Abstract
It has been proposed that a multivalent malaria vaccine is necessary to mimic the naturally acquired resistance to this disease observed in humans. A major experimental challenge is to identify the optimal components to be used in such a multivalent vaccine. Expression library immunization (ELI) is a method for screening genomes of a pathogen to identify novel combinations of vaccine sequences. Here we describe immune responses associated with, and the protective efficacy of, genomic Plasmodium chabaudi adami DS expression libraries constructed in VR1020 (secretory), monocyte chemotactic protein-3 (chemoattractant), and cytotoxic T lymphocyte antigen 4 (lymph node-targeting) DNA vaccine vectors. With splenocytes from vaccinated mice, specific T-cell responses, as well as gamma interferon and interleukin-4 production, were observed after stimulation with P. chabaudi adami-infected erythrocytes, demonstrating the specificity of genomic library vaccination for two of the three libraries constructed. Sera obtained from mice vaccinated with genomic libraries promoted the opsonization of P. chabaudi adami-infected erythrocytes by murine macrophages in vitro, further demonstrating the induction of malaria-specific immune responses following ELI. Over three vaccine trials using biolistic delivery of the three libraries, protection after lethal challenge with P. chabaudi adami DS ranged from 33 to 50%. These results show that protective epitopes or antigens are expressed within the libraries and that ELI induces responses specific to P. chabaudi adami malaria. This study further demonstrates that ELI is a suitable approach for screening the malaria genome to identify the components of multivalent vaccines.
Malaria is one of the major causes of death in the developing world and is an important barrier to economic progress in countries where the disease is endemic (reviewed in reference 37). Measures to contain malaria such as vector control by use of insecticides and drug treatment of active infection have become decreasingly effective, and new methods such as the use of vaccines against malaria-causing parasites at various life cycle stages are needed to control this disease (reviewed in reference 14). Although there are several candidate antigens under development to combat malaria, there is no effective single-stage malarial vaccine yet available (reviewed in reference 37). There is a general consensus that a multivalent vaccine is necessary to mimic naturally acquired resistance in humans. A major challenge is to identify the best antigen components to be used in such a multivalent vaccine (12, 20).
Protection in humans against asexual blood stages of malaria-causing parasites is believed to include mechanisms such as antibodies that block merozoite entry into erythrocytes and inhibit parasite development (reviewed in reference 20). In order to help resolve blood-stage malarial infection, induction of antibody and the activation of CD4+ T cells are required when vaccinating with recombinant candidate blood-stage antigens such as apical membrane antigen-1 (18, 40) and merozoite surface protein-119 (10, 16). In addition, other studies have shown that malaria parasite-specific T cells can adoptively transfer resistance in mice against challenge with Plasmodium yoelii and P. chabaudi, indicating that CD4+ T cells alone may also play a role in vaccine-induced immunity (39).
Improvements in the immunogenicity of malarial antigens delivered by DNA vaccines are important if there is to be any possible practical application of this technology to provide protection against blood-stage malaria. It has been suggested that DNA vaccines alone may not be adequate to protect against malaria (reviewed in reference 19). Antigen-presenting cells, in particular dendritic cells, have been shown to be potent initiators of immune responses following DNA vaccination and are important in the uptake of antigen expressed from cells transfected by a DNA vaccine (reviewed in reference 27). Improvement of DNA vaccine efficacy by targeting antigen to antigen-presenting cells may be required if DNA priming alone is to be sufficient to provide a significant level of protection against malaria. The use of the targeting ligand cytotoxic T lymphocyte antigen 4 (CTLA4) in DNA vaccination has been shown to improve the magnitude and speed of the antibody response (6, 13, 22, 29). Fusion of antigens to the chemokine monocyte chemotactic protein-3 (MCP-3) has also been shown to enhance protective efficacy after DNA vaccination in a tumor challenge model (5), to induce cytotoxic T lymphocytes and neutralizing antibodies to human immunodeficiency virus type 1 envelope proteins (4), and to induce antibody responses to the P. chabaudi adami merozoite surface protein 4/5 (MSP4/5) antigen in mice (29); in the latter case, the MCP-3/MSP4/5 DNA vaccine protected mice against lethal challenge with P. chabaudi adami.
Expression library immunization (ELI) enables screening of a pathogen's genome and eventual discovery of potential vaccine candidates. ELI has been applied to bacterial (3, 7) and parasitic (1, 24, 28, 31) infections and recently to simian immunodeficiency virus infection (32). Genomic ELI libraries encode antigens from all stages of the life cycle, potentially allowing the discovery of antigens from particular stages of a life cycle. A primary focus for malaria vaccine development is the blood stage, which is responsible for the morbidity and mortality associated with malaria (reviewed in reference 37).
It was previously reported that ELI with a P. chabaudi adami genomic library significantly protects mice against blood-stage malaria caused by the lethal P. chabaudi adami DS strain (31), although the protective mechanism(s) induced by this multivalent genomic vaccine remained to be elucidated. Here we extend these observations and report the DNA vaccination of mice with three different libraries of P. chabaudi adami genomic DNA, expressed in the VR1020, MCP-3, or CTLA4 vector. Protection after library vaccination was observed only with the VR1020 and MCP-3 libraries and was found to be associated with T-cell responses of splenocytes from vaccinated mice that were specific to native malarial antigens or epitopes produced in P. chabaudi adami DS-infected blood. Murine macrophages incubated with sera, obtained after genomic library vaccination, also possessed the ability to opsonize P. chabaudi adami DS-infected red blood cells (P. chabaudi adami DS-IRBCs) in vitro, providing evidence that genomic library vaccination enhances humoral effector responses.
MATERIALS AND METHODS
Creation of plasmid pools.
