Abstract
Members of the genus Bartonella have historically been connected with human disease, such as cat scratch disease, trench fever, and Carrion's disease, and recently have been recognized as emerging pathogens causing other clinical manifestations in humans. However, because little is known about the antigens that elicit antibody production in response to Bartonella infections, this project was undertaken to identify and molecularly characterize these immunogens. Immunologic screening of a Bartonella vinsonii subsp. berkhoffii genomic expression library with anti-Bartonella antibodies led to the identification of the sucB gene, which encodes the enzyme dihydrolipoamide succinyltransferase. Antiserum from a mouse experimentally infected with live Bartonella was reactive against recombinant SucB, indicating the mounting of an anti-SucB response following infection. Antigenic cross-reactivity was observed with antiserum against other Bartonella spp. Antibodies against Coxiella burnetti, Francisella tularensis, and Rickettsia typhi also reacted with our recombinant Bartonella SucB. Potential SucB antigenic cross-reactivity presents a challenge to the development of serodiagnostic tests for other intracellular pathogens that cause diseases such as Q fever, rickettsioses, brucelloses, tularemia, and other bartonelloses.
There is an increasing awareness of Bartonella spp. as causative agents of emerging diseases of human and veterinary importance. The genus Bartonella is comprised of several species of human and animal pathogens causing various zoonotic-related diseases (4, 9). In humans, B. bacilliformis causes Carrion's disease, seen mainly in the Andes mountain region of South America. The acute form of the illness is a severe hemolytic anemia, with the chronic form characterized by vascular proliferative lesions of the skin (21). B. henselae is responsible for cat scratch disease (CSD) and bacillary angiomatosis (28, 41). B. quintana is also associated with bacillary angiomatosis (28) but is more widely recognized as the causative agent of trench fever (26). Human endocarditis cases have been described involving B. henselae, B. quintana, B. elizabethae (16), and B. vinsonii subsp. berkhoffii (42). B. vinsonii subsp. arupensis has recently been described as causing a human febrile bacteremia (45), and B. grahamii has been associated with human neuroretinitis (24).
Bartonella infections are associated with arthropod vector transmission. B. bacilliformis is transmitted by sand flies (21), and B. henselae has been demonstrated in cat fleas (18, 22), with the human body louse (Pediculus humanus) instrumental in the transmission of B. quintana (43). Domestic cats are considered a reservoir host for B. henselae (14, 27), but some Bartonella are carried asymptomatically in a variety of wild rodents worldwide (5, 7, 31). Additionally, there is serological and molecular evidence of California coyotes serving as reservoir hosts (10, 13), and PCR data have implicated Ixodid ticks in harboring Bartonella (11, 12).
The current state of diagnostics for the determination of infection is underdeveloped, but serology by the indirect fluorescence assay and enzyme immunoassay against whole cells are the predominant methodologies being applied, mostly for CSD (15, 41). However, problems with cross-reactivity among Bartonella species and variable sensitivities and specificities observed among laboratories have led to caution when interpreting the serologic-based results (1, 6, 19, 23, 32). Furthermore, diagnostic assays for bartonelloses caused by organisms other than B. henselae or B. quintana are underdeveloped.
Little is known regarding antigens that induce an antibody response following Bartonella infection. Several immunogenic proteins associated with Bartonella infections have been noted by Western blot banding patterns (20, 35, 36-39), but only the B. henselae 17-kDa antigen and HtrA stress response protein and the B. bacilliformis Bb65 antigen have been characterized (2, 3, 25). Bartonella-specific monoclonal antibodies have been described, but the molecular identities of the corresponding antigens have not yet been elucidated (33, 34). The goal of this project was to identify immunogens associated with Bartonella infections. As a first step, we screened Bartonella genomic libraries with polyclonal antiserum against whole-cell lysates of various Bartonella isolates. In this report, we describe an immunoreactive Bartonella gene product as being dihydrolipoamide succinyltransferase expressed by the sucB gene, which is part of the α-ketoglutarate dehydrogenase complex that has been described in several prokaryotes.
Identification of the sucB gene from genomic libraries.
