bacteriophages that infect E. coli O157:H7 and that carry the Shiga toxin 2 gene. Appl. Environ. Microbiol. 64:2443-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muniesa, M., J. Recktenwald, M. Bielaszewska, H. Karch, and H. Schmidt. 2000. Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect. Immun. 68:4850-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nataro, J. P., and J. J. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neely, M. N., and D. I. Friedman. 1998. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 28:1255-1268. [DOI] [PubMed] [Google Scholar]
- 21.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. Williams Smith, and S. B. Formal. 1984. Shiga like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 22.Paton, A. W., and J. C. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plunkett, G., D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933 W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rüssman, H., H. Schmidt, A. Caprioli, and H. Karch. 1994. Highly conserved B subunits genes of Shiga-like toxin II variants found in Escherichia coli O157 strains. FEMS Microbiol. Lett. 118:335-340. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga-toxin 2-encoding bacteriophage φ3538 isolated from E. coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart, D. S., M. L. Tortorello, and S. M. Gendel. 1998. Evaluation of DNA preparation techniques for detection of the SLT-1 gene of Escherichia coli O157:H7 in bovine faeces using the polymerase chain reaction. Lett. Appl. Microbiol. 26:93-97. [DOI] [PubMed] [Google Scholar]
- 28.Strauch, E., R. Lurz, and L. Beutin. 2001. Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei. Infect. Immun. 69:7588-7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teel., L. D., A. R. Melton-Celsa, C. K. Schmitt, and A. D. O'Brien. 2002. One of two copies of the gene for the activatable Shiga toxin type 2d in Escherichia coli O91:H21 strain B2F1 is associated with an inducible bacteriophage. Infect. Immun. 70:4282-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner, P. L., D. W. K. Acheson, and M. K. Waldor. 1999. Isogenic lysogens of diverse Shiga toxin 2-encoding bacteriophages produce markedly different amounts of Shiga toxin. Infect. Immun. 67:6710-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner, P. L., M. N. Neely, X. Zhang, D. W. K. Acheson, M. K. Waldor, and D. I. Friedman. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watarai, M., T. Sato, M. Kobayashi, T. Shimizu, S. Yamakasi, T. Tobe, C. Sasakawa, and W. Takeda. 1998. Identification and characterization of a newly isolated Shiga toxin 2-converting phage from Shiga toxin-producing Escherichia coli. Infect. Immun. 66:4100-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yokoyama, K., K. Makino, Y. Kubota, et al. 2000. Complete nucleotide sequence of the prophage VT1-Sakai carrying the Shiga toxin 1 genes of the enterohemorrhagic Escherichia coli O157:H7 strain derived from the Sakai outbreak. Gene 258:127-139. [DOI] [PubMed] [Google Scholar]