The creation of a VR1020 genomic expression library has been described previously (31). Briefly, P. chabaudi adami DS genomic DNA was isolated from Ficoll-purified erythrocytes of infected BALB/c mice (parasitemia, 20 to 30%). The purified DNA was partially digested with Tsp509I (5 U for 90 s at 65°C), and the digested fraction between 1 and 3 kbp was isolated by gel elution. Vector VR1020 (VICAL, San Diego, Calif.) was prepared by digestion with BamHI and BglII and the insertion of an EcoRI linker constructed from the oligonucleotides 5′GATCCGGGAATTCAA and 5′GATCTTGAATTCCCG. Once obtained, the new vector was digested with EcoRI, treated with alkaline phosphatase, and ligated with Tsp509I-digested P. chabaudi adami genomic DNA. Ligation mixes were transformed into Escherichia coli DH5α, and colonies were selected on solid medium containing 50 μg of kanamycin/ml. After overnight growth, E. coli colonies were combined into pools. A total of 10 pools (termed 3KA to 3KJ), each comprising approximately 3,000 individual clones, were constructed and stored at −80°C as glycerol stocks. The pool of 30,000 clones (termed 30K) was obtained by combining each of the 10 pools of 3,000 clones. Murine cDNA was supplied by Harshall Nandurkar (Monash University, Clayton, Australia). The oligonucleotides 5′TATTATTAGCGGCCGCATGAGGATCTCTGCCACGCTT (containing a NotI site) and 5′TATGGATCCTCCACCTCCACCTCCAGGCTTTGGAGTTGGGGT (encoding six glycine residues followed by a BamHI site) were used to amplify the MCP-3 cDNA by PCR. The MCP-3 PCR fragment was digested with NotI and BamHI and inserted into the VR1012 vector (VICAL) digested with the same enzymes to produce the MCP-3 vector (29). The CTLA4 vector (6) was a gift from Andrew Lew (Walter and Eliza Hall Institute, Melbourne, Australia). Libraries in MCP-3 and CTLA4 vectors were constructed as described for the VR1020 vector by using the same stock of P. chabaudi adami genomic DNA. Ligation reaction mixtures were transformed into E. coli DH5α, and colonies were selected on solid media containing 50 μg of kanamycin/ml for the MCP-3 vector and 100 μg of ampicillin/ml for the CTLA4 vector.
Isolation of plasmid DNA and construction of vaccination cartridges.
Library pools stored as glycerol stocks were grown to confluence on solid medium prior to inoculation into liquid medium. Bacteria from five to ten confluent plates were used to inoculate 1 liter of Luria broth and grown with shaking at 37°C for 6 h prior to harvest. Plasmid preparation and endotoxin removal were performed by using an endotoxin-free plasmid giga kit according to the instructions of the manufacturer (QIAGEN Inc, Valencia, Calif.). DNA purified under endotoxin-free conditions was precipitated onto gold microcarriers which were attached to plastic supports as per the recommendations of the manufacturer (Bio-Rad Laboratories, Hercules, Calif.). DNA was combined with carriers at a ratio of 100 μg of DNA/50 mg of carriers. Each projectile used for vaccination contained approximately 1 μg of DNA.
Mice and vaccinations.
All mice were BALB/c, female, and 4 to 6 weeks of age at the time of first vaccination. For vaccination, the abdominal regions were shaved and particles were delivered via the intraepidermal (i.d.) route by using a Helios gene gun (Bio-Rad Laboratories) with a pulse of helium gas at 400 lb/in2.
Infection of mice, blood sampling, and parasitemia measurements.
Blood from an infected mouse with a known parasitemia level (1 to 10%) was taken and immediately diluted in phosphate-buffered saline (PBS) to give the required dosage (105 IRBCs/dose). Mice were infected by the intraperitoneal route at day 0, and parasitemia was assessed from day 6 through the period of crisis until the resolution of parasitemia. Infection levels were assessed by Giemsa staining of tail smears. Mean peak parasitemia levels and the numbers of days to peak parasitemia were compared by using the Student t test. The Mantel-Haenszel (or log rank) test was used to measure the parameter of survival. This method involves calculating a P value and testing the null hypothesis that the survival curves are identical between vaccinated and control groups. This analysis was performed with Prism 3 software (GraphPad, San Diego, Calif.).
In vitro spleen cell proliferation.
Ten days after the final DNA vaccination, mice were sacrificed and single-cell suspensions of splenocytes were obtained by crushing whole spleens with a 5-ml syringe barrel. The splenocytes were then resuspended in complete RPMI 1640 medium (Invitrogen Corporation, Grand Island, N.Y.) supplemented with 10% fetal calf serum, 2 mM glutamine (Invitrogen), 0.05 mM 2-mercaptoethanol (Sigma Chemical Co., St Louis, Mo.), and penicillin-streptomycin (100 U/ml; Invitrogen). Suspensions were then passed through a 100-μm-pore-size nylon cell strainer (Becton Dickinson, Franklin Lakes, N.J.) and exposed for 3 min to erythrocyte lysis buffer (155 mM ammonium chloride, 10 mM potassium hydrogen carbonate, 0.1 mM EDTA [pH 7.4]). Splenocytes were again washed and resuspended in RPMI 1640 medium. Cell viability was greater than 80% as determined by trypan blue exclusion (Invitrogen). Splenocytes were stimulated with 2 × 106 P. chabaudi adami DS-IRBCs, freshly extracted from an infected mouse, or 2 × 106 freshly extracted red blood cells (RBCs) as a control. As a control for cell viability, splenocytes were stimulated with concanavalin A (Sigma) at a final concentration of 2.5 μg/ml. Splenocytes were cultured for 96 h in flat-bottom microtiter plates in triplicate at a final concentration of 5 × 106 cells/ml (106 cells/well) and pulsed with 1 μCi of [3H]thymidine (Amersham Biosciences Corp., Piscataway, N.J.)/well 18 h before harvesting. The splenocytes were harvested onto fiberglass filter mats (Saktron Instruments Inc., Sterling, Va.) with an automated cell harvester (Saktron), and incorporated radioactivity was measured with a liquid scintillation counter (Perkin Elmer Life Sciences, Wellesley, Mass.).