Bartonella strains used in this study for DNA manipulations, immunoblotting, and antibody production are listed in Table 1. B. quintana and B. vinsonii subsp. berkhoffii were cultivated on brain heart infusion agar medium supplemented with 5% rabbit blood (BBL Becton Dickinson Microbiology Systems, Cockeysville, Md.) and harvested as previously described (31). Genomic DNA was purified from thawed cell suspensions by a phenol-chloroform extraction procedure followed by ethanol precipitation according to standard procedures. For genomic cloning, purified Bartonella DNA was subjected to partial Sau3AI restriction enzyme digestion and was ligated into the ZapExpress BamHI-predigested bacteriophage lambda cloning vector (Stratagene, La Jolla, Calif.) with subsequent packaging of the ligated DNA with the GigaPack III Gold packaging extract (Stratagene) as directed by the manufacturer. Recombinant lambda plaques were plated, titrated, and amplified according to the manufacturer's instruction manual.
TABLE 1.
Bartonella strains used in this study
| Strain | Description |
|---|---|
| B. quintana Fuller ATCC VR-358 | Human isolate |
| B. vinsonii subsp. berkhoffii ATCC 51672 | Dog isolate |
| B. vinsonii subsp. vinsonii ATCC VR-152 | Vole isolate |
| B. henselae Houston | Type strain |
| B. elizabethae ATCC 49927 | Type strain |
| A1, strain Sh6397ga | Cotton rat (Sigmodon hispidus) isolate (31) |
| B1, strain Sh6396ga | Cotton rat (S. hispidus) isolate (31) |
| C1, strain Sh6537ga | Cotton rat (S. hispidus) isolate (31) |
| Neotoma albigula strain Na18985nm | Wood rat isolate; New Mexico |
| Peromyscus maniculatus strain Pm15590co | Deer mouse (P. maniculatus) isolate; Colorado |
| Spermophilus beecheyi strain Sb944nv | California ground squirrel (S. beecheyi) isolate; Nevada |
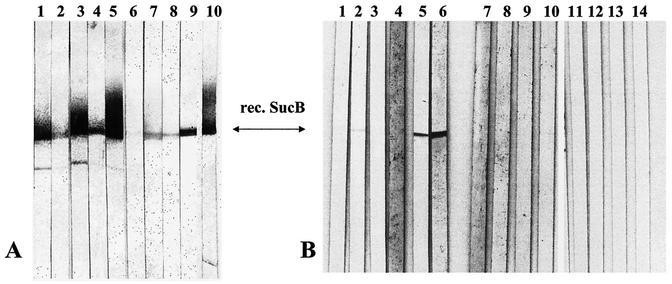
Thirty-five immunopositive plaques from the B. vinsonii subsp. berkhoffii genomic library were recognized by a pool of mouse polyclonal antibodies raised against heat-killed Bartonella sp. cotton rat isolates A1, strain Sh6397ga, B1, strain Sh6396ga, and C1, strain Sh6537ga, and selected for further analysis. Western blotting of the recombinant products revealed one clone that consistently produced a stable protein that was strongly reactive to the screening antibody. The insert of this clone was 3.36 kb, and DNA sequence analysis revealed three open reading frames. According to the GenBank database, these genes were homologs of three components of the α-ketoglutarate dehydrogenase operon complex present in several prokaryotes: sucA, sucB, and lpdA, which encode the α-ketoglutarate dehydrogenase (E1o), dihydolipoamide succinyltransferase (E2o), and dihydrolipoamide dehydrogenase enzymes, respectively. The sucA and lpdA genes are truncated within this insert, and sucB is represented in its entirety. The sucB coding sequence consists of 1,233 bp with a calculated molecular mass of 43.8 kDa from the deduced amino acid sequence. The sucB gene coding sequence was subcloned into a plasmid expression vector and transformed into Escherichia coli, and the gene product was synthesized. The recombinant SucB immunoblotted positively with the anti-Bartonella antibodies used to screen the library (Fig. 1A, lanes 1 to 3).
FIG. 1.
Immunoblots of recombinant B. vinsonii subsp. berkhoffii SucB reacted with various polyclonal antibodies. (A) Mouse anti-Bartonella diluted 1:200 raised against the following isolates: cotton rat strain A1 (lane 1); cotton rat strain B1 (lane 2); cotton rat strain C1 (lane 3); B. vinsonii subsp. berkhoffii (lane 4); B. vinsonii subsp. vinsonii (lane 5); B. quintana (lane 6); B. elizabethae (lane 7); B. henselae (lane 8); N. albigula (wood rat) strain Na18985 nm (lane 9); BALB/c mouse experimentally infected with Bartonella strain Pm15590co and boosted with Bartonella ground squirrel strain Sb944nv (lane 10). (B) Polyclonal antiserum samples against the following various pathogens diluted 1:500: anti-L. pneumophila (lane 1); anti-R. typhi (lane 2); anti-R. prowazekii (lane 3); anti-R. rickettsii (lane 4); anti-C. burnetti (lane 5); anti-F. tularensis (lane 6); anti-Y. pestis (lane 7); anti-B. burgdorferi (lane 8); anti-Leptospira spp. (lane 9); anti-T. pallidum (lane 10); mouse preimmunized negative control serum (lanes 11 to 14).
The sucB gene was amplified by PCR from B. quintana DNA using primers derived from the B. vinsonii subsp. berkhoffii sequence, which encompassed the entire coding sequence of the gene. The B. quintana sucB gene was determined to have 87.8% amino acid sequence identity to the SucB of B. vinsonii subsp. berkhoffii. A BLAST search of the protein database with SucB found the closest matches were to Mesorhizobium loti, Agrobacterium tumefaciens, Sinorhizobium meliloti, and Brucella melitensis. Not surprisingly, these organisms are closely related phylogenetically to Bartonella. The amino acid sequence identity comparisons are shown in Table 2.
TABLE 2.
SucB amino acid identity between related organisms
| Organism | Identity (%)
|
|||||
|---|---|---|---|---|---|---|
| B. vinsonii subsp. berkhoffii | B. quintana | B. melitensis | A. tumefaciens | M. loti | S. meliloti | |
| B. vinsonii subsp. berkhoffii | 100 | 87.8 | 72.4 | 70.2 | 72.0 | 69.6 |
| B. quintana | 100 | 71.6 | 69.3 | 71.5 | 68.0 | |
| B. melitensis | 100 | 81.7 | 80.2 | 78.0 | ||
| A. tumefaciens | 100 | 77.3 | 83.2 | |||
| M. loti | 100 | 79.6 | ||||
| S. meliloti | 100 | |||||
Expression of sucB and antibody reactivities.
The coding sequence of the B. vinsonii subsp. berkhoffii sucB gene was amplified by PCR using the primers BvSucB-F (5′ ATGACTACTGAAATCCGTGTTCC 3′) and BvSucB-R (5′ CAAGTCAAGAACAAGGCGTTC 3′) under the following conditions: 10 mM Tris (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, a 200 μM concentration of each deoxynucleoside triphosphate, a 0.5 μM concentration of each primer, and 2.5 U of AmpliTaq DNA polymerase (Applied Biosystems, Branchburg, N.J.). Approximately 10 ng of the original recombinant plasmid containing the sucB gene was amplified using the thermocycler parameters 94°C for 30 s, 50°C for 30 s, and 72°C for 60 s for 35 cycles. Purified sucB amplicon was subcloned into the expression vector pBAD/thio TOPO (Invitrogen) and transformed into E. coli strain TOP10 (Invitrogen). A colony containing the sucB gene was inoculated into Luria-Bertani broth and incubated at 37°C until growth reached mid-log phase, whereby protein expression was induced by the addition of 0.02% arabinose. The culture was incubated another 2 to 4 h, and then the cells were harvested, pelleted, and frozen at −20°C until needed. Recombinant SucB was purified from E. coli proteins by His-tag affinity chromatography using His-Bind Quick 300 cartridges (Novagen, Madison, Wis.) following lysis of the E. coli cells by suspension in B-PER bacterial protein extraction reagent (Pierce, Rockland, Ill.) with the addition of the protease inhibitor Pefabloc SC (Boehringer GmbH, Mannheim, Germany).
SucB reactivity was tested against polyclonal antibodies raised against various Bartonella species and strains. Cross-reactivity was observed against all anti-Bartonella antibodies tested (Fig. 1A). Although reactivity against anti-B. quintana and anti-B. henselae was weaker, this may simply reflect the strength of the antiserum used. Four samples of preimmunized mouse serum showed no reactivity to SucB (Fig. 1B, lanes 11 to 14). Significantly, there was robust reactivity with the recombinant SucB against antibodies from an experimentally infected mouse injected with live Bartonella isolated from P. maniculatus and boosted with an S. beecheyi strain (Fig. 1A, lane 10). This result demonstrated that an antibody response is mounted against SucB in response to a Bartonella infection, as well as showing the strong cross-reactivity between B. vinsonii subsp. berkhoffii and rodent-isolated Bartonella.