ELISA for IFN-γ and IL-4.
Spleen cells from vaccinated and control mice were incubated for 72 h in the presence of 2 × 106 P. chabaudi adami-IRBCs or 2 × 106 control RBCs, and supernatants were harvested after 72 h and stored at −20°C until cytokine levels were measured by enzyme-linked immunosorbent assay (ELISA). Gamma interferon (IFN-γ) and interleukin-4 (IL-4) ELISAs were carried out according to the instructions of the manufacturer (Endogen, Woburn, Mass.). Briefly, Maxisorp ELISA plates (Nunc, Kamstrupvej, Denmark) were coated with anti-mouse IFN-γ monoclonal antibody (Endogen) or anti-mouse IL-4 monoclonal antibody (Endogen) and incubated overnight at room temperature. The ELISA plates were then blocked in assay buffer (PBS with 4% bovine serum albumin, pH 7.4). The supernatants obtained from the splenocyte cultures were tested in duplicate against serial dilutions of recombinant IFN-γ (starting at 5 ng) and IL-4 (starting at 1 ng) standards. After incubation at room temperature for 1 h, anti-mouse IL-4 biotin-labeled monoclonal antibodies (Endogen) or anti-mouse IFN-γ biotin-labeled monoclonal antibodies (Endogen) were added, and the plates were incubated for 1 h at room temperature. After the plates were washed three times, horseradish peroxidase-conjugated streptavidin (Amersham) was added at 1:16,000, and the plates were incubated for 30 min at room temperature. The plates were again washed three times. The reaction was developed with 3,3′,5,5′-tetramethylbenzidine substrate solution (Sigma) and stopped with 0.18 M H2SO4, and the absorbance was read at 450 nm.
Phagocytosis assays.
Phagocytosis assays were performed according to the method described in reference 26, with the following modifications. Macrophages were obtained from BALB/c mice by peritoneal lavage with 9 ml of ice-cold 0.34 M sucrose. The cells were then centrifuged at 1,200 rpm (Beckman Coulter Allegra 25R centrifuge) for 10 min at 4°C. Peritoneal cell exudates were then resuspended in complete RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum, 2 mM glutamine (Invitrogen), 0.05 mM 2-mercaptoethanol (Sigma), and penicillin-streptomycin (100 U/ml; Invitrogen) to a final concentration of 2 × 106 cells/ml. Eight-well chamber slides (Nalge Nunc International Corp, Naperville, Ill.) were used, with 4 × 105 macrophages added to each well. The macrophages were allowed to adhere for 2 h at 37°C with 5% CO2. After 2 h, nonadherent cells were removed by careful washing with 1 ml of 37°C RPMI 1640 medium. Fresh complete RPMI 1640 medium was added, and macrophages were left to adhere for a further 2 h. During this time, fresh P. chabaudi adami DS-IRBCs (108 IRBCs/ml containing trophozoites and schizonts at approximately 40 to 50% parasitemia) in complete RPMI 1640 medium were purified with Ficoll (Amersham). The IRBCs were washed twice with complete RPMI 1640 medium. After washing, IRBC pellets were placed into 1.5-ml centrifuge tubes in 15-μl aliquots. PBS (30 μl) and 1 μl of sera obtained from groups vaccinated with the genomic libraries, or sera from mice vaccinated with empty vector, were then added to the IRBC pellets and incubated for 1.5 h at 37°C with shaking. After 1.5 h, IRBCs and sera were added to adherent macrophages and incubated for 2 h at 37°C with 5% CO2. The eight-well slides were then washed four times with PBS to remove nonadherent macrophages and noningested IRBCs. Noningested but adherent IRBCs were then lysed by incubation of the slides with cold water for 20 s, followed by washing with PBS. The eight-well slides were then fixed and stained with Kwik Diff staining solution (Terno Shandon, Pittsburgh, Pa.). The number of IRBCs taken up by 200 macrophages per individual sample was then quantified by using light microscopy.
Blockage of macrophage Fcγ receptors to determine opsonization specificity.
Macrophages from BALB/c mice were prepared in eight-well chamber slides (Nalge Nunc International Corp.) as described for the phagocytosis assay. Macrophages were washed three times with warm RPMI 1640 medium (Invitrogen) to remove nonadherent cells. The cells were incubated in 200 μl of complete RPMI 1640 medium plus rat anti-mouse CD16/32 (Fcγ receptor)-blocking antibody (clone 93; Southern Biotech, Birmingham, Ala.) at a concentration of 100 μg/ml for 60 min at 37°C. The excess blocking antibody was removed by two washes with warm RPMI 1640 medium. IRBCs and sera from all three genomic libraries were prepared and incubated with macrophages (treated with and without the blocking antibody) as described for the phagocytosis assay.
RESULTS
Characterization of library clones.
All libraries were constructed by using the same stock of a partial digest of P. chabaudi adami DS genomic DNA in order to minimize any bias among the libraries in insert sizes after enzymatic digestion of the genomic DNA. A partial enzymatic digest of DNA (described in Materials and Methods and in reference 31) with Tsp509I was required as this enzyme recognizes AATT sequences and the P. chabaudi adami genome is more than 80% adenosine and thymidine. Table 1 shows the characteristics of random samples of 32 to 58 plasmid clones taken from the 30,000-clone VR1020, MCP-3, and CTLA4 (hereafter referred to as VR1020/30K (31), MCP-3/30K, and CTLA4/30K, respectively) genomic libraries. The DNA inserts in selected clones were sequenced, and the sequences were compared against P. falciparum and P. yoelii genomic databases found at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov/PMGifs/Genomes/plasmodium.html), which confirmed the presence of P. chabaudi adami genomic DNA (data not shown). The DNA sequences were also translated and compared against sequences in P. yoelii and P. falciparum protein databases (data not shown), which revealed in-frame peptides predicted to be encoded by the insert DNA, as shown in Table 1. The construction method resulted in three libraries that were almost uniform in terms of numbers of recombinant plasmids, mean sizes of DNA inserts and encoded peptides, and percentages of encoded peptides of >50 amino acids (aa) in length. This uniformity was important in order to make comparisons between the three different vectors. The average size of the inserts among the three libraries was 1.43 kbp. The P. chabaudi adami genome is predicted to be approximately 23 Mbp in size (see the National Center for Biotechnology Information website at www.ncbi.nlm.nih.gov/projects/Malaria/Rodent/chabaudi.html). Therefore, the 30,000 plasmids contained in each of the genomic libraries represent 43 Mbp of DNA, resulting in an overall coverage of approximately two times the size of the P. chabaudi adami genome for each library.