Antibodies against other bacterial pathogens were tested for cross-reactivity against SucB. Polyclonal antisera against various bacterial pathogens were obtained from the following sources: rabbit anti-Legionella pneumophila from Yousef Abu Kwaik (University of Kentucky); rabbit anti-Coxiella burnetti phase II from Bob Heinzen (University of Wyoming); rabbit anti-Rickettsia rickettsii, Rickettsia typhi, and Rickettsia prowazekii generated at the Rocky Mountain Laboratories, Hamilton, Mont.; rabbit anti-Francisella tularensis and human anti-Yersinia pestis from the Diagnostic and Reference Section, Division of Vector-Borne Infectious Diseases (DVBID), Centers for Disease Control and Prevention (CDC); human anti-Borrelia burgdorferi, Leptospira spp., and Treponema pallidum from the Molecular Bacteriology Section, DVBID, CDC. SucB seroreactivity was seen with antisera specific to C. burnetti and F. tularensis, while weaker reactivity was observed against R. typhi (Fig. 1B). No cross-reactivity was observed with antibodies against the other microbes tested. Anti-Brucella spp. antibodies were unavailable for testing.
Recent serological testing of patients with a febrile illness of unexplained origin from New Mexico by this laboratory had suggested possible infections with rodent-associated Bartonella (M. Y. Kosoy et al., Abstr. Am. Soc. Rickettsiology-Bartonella Emerg. Pathogen Group 2001 Joint Conf., abstr. 108, 2001; F. Koster et al., Abstr. Am. Soc. Rickettsiology-Bartonella Emerg. Pathogen Group 2001 Joint Conf., abstr. 133, 2001). This observation and the discovery that rodent species in the western United States harbor Bartonella led us to investigate whether these organisms could be the causative agents of illnesses in humans having exposure to wild rodents. Concomitantly, this laboratory recently discovered that Bartonella isolates obtained from ground squirrels in Nevada had gltA (citrate synthase), 16S rRNA, and groEL gene sequences identical to those of B. washoensis isolated from a cardiac patient from the same area, providing evidence of Bartonella rodent-to-human transmission (30). In conjunction with these observations, we sought to expand our understanding of Bartonella infection-associated immunogens, particularly those putatively causing non-CSD illnesses, by screening genomic libraries using antibodies generated against rodent-isolated Bartonella.
B. vinsonii subsp. berkhoffii and B. quintana were initially chosen as genomic library representatives for Bartonella, as we were interested in investigating antigens from other species besides B. henselae. Although B. vinsonii subsp. berkhoffii is associated with infection in dogs (8, 29), coyotes and ticks have been implicated as reservoir and vector hosts, respectively, in the western United States (11, 13), and there has been one documented human case infection (42). In addition, a related organism, B. vinsonii subsp. arupensis, was recently isolated from a human patient in Wyoming (45).
The genomic libraries were screened initially with antibodies specific to rodent isolates. The rationale was to recognize any putative gene products reactive against antibodies to rodent-borne Bartonella and subsequently to use comparative genomics to determine the extent of cross-reactive, homologous genes between genus and species. Although in this study we screened genomic libraries from only two Bartonella species, we have purified genomic DNA from several Bartonella strains and generated libraries to other rodent Bartonella isolates for future genetic comparisons. We have indeed been successful in amplifying sucB by PCR from several Bartonella spp. and isolates, indicating the presence of this gene as expected (data not shown).
At the time this study began, Bartonella isolates from clinically defined human cases in the western United States were not available and, accordingly, neither were antiserum samples from culture-confirmed patients. In addition, we could not utilize antiserum from the rodent reservoir hosts from which the Bartonella organisms were isolated, as these naturally infected animals do not seem to mount a detectable antibody response (31). Antigens identified by polyclonal antibodies prepared against killed whole-cell lysates do not necessarily correlate with immunogens associated with Bartonella infections; however, this approach does identify candidate antigens that can be assayed for their reactivity against antiserum raised in an infected host. Indeed, SucB proved to be an infection-associated immunogen, as it was detected by antiserum from the experimentally infected mouse shown in Fig. 1A.
Molecular analysis of the expression library clone indicated that the gene encoding the protein reactive against anti-Bartonella antibodies was sucB, which encodes dihydrolipoamide succinyltransferase (E2o), an enzyme that is one part of three components forming the α-ketoglutarate dehydrogenase complex found in several eukaryotes and prokaryotes. This enzyme complex is also composed of α-ketoglutarate dehydrogenase (E1o) and dihydrolipoamide dehydrogenase (E3) and is encoded by the genes sucA and lpd, respectively. The DNA sequence of the B. vinsonii subsp. berkhoffii cloned insert showed a similar gene arrangement as that described in Rhodobacter capsulatus and E. coli (17, 44).
To our knowledge, only three other immunoreactive Bartonella antigens have been molecularly characterized. They are the 17-kDa antigen of B. henselae that elicits a strong humoral response in patients with CSD (2), the B. henselae HtrA stress response protein (3), and a GroEL class of heat shock protein from B. bacilliformis termed Bb65 (25). Other Bartonella antigens have been identified by Western blot banding patterns, but they have not been cloned and sequenced. Several groups have shown that anti-Bartonella serum samples are reactive on immunoblots with proteins of molecular masses of 45 to 50 kDa. These bands may correspond to the SucB antigen (20, 35, 39).
Antigenic cross-reactivity was seen when recombinant SucB was assayed with antibodies raised against various Bartonella strains. Significantly, the SucB was reactive against antiserum from a mouse experimentally infected with live Bartonella. The antigenic cross-reactivity was evidenced by the fact that this antibody was directed against Bartonella obtained from deer mice (P. maniculatus) and California ground squirrels (S. beecheyi). This result suggested that SucB can be a broad indicator of infection against different Bartonella species. Importantly, antigenic cross-reactivity was observed when SucB was immunoblotted against antibodies to C. burnetti and F. tularensis, with slight reactivity against anti-R. typhi. SucB may therefore be one of the antigens responsible for the serological cross-reactions that have been noted in Western blotting and indirect fluorescence assays by other researchers (32, 39).
SucB has been shown to be an immunogenic protein during infections by two other intracellular pathogens, B. melitensis, which causes ovine and caprine brucellosis and can be transmitted to humans (46), and C. burnetti, the causative agent of Q fever (40). Noting the immunogenicity of SucB from these organisms that are close phylogenetic relatives of Bartonella and the cross-reactivity observed in serological assays between them, one must be cautious when correlating antibody reactivity against SucB with a Bartonella infection and vice versa. Also confounding is that the clinical manifestations of bartonelloses, Q fever, and brucelloses are similar and could be confused. However, a broadly cross-reactive antigen such as SucB could potentially be used as an identifier of these diseases, with more specific diagnostic tools to differentiate the infectious agents. Moreover, as serologic diagnostic assays for these diseases are explored and become more developed, researchers should be aware of the potential cross-reactivity of Bartonella SucB with antibodies against other organisms.
Nucleotide sequence accession numbers. The DNA sequence of the B. vinsonii subsp. berkhoffii clone, which includes the sequence encoding the sucB gene and the partial sequence encoding the sucA and lpdA genes, has been submitted to GenBank under accession number AY160679. The DNA sequence of the B. quintana sucB gene has been submitted to GenBank under accession number AY160680.
Acknowledgments
We thank Treasa Burke, Heather Stevenson, Lori Phelan, Jennifer Lowell, Travis Bellville, and Steve Sviat for their assistance. We acknowledge Fred Koster (University of New Mexico) for his involvement in obtaining clinical samples and Ned Hayes of the Epidemiology Section of DVBID's Bacterial Zoonoses Branch. We appreciate the expert insight of Jane Koehler (University of California at San Francisco) and her time to discuss the issues. We thank Tom Burkot for a critical reading of the manuscript along with Barbara J. B. Johnson for helpful suggestions and discussions about this project. We thank Bob Heinzen (University of Wyoming), Yousef Abu Kwaik (University of Kentucky), Rendi Bacon, and Jeanine Petersen for sharing antiserum samples with us. We gratefully acknowledge Duane Gubler, Director of the DVBID, for his support of this project.