TABLE 1.
Features of P. chabaudi adami library clones
| Librarya | No. of plasmid clones examined by restriction digestionb | No. (%) with inserts | No. of plasmid clones examined by sequencing | Avg insert size (kb) | No. (%) with reading frames expressing peptidesc | Size range (aa) of encoded peptides | No. (%) of peptides longer than:
|
Avg size (aa) of encoded peptides | |
|---|---|---|---|---|---|---|---|---|---|
| 20 aa | 50 aa | ||||||||
| VR1020/30Kd | 58 | 58 (100) | 24 | 1.5 | 18 (75) | 1-115 | 10 (42) | 3 (13) | 25 |
| MCP-3/30K | 32 | 32 (100) | 22 | 1.3 | 20 (90) | 3-113 | 8 (36) | 3 (14) | 32 |
| CTLA4/30K | 45 | 41 (93) | 33 | 1.5 | 23 (70) | 2-139 | 11 (33) | 6 (18) | 36 |
Ten plasmid pools, designated 3KA to 3KJ and each containing approximately 30,000 individual clones, were combined to create pool 30K, containing 30,000 clones.
All clones examined were from pool 3KA from each library.
Number in frame with the tissue plasminogen activator signal sequence (VR1020/30K), the MCP-3 sequence (MCP-3/30K), and the CTLA4 sequence (CTLA4/30K).
The VR1020/30K library is described in reference 31.
We also estimated the proportion of clones that are predicted to encode peptides. Out of the 30,000 clones, about 50% will include a reading frame (since 50% of the genome comprises exons) (8). Of the remaining 15,000 clones, only 50% will be oriented correctly (since cloning into the vector is not directional), and of these 7,500 clones, only 1:3 will be in frame with the signal sequence: this leaves approximately 2,500 clones (or about 8%) potentially encoding an in-frame peptide. From the sequence analysis, it was found that 13 to 18% of the clones from all the libraries actually encoded in-frame peptides longer than 50 aa (we assume anything longer than 50 aa is likely to be a real peptide due to the high AT content of malaria parasite DNA, which creates stop codons at high frequency). Thus, in the 30K genomic vaccines, approximately 13 to 18% of clones (3,900 to 5,400) should potentially be able to encode peptides.
Antigen-specific cellular immune responses induced by ELI DNA vaccination with three different vectors.
Groups of mice were vaccinated i.d. with either the VR1020/30K, MCP-3/30K, or CTLA4/30K genomic library or with the corresponding empty plasmid DNA control vectors, according to the procedure described in Materials and Methods. To evaluate cellular responses to native P. chabaudi adami antigens induced by each of the genomic libraries (and responses of corresponding empty vector-vaccinated control mice), spleens were removed from mice 10 days after the final i.d. vaccination and cell suspensions from individual mice were stimulated by using 2 × 106 P. chabaudi adami-IRBCs or 2 × 106 RBCs (prepared from naïve BALB/c mice) as a control.
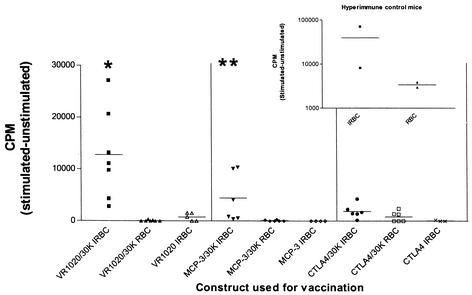
Significant proliferation of spleen cells taken from VR1020/30K-vaccinated mice (P < 0.01) and MCP-3/30K-vaccinated mice (P < 0.05) was detected upon stimulation with P. chabaudi adami-IRBCs compared to that of spleen cells taken from VR1020/30K- and MCP-3/30K-vaccinated mice and stimulated with control RBCs (Fig. 1). In contrast, significant levels of proliferation were not detected with spleen cells obtained from CTLA4/30K-vaccinated mice upon stimulation with IRBCs (P = 0.17) compared to that of spleen cells obtained from CTLA4/30K-vaccinated mice and stimulated with control RBCs. Spleen cells obtained from control mice vaccinated with the MCP-3 or CTLA4 vector alone and stimulated with IRBCs (Fig. 1) or RBCs (data not shown) showed no detectable proliferation. However, spleen cells obtained from VR1020 vector-vaccinated control mice did proliferate to a very low extent above background levels upon stimulation with IRBCs (but not RBCs), but the levels of proliferation observed were significantly lower than those of IRBC-stimulated spleen cells from VR1020/30K-vaccinated mice (P = 0.0128) (Fig. 1).
FIG. 1.
In vitro proliferation of splenocytes from individual BALB/c mice vaccinated i.d. with the gene gun. Mice were vaccinated with VR1020/30K, MCP-3/30K, and CTLA4/30K three times at 2-week intervals. Splenocytes were harvested 10 days after the final vaccination. Following 72 h of stimulation with 2 × 106 IRBCs or 2 × 106 RBCs, [3H]thymidine was added for 18 h. Numbers of counts per minute of radioactivity incorporated by cells were then recorded. Splenocytes vaccinated with empty vector control DNA (VR1020, MCP-3, or CTLA4) were also stimulated with 2 × 106 RBCs, but no incorporation of [3H]thymidine was observed (data not shown). Splenocytes from all individual splenocyte cultures responded to concanavalin A stimulation (data not shown). The mean number of counts per minute (bars) is shown. ∗, P < 0.01; ∗∗, P < 0.05. The insert shows the response of splenocytes from hyperimmune control mice (mice that had survived P. chabaudi adami infection) that were used as a positive control for IRBCs.