Editor: V. J. DiRita
REFERENCES
- 1.Amerein, M. P., D. De Briel, B. Jaulhac, P. Meyer, H. Monteil, and Y. Piemont. 1996. Diagnostic value of the indirect immunofluorescence assay in cat scratch disease with Bartonella henselae and Afipia felis antigens. Clin. Diagn. Lab. Immunol. 3:200-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, B., E. Lu, D. Jones, and R. Regnery. 1995. Characterization of a 17-kilodalton antigen of Bartonella henselae reactive with sera from patients with cat scratch disease. J. Clin. Microbiol. 33:2358-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, B., D. Jones, and A. Burgess. 1996. Cloning, expression and sequence analysis of the Bartonella henselae gene encoding the HtrA stress-response protein. Gene 178:35-38. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, B. E., and M. A. Neuman. 1997. Bartonella spp. as emerging human pathogens. Clin. Microbiol. Rev. 10:203-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai, Y., M. Y. Kosoy, G. O. Maupin, K. R. Tsuchiya, and K. L. Gage. 2002. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am. J. Trop. Med. Hyg. 66:622-627. [DOI] [PubMed] [Google Scholar]
- 6.Bergmans, A. M., M. F. Peeters, J. F. Schellekens, M. C. Vos, L. J. Sabbe, J. M. Ossewaarde, H. Verbakel, H. J. Hooft, and L. M. Schouls. 1997. Pitfalls and fallacies of cat scratch disease serology: evaluation of Bartonella henselae-based indirect fluorescence assay and enzyme-linked immunoassay. J. Clin. Microbiol. 35:1931-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birtles, R. J., T. G. Harrison, and D. H. Molyneux. 1994. Grahamella in small woodland mammals in the U.K.: isolation, prevalence and host specificity. Ann. Trop. Med. Parasitol. 88:317-327. [DOI] [PubMed] [Google Scholar]
- 8.Breitschwerdt, E. B., C. E. Atkins, T. T. Brown, D. L. Kordick, and P. S. Snyder. 1999. Bartonella vinsonii subsp. berkhoffii and related members of the alpha subdivision of the Proteobacteria in dogs with cardiac arrhythmias, endocarditis, or myocarditis. J. Clin. Microbiol. 37:3618-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitschwerdt, E. B., and D. L. Kordick. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin. Microbiol. Rev. 13:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, C., K. Yamamoto, B. B. Chomel, R. W. Kasten, D. C. Simpson, C. R. Smith, and V. L. Kramer. 1999. Seroepidemiology of Bartonella vinsonii subsp. berkhoffii infection in California coyotes, 1994-1998. Emerg. Infect. Dis. 5:711-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, C. C., B. B. Chomel, R. W. Kasten, V. Romano, and N. Tietze. 2001. Molecular evidence of Bartonella spp. in questing adult Ixodes pacificus ticks in California. J. Clin. Microbiol. 39:1221-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, C. C., H. Hayashidani, N. Pusterla, R. W. Kasten, J. E. Madigan, and B. B. Chomel. 2002. Investigation of Bartonella infection in ixodid ticks from California. Comp. Immunol. Microbiol. Infect. Dis. 25:229-236. [DOI] [PubMed] [Google Scholar]
- 13.Chang, C. C., R. W. Kasten, B. B. Chomel, D. C. Simpson, C. M. Hew, D. L. Kordick, R. Heller, Y. Piemont, and E. B. Breitschwerdt. 2000. Coyotes (Canis latrans) as the reservoir for a human pathogenic Bartonella sp.: molecular epidemiology of Bartonella vinsonii subsp. berkhoffii infection in coyotes from central coastal California. J. Clin. Microbiol. 38:4193-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Childs, J. E., J. A. Rooney, J. L. Cooper, J. G. Olson, and R. L. Regnery. 1994. Epidemiologic observations on infection with Rochalimaea species among cats living in Baltimore, Md. J. Am. Vet. Med. Assoc. 204:1775-1778. [PubMed] [Google Scholar]
- 15.Dalton, M. J., L. E. Robinson, J. Cooper, R. L. Regnery, J. G. Olson, and J. E. Childs. 1995. Use of Bartonella antigens for serologic diagnosis of cat-scratch disease at a national referral center. Arch. Intern. Med. 155:1670-1676. [PubMed] [Google Scholar]