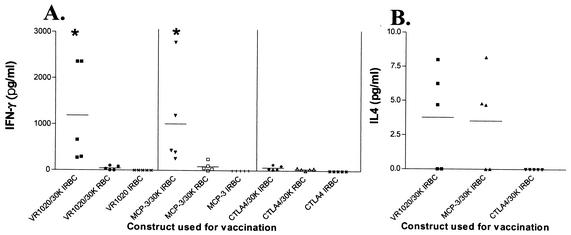
Splenocytes taken from VR1020/30K-vaccinated mice and stimulated with IRBCs produced significantly higher levels of IFN-γ than splenocytes from control mice vaccinated with vector DNA (P < 0.05) (Fig. 2A). When stimulated with IRBCs, spleen cells obtained from MCP-3/30K-vaccinated mice also produced detectable levels of IFN-γ in the culture supernatant (P < 0.05) (Fig. 2A). No IFN-γ was detected in culture supernatants of splenocytes taken from CTLA4/30K-vaccinated mice and stimulated with IRBCs (Fig. 2A) or in culture supernatants of splenocytes taken from control mice vaccinated with VR1020, MCP-3, and CTLA4 vector DNA and stimulated with IRBCs (Fig. 2A) or RBCs (data not shown).
FIG. 2.
IFN-γ (A) and IL-4 (B) production by splenocytes from BALB/c mice vaccinated with the VR1020/30K, MCP-3/30K, and CTLA4/30K libraries. Splenocytes from individual mice were cultured as described in the legend to Fig. 1. After 72 h of stimulation with 2 × 106 IRBCs or 2 × 106 RBCs, supernatants were harvested and analyzed by ELISA for the presence of IFN-γ and IL-4. Splenocytes from mice vaccinated with empty vector control DNA (VR1020, MCP-3, or CTLA4) were also stimulated with 2 × 106 RBCs, but IFN-γ and IL-4 responses were not observed (data not shown). Unstimulated splenocytes did not produce detectable IFN-γ or IL-4. The mean level of IFN-γ or IL-4 production is shown (bars). The raw data were log10 transformed and compared by using the Student t test. ∗, P < 0.05.
IL-4 was detected in three out of five culture supernatants from spleen cells primed with the VR1020/30K and MCP-3/30K genomic libraries and stimulated with IRBCs (Fig. 2B). No IL-4 was detected with CTLA4/30K-primed IRBC-stimulated splenocytes or in supernatants of splenocytes from control mice vaccinated with VR1020, MCP-3, and CTLA4 vector DNA that were stimulated with IRBCs or RBCs (data not shown).
Phagocytosis by macrophages of P. chabaudi adami DS-IRBCs after incubation with sera from mice vaccinated with each of the genomic libraries.
i.d. DNA vaccination with any of the genomic expression libraries did not produce humoral responses that were detectable when sera were evaluated by ELISA using soluble blood-stage P. chabaudi adami lysate as the antigen (data not shown). However, the method of gene gun vaccination has been shown to produce a strong humoral response after delivery of plasmids containing known antigens in many different vaccine models (reviewed in reference 27). Antibody-mediated opsonization of IRBCs, and subsequent internalization and destruction by macrophages, has been shown to be a factor contributing to a reduction in parasitemia during crisis in the P. chabaudi mouse model (25, 26).
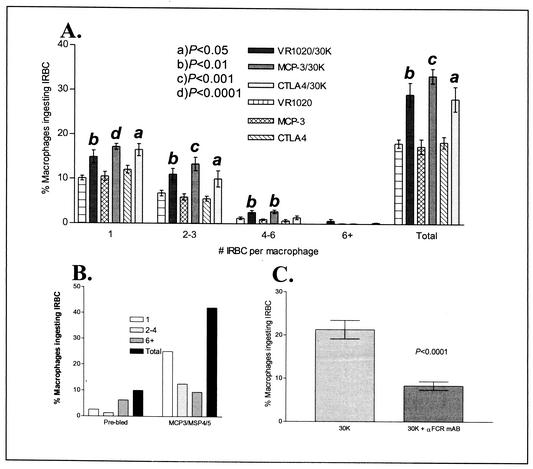
To determine whether vaccination with the P. chabaudi adami genomic expression libraries could induce opsonizing antibodies, sera were taken before challenge from mice vaccinated with each library. The prechallenge sera were tested for the ability to promote opsonization of P. chabaudi adami-IRBCs by BALB/c macrophages. Figure 3 shows that sera obtained from individual mice vaccinated with each of the genomic libraries have the ability to enhance uptake and promote destruction of IRBCs by macrophages. The percentage of macrophages ingesting IRBCs was significantly higher in incubation mixtures containing macrophages treated with sera taken from mice vaccinated with VR1020/30K (P < 0.01), MCP-3/30K (P < 0.001), and CTLA4/30K (P < 0.05) than in incubation mixtures containing sera from control mice vaccinated with vector DNA (Fig. 3A). For comparison, Fig. 3B shows the percentages of macrophages ingesting IRBCs after incubation with a pool of sera from six mice vaccinated with the malarial blood-stage antigen MSP4/5 (fused to the MCP-3 vector) used as a positive control. We have shown that this construct can significantly enhance antibody responses to MSP4/5 (29).
FIG. 3.