- 16.Daly, J. S., M. G. Worthington, D. J. Brenner, C. W. Moss, D. G. Hollis, R. S. Weyant, A. G. Steigerwalt, R. E. Weaver, M. I. Daneshvar, and S. P. O'Connor. 1993. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 31:872-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dastoor, F. P., M. E. Forrest, and J. T. Beatty. 1997. Cloning, sequencing, and oxygen regulation of the Rhodobacter capsulatus alpha-ketoglutarate dehydrogenase operon. J. Bacteriol. 179:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foil, L., E. Andress, R. L. Freeland, A. F. Roy, R. Rutledge, P. C. Triche, and K. L. O'Reilly. 1998. Experimental infection of domestic cats with Bartonella henselae by inoculation of Ctenocephalides felis (Siphonaptera: Pulicidae) feces. J. Med. Entomol. 35:625-628. [DOI] [PubMed] [Google Scholar]
- 19.Fournier, P. E., J. L. Mainardi, and D. Raoult. 2002. Value of microimmunofluorescence for diagnosis and follow-up of Bartonella endocarditis. Clin. Diagn. Lab. Immunol. 9:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeland, R. L., D. T. Scholl, K. R. Rohde, L. J. Shelton, and K. L. O'Reilly. 1999. Identification of Bartonella-specific immunodominant antigens recognized by the feline humoral immune system. Clin. Diagn. Lab. Immunol. 6:558-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray, G. C., A. A. Johnson, S. A. Thornton, W. A. Smith, J. Knobloch, P. W. Kelley, L. Obregon Escudero, M. Arones Huayda, and F. S. Wignall. 1990. An epidemic of Oroya fever in the Peruvian Andes. Am. J. Trop. Med. Hyg. 42:215-221. [DOI] [PubMed] [Google Scholar]
- 22.Higgins, J. A., S. Radulovic, D. C. Jaworski, and A. F. Azad. 1996. Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera: Pulicidae). J. Med. Entomol. 33:490-495. [DOI] [PubMed] [Google Scholar]
- 23.Hollingdale, M. R., J. E. Herrmann, and J. W. Vinson. 1978. Enzyme immunoassay of antibody to Rochalimaea quintana: diagnosis of trench fever and serologic cross-reactions among other rickettsiae. J. Infect. Dis. 137: 578-582. [DOI] [PubMed] [Google Scholar]
- 24.Kerkhoff, F. T., A. M. Bergmans, A. van Der Zee, and A. Rothova. 1999. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J. Clin. Microbiol. 37:4034-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knobloch, J., and M. Schreiber. 1990. Bb65, a major immunoreactive protein of Bartonella bacilliformis. Am. J. Trop. Med. Hyg. 43:373-379. [DOI] [PubMed] [Google Scholar]
- 26.Koehler, J. E. 1996. Bartonella infections. Adv. Pediatr. Infect. Dis. 11:1-27. [PubMed] [Google Scholar]
- 27.Koehler, J. E., C. A. Glaser, and J. W. Tappero. 1994. Rochalimaea henselae infection. A new zoonosis with the domestic cat as reservoir. JAMA 271:531-535. [DOI] [PubMed] [Google Scholar]
- 28.Koehler, J. E., F. D. Quinn, T. G. Berger, P. E. LeBoit, and J. W. Tappero. 1992. Isolation of Rochalimaea species from cutaneous and osseous lesions of bacillary angiomatosis. N. Engl. J. Med. 327:1625-1631. [DOI] [PubMed] [Google Scholar]
- 29.Kordick, D. L., B. Swaminathan, C. E. Greene, K. H. Wilson, A. M. Whitney, S. O'Connor, D. G. Hollis, G. M. Matar, A. G. Steigerwalt, G. B. Malcolm, P. S. Hayes, T. L. Hadfield, E. B. Breitschwerdt, and D. J. Brenner. 1996. Bartonella vinsonii subsp. berkhoffii subsp. nov., isolated from dogs; Bartonella vinsonii subsp. vinsonii; and emended description of Bartonella vinsonii. Int. J. Syst. Bacteriol. 46:704-709. [DOI] [PubMed] [Google Scholar]
- 30.Kosoy, M., M. Murray, R. D. Gilmore, Jr., B. Ying, and K. L. Gage. 2003. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J. Clin. Microbiol. 41:645-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosoy, M. Y., R. L. Regnery, T. Tzianabos, E. L. Marston, D. C. Jones, D. Green, G. O. Maupin, J. G. Olson, and J. E. Childs. 1997. Distribution, diversity, and host specificity of Bartonella in rodents from the southeastern United States. Am. J. Trop. Med. Hyg. 57:578-588. [DOI] [PubMed] [Google Scholar]
- 32.La Scola, B., and D. Raoult. 1996. Serological cross-reactions between Bartonella quintana, Bartonella henselae, and Coxiella burnetii. J. Clin. Microbiol. 34:2270-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang, Z., B. La Scola, H. Lepidi, and D. Raoult. 2001. Production of Bartonella genus-specific monoclonal antibodies. Clin. Diagn. Lab. Immunol. 8:847-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang, Z., and D. Raoult. 2000. Species-specific monoclonal antibodies for rapid identification of Bartonella quintana. Clin. Diagn. Lab. Immunol. 7:21-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang, Z., and D. Raoult. 2000. Differentiation of Bartonella species by a microimmunofluorescence assay, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western immunoblotting. Clin. Diagn. Lab. Immunol. 7:617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Litwin, C. M., T. B. Martins, and H. R. Hill. 1997. Immunologic response to Bartonella henselae as determined by enzyme immunoassay and Western blot analysis. Am. J. Clin. Pathol. 108:202-209. [DOI] [PubMed] [Google Scholar]
- 37.Mallqui, V., E. C. Speelmon, M. Verastegui, C. Maguina-Vargas, P. Pinell-Salles, R. Lavarello, J. Delgado, M. Kosek, S. Romero, Y. Arana, and R. H. Gilman. 2000. Sonicated diagnostic immunoblot for bartonellosis. Clin. Diagn. Lab. Immunol. 7:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurin, M., V. Roux, A. Stein, F. Ferrier, R. Viraben, and D. Raoult. 1994. Isolation and characterization by immunofluorescence, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blot, restriction fragment length polymorphism-PCR, 16S rRNA gene sequencing, and pulsed-field gel electrophoresis of Rochalimaea quintana from a patient with bacillary angiomatosis. J. Clin. Microbiol. 32:1166-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGill, S. L., R. L. Regnery, and K. L. Karem. 1998. Characterization of human immunoglobulin (Ig) isotype and IgG subclass response to Bartonella henselae infection. Infect. Immun. 66:5915-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen, S. V., H. To, T. Yamaguchi, H. Fukushi, and K. Hirai. 1999. Characterization of the Coxiella burnetti sucB gene encoding an immunogenic dihydrolipoamide succinyltransferase. Microbiol. Immunol. 43:743-749. [DOI] [PubMed] [Google Scholar]
- 41.Regnery, R. L., J. G. Olson, B. A. Perkins, and W. Bibb. 1992. Serological response to “Rochalimaea henselae” antigen in suspected cat-scratch disease. Lancet 339:1443-1445. [DOI] [PubMed] [Google Scholar]
- 42.Roux, V., S. J. Eykyn, S. Wyllie, and D. Raoult. 2000. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J. Clin. Microbiol. 38:1698-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roux, V., and D. Raoult. 1999. Body lice as tools for diagnosis and surveillance of reemerging diseases. J. Clin. Microbiol. 37:596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spencer, M. E., and J. R. Guest. 1985. Transcription analysis of the sucAB, aceEF and lpd genes of Escherichia coli. Mol. Gen. Genet. 200:145-154. [DOI] [PubMed] [Google Scholar]
- 45.Welch, D. F., K. C. Carroll, E. K. Hofmeister, D. H. Persing, D. A. Robison, A. G. Steigerwalt, and D. J. Brenner. 1999. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J. Clin. Microbiol. 37:2598-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zygmunt, M. S., M. A. Diaz, A. P. Teixeira-Gomes, and A. Cloeckaert. 2001. Cloning, nucleotide sequence, and expression of the Brucella melitensis sucB gene coding for an immunogenic dihydrolipoamide succinyltransferase homologous protein. Infect. Immun. 69:6537-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]