Phagocytosis of P. chabaudi adami-IRBCs preincubated with sera from mice vaccinated with VR1020/30K, MCP-3/30K, and CTLA4/30K (and control vector DNA). (A) Percentage of macrophages ingesting IRBCs after preincubation with sera. All six groups contained 10 BALB/c mice each. The values are expressed as percentages of macrophages containing IRBCs out of a total of 200 counted. The number of IRBCs contained within an individual macrophage for each vaccine group is also shown. Data are expressed as means ± standard errors of the means. a, P < 0.05; b, P < 0.01; c, P < 0.001; d, P < 0.0001. (B) Percentage of macrophages ingesting IRBCs after incubation with pooled sera from naive (unvaccinated) mice (n = 6) and pooled sera from mice vaccinated with MCP-3/MSP4/5 DNA (n = 6) as a positive control (29). (C) Incubation of macrophages with a monoclonal antibody specific for Fcγ II and Fcγ III receptors inhibits phagocytosis of IRBCs. Macrophages were treated with a α-Fcγ monoclonal antibody, followed by incubation with IRBCs previously exposed to sera from mice vaccinated with each genomic library (30K + α-FCR mAB), or they were left untreated and incubated with IRBCs exposed to sera from vaccinated mice as a positive control (30K). The data shown are expressed as means ± standard errors of the means of the percentages of macrophages ingesting IRBCs. Five serum samples from mice vaccinated with each of the VR1020/30K, MCP-3/30K, and CTLA4/30K libraries were tested, giving a total of 15 individual samples. Treated (30K + α-FCR mAB) and untreated (30K) groups were compared by using the Student t test.
To confirm that macrophage uptake of IRBCs was antibody-mediated, CD16/Fcγ II and CD32/Fcγ III receptors on macrophages were blocked by incubation of macrophages with the specific monoclonal antibody MAb93 (as described in Materials and Methods); antibodies directed against the CD16/Fcγ II and CD32/Fcγ III receptors have been shown to block Fc receptor binding by immunoglobulin G (IgG) antibodies and inhibit effector functions (17, 34, 35). Figure 3C shows that incubation of macrophages with the anti-Fcγ antibody MAb93 significantly inhibits the percentage of macrophages ingesting IRBCs opsonized with sera from mice vaccinated with the genomic libraries (VR1020/30K, MCP-3/30K, and CTLA4/30K). This result demonstrates that opsonizing antibodies are indeed present in sera of mice after genomic library vaccination and that these antibodies are the major component in the sera responsible for macrophage uptake of IRBCs.
Efficacy of protection induced by DNA vaccination with genomic expression libraries against lethal challenge with blood-stage P. chabaudi adami DS.
The P. chabaudi adami DS mouse model is a stringent test for vaccine efficacy due to the high virulence of the DS strain (2, 9, 31). In order to determine the protective efficacy of the VR1020/30K, MCP-3/30K, and CTLA4/30K genomic libraries against a virulent challenge, three separate challenge trials were conducted with BALB/c mice (Table 2). It has been previously shown with the P. chabaudi adami DS BALB/c mouse model that the VR1020/30K library induces significant (albeit partial) protection against lethal challenge (31).
TABLE 2.
Protocol and parasitemia measurements for mice challenged with lethal P. chabaudi adami DS and percentages of survivors over three challenge trials
| Group | No. in group | Mean peak parasitemia (%) ± SD | No. of survivors (%) |
|---|---|---|---|
| Trial 1 | |||
| CTLA4 | 10 | 42 ± 6 | 3 (30) |
| CTLA4/30K | 10 | 35 ± 7a | 3 (30) |
| VR1020/30K | 10 | 35 ± 4a | 5 (50) |
| Nonsurvivors | 19 | 36 ± 8 | |
| Survivors | 11 | 37 ± 5 | |
| Trial 2 | |||
| MCP-3 | 6 | 40 ± 13 | 0 (0) |
| MCP-3/30K | 6 | 31 ± 15 | 2 (33)b |
| Nonsurvivors | 10 | 39 ± 12 | |
| Survivors | 2 | 34 ± 2 | |
| Trial 3 | |||
| VR1020 | 6 | 45 ± 11 | 0 (0) |
| VR1020/30K | 6 | 37 ± 5 | 3 (50)c |
| MCP-3 | 6 | 43 ± 7 | 0 (0) |
| MCP-3/30K | 6 | 29 ± 18 | 3 (50) |
| CTLA4 | 6 | 41 ± 7 | 0 (0) |
| CTLA4/30K | 6 | 38 ± 6 | 0 (0) |
| Nonsurvivors | 30 | 39 ± 11 | |
| Survivors | 6 | 25 ± 14a |
P < 0.05; Student's t test.
P = 0.05; Mantel-Haenszel test for comparison with MCP-3 vector.
P < 0.01; Mantel-Haenszel test for comparison with VR1020 vector.
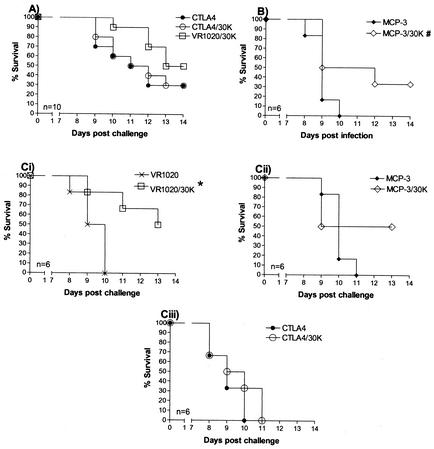
The first challenge trial included 10 mice per group. Mice were vaccinated i.d. with a gene gun a total of three times at 2-week intervals and challenged 2 weeks after the final vaccination with 105 P. chabaudi adami-IRBCs. Trial 1 involved a comparison of the CTLA4/30K and VR1020/30K libraries. The VR1020/30K library was used as a positive control based on previous experiments where this vaccine has induced 30 to 60% protection against lethal challenge (31). Although there was a significant reduction in peak parasitemia in mice vaccinated with both the VR1020/30K and CTLA4/30K libraries compared to those vaccinated with the empty CTLA4 control vector (Table 2), this result was not reproduced in trial 3 (see below). While there were no significant differences between survival curves (i.e., delays in death) of control and vaccinated mice, increased survival was observed in the group vaccinated with the VR1020/30K library (Fig. 4A), and this result was repeated in trial 3 (Fig. 4Ci). In contrast, the CTLA4/30K vaccine did not induce increased survival (Fig. 4A).
FIG. 4.
Survival curves of mice vaccinated with VR1020/30K, MCP-3/30K, and CTLA4/30K genomic libraries and challenged with virulent P. chabaudi adami. Mice were vaccinated three times at 2-week intervals via the gene gun, followed by challenge with 100,000 P. chabaudi adami-IRBCs 2 weeks later. (A) Evaluation of efficacies of CTLA4/30K and VR1020/30K libraries. (B) Evaluation of efficacy of MCP-3/30K library. (C) Comparative evaluations of all three genomic libraries. #, P = 0.05; ∗, P < 0.05 compared to empty control vectors (Mantel-Haenszel test).
The second trial involved the evaluation of the MCP-3/30K genomic library. Although there was a drop in mean peak parasitemia level in the MCP-3/30K group compared to that in the MCP-3 vector control group, the difference was not statistically significant due to the large variation in parasitemia levels between individual mice (Table 2). A significant delay in death was observed in mice vaccinated with the MCP-3/30K library compared to that in mice vaccinated with the MCP-3 control vector (P = 0.05) (Fig. 4B). Half of the mice vaccinated with the MCP-3/30K library survived until day 12 postinfection, with two mice surviving to the conclusion of the trial at day 14. No MCP-3 vector control mice survived, with all mice dying by day 10 postinfection.
The third challenge trial directly compared the efficacy of the VR1020/30K, MCP-3/30K, and CTLA4/30K genomic libraries. Although there was a reduction in the mean peak parasitemia level for all mice vaccinated with the genomic libraries compared with that of their respective controls (mice vaccinated with vector-only DNA), the difference among groups was not statistically significant (Table 2). However, there was a significant reduction in peak parasitemia in surviving mice (in the MCP-3/30K- and VR1020/30K-vaccinated groups) compared with that in nonsurviving mice (Table 2). Of the mice vaccinated with the VR1020/30K and MCP-3/30K libraries, 50% survived, and there was a significant delay in death (P < 0.05) in the VR1020/30K-vaccinated group versus that in the VR1020 vector control group (Fig. 4Ci and Cii). In contrast, the CTLA4/30K library did not protect mice against lethal P. chabaudi adami challenge (Fig. 4Ciii).
DISCUSSION
Protection of mice against lethal P. chabaudi adami DS challenge by using ELI has been previously demonstrated (31). The aim of the present study was to determine whether the efficacy of ELI could be enhanced by using targeting vectors and to assess the immune responses induced by ELI in mice in order to confirm that ELI induces an antigen-specific response. Both humoral and cellular responses have been found to be required for resolution of infection with P. chabaudi in mice (reviewed in references 21 and 36). The data presented demonstrate that genomic ELI can induce cellular as well as humoral responses to native P. chabaudi adami DS antigens following vaccination. The data also show that protection against lethal P. chabaudi adami DS challenge is dependent on the type of DNA expression vector used, with survival correlating with in vitro cellular responses that were induced by vaccination with the VR1020 and MCP-3 libraries. The ELI vaccines promote significant survival, rather than reductions in parasitemia, in challenged mice, which is an important observation due to the high lethality associated with a P. chabaudi adami DS challenge (2, 21).
Vaccination of BALB/c mice with the VR1020/30K or MCP-3/30K library induced a specific cellular immune response to native antigens shed by IRBCs which was characterized by a significant increase in splenocyte proliferation, as well as by detectable levels of IFN-γ and IL-4 secretion. The ability of the VR1020/30K and MCP-3/30K genomic libraries to promote proliferation of splenocytes in response to live P. chabaudi adami-IRBCs demonstrates that the genomic approach to vaccination is specific to epitopes or antigens produced by the parasite and not an artifact produced by nonspecific stimulation of the immune system by foreign DNA. The presence of cellular responses in vitro induced by P. chabaudi adami ELI correlates with the protection observed in vivo after lethal challenge with blood-stage parasites, as significant splenocyte proliferation was not observed after vaccination with the nonprotective CTLA4/30K library. The production of opsonizing antibodies to promote phagocytosis in vitro (regardless of the vector used) revealed another possible mechanism by which parasite clearance after ELI may occur.
The nature of the epitopes or antigens that are recognized by splenocytes from vaccinated mice is clearly of interest, as this may identify sequences in the library encoding protective antigens. Sequencing of a sample of 664 plasmids derived from the VR1020/30K pool showed that open reading frames, predicted to encode peptides of various sizes (with many homologous to P. falciparum sequences), are indeed present in the library (unpublished data). Recently it has been shown that peptides of 20 aa synthesized to span the length of MSP1 (38) could induce T-cell proliferation in mice. Multiple epitopes were recognized in two strains of mice, with two epitopes discovered that were able to induce effector T cells capable of delaying growth of lethal P. yoelii YM following adoptive transfer into immunodeficient mice without inducing detectable antibody responses (38). In addition, peptides were able to protect mice against P. yoelii, suggesting that the T-cell epitopes may be useful as a vaccine against P. yoelii (38) Given that the plasmids in the genomic library will encode a variety of in-frame peptides, a major contributory factor to protection observed with ELI is possibly due to T-cell help delivered by the library.
Although there was a response to IRBC stimulation in the VR1020/30K- and MCP-3/30K-vaccinated mice, significant levels of splenocyte proliferation did not occur when mice were vaccinated with the CTLA4/30K library. This result is curious as all three libraries were constructed by the same method with the same stock of P. chabaudi adami genomic DNA and show similar properties in terms of coding capacities. One possibility is that the nature of the CTLA4 vector itself may be contributing to the lack of proliferation. The CTLA4 vector expresses proteins as fusions with the CTLA4-human Ig moiety (approximately 60 kDa), and the fusion partner is thus considerably larger than the fused P. chabaudi adami sequences, based on a peptide of size 20 to 100 aa. It is possible that many of the epitopes contained within the CTLA4/30K pool may have been obscured upon folding of the large CTLA4 moiety and poorly processed or presented to the immune system. CTLA4 in vivo is a T-cell receptor involved in cell signaling and is known to be a negative regulator of T-cell activation (reviewed in reference 30); however, very high doses of CTLA4 (50 μg) are required to completely suppress the immune response in mice (23). DNA vaccines producing CTLA4 fusion proteins have previously been reported to enhance antibody titers in mice and sheep (6, 11, 13, 22, 29). However, not all cases of the use of CTLA4 fusion vaccines have resulted in enhanced cellular responses (11), as was found in the present experiment. Overall, although the reason underlying the poor responsiveness induced by the CTLA4 library is not clear, the result shows that this vector is not useful for delivering malaria ELI vaccines in mice.
Gene gun vaccination with the genomic libraries did not produce humoral responses detectable upon ELISA analysis of sera, presumably due to the diverse repertoire of P. chabaudi adami sequences delivered in the libraries, as seen previously (31). Therefore, another method, opsonization, was used to establish the presence of antibody after vaccination and to further explore protective mechanisms mediated by genomic vaccination. Vaccination with all the genomic libraries constructed produced sera with significantly enhanced uptake of IRBCs by macrophages in vitro compared with that of IRBCs incubated with sera from empty vector-vaccinated control animals. In P. chabaudi infections, it has been shown that the antibody-mediated immune response is directed primarily to epitopes on the surface of IRBCs, with these antibodies enhancing phagocytosis and subsequent destruction of IRBCs in vitro (25, 26). In infected mice, IgG2a and IgG3 antibodies are dominant during ascending primary parasitemia and help to promote phagocytosis and opsonization of IRBCs (reviewed in reference 37). Opsonizing activity found in sera from immune individuals has also been associated with protection against P. falciparum (15). Our results show that ELI is able to induce opsonizing antibodies that may contribute to the efficacy of the VR1020/30K and MCP-3/30K vaccines.
The MCP-3 and CTLA4 vectors were used in an attempt to enhance the efficacy of genomic vaccination above the levels obtained by using the secretory VR1020 expression vector (31), since this enhancement would have made subsequent devolution of the libraries an easier task. Recently, we have shown that i.d. DNA vaccination with MSP4/5 fused to MCP-3 can significantly enhance survival in mice after challenge with blood-stage P. chabaudi adami, although there was no significant reduction in peak parasitemia compared to that in control animals (29). This construct also produced significant increases in antibody compared to a VR1020/MSP4/5 construct. Comparison of the efficacies of the VR1020/30K (secretory), MCP-3/30K (chemoattractant), and CTLA4/30K (lymph node-targeting) libraries emphasized the need for cellular as well as humoral responses to protect against lethal P. chabaudi adami challenge. However, vaccination with the MCP-3/30K library did not significantly enhance any of the parameters measured (protection, splenocyte proliferation, opsonization, and IFN-γ and IL-4 production) compared to vaccination with the VR1020/30K library. The use of MCP-3 as a fusion partner was not detrimental to the library's protective efficacy, but this was not the case when mice were vaccinated with the CTLA4/30K library. Similarly, i.d. DNA vaccination with MSP4/5 fused to CTLA4 did not protect mice after challenge with blood-stage P. chabaudi adami (29). Clearly, the choice of fusion partner is important when utilizing targeting vectors in DNA vaccine studies. The results of the present study demonstrate that standard secretory vectors are sufficient to promote protective immune responses by using a malarial genomic library.
Priming of mice with the CTLA4/30K library did not promote significant splenocyte proliferation specific to P. chabaudi adami-IRBCs, which may have contributed to the lack of protection observed in the present study. Sera obtained after vaccination with CTLA4/30K did however promote phagocytosis of IRBCs in vitro. This suggests the possibility that CTLA4/30K vaccination primed the immune system with enough T- and B-cell epitopes to produce antibody and promote phagocytosis, but this response was not sufficient to protect mice from death.
The combination of both specific cellular immune responses and opsonization of IRBCs by macrophages are two possible mechanisms which may be responsible for the protection observed with the VR1020/30K and MCP-3/30K libraries. The detection of IFN-γ and IL-4 in splenocyte cultures of mice vaccinated with these two libraries suggests priming of Th1 and Th2 cell subsets within the spleen. With P. chabaudi it has been shown that splenic CD4+ T cells purified from immunologically intact mice during ascending parasitemia produce high levels of IFN-γ and IL-2 to limit infection, while IL-4 and IL-10 are produced by splenic CD4+ T cells during descending primary parasitemia (33). B-cell-deficient mice, however, are not able to mount a Th2 response sufficient to completely resolve the primary parasitemia (33). Acute P. chabaudi adami infections in B-cell-deficient mice are suppressed at the same rate as those in normal mice, but complete depletion of CD4+ T cells results in the inability to control parasite growth, emphasizing the importance of CD4+-T-cell activation during blood-stage infection (reviewed in reference 37).
With these results, a mechanism by which ELI might induce a protective response has been established. The results suggest the possibility that the genomic libraries encode previously undiscovered combinations of malarial epitopes or whole antigens which are protective. We have now begun exploiting the synergistic properties of the VR1020/30K library, in combination with known antigens, by codelivery in bicistronic vectors. This has resulted in a significant reduction in peak parasitemia in challenged mice covaccinated with the 30K vaccine and known antigens, and this reduction is greater than that observed with the VR1020/30K library alone. Thus, the 30K vaccine may allow us to identify new unknown antigens that synergize with known antigens. The ELI vaccine may thus represent a method to mimic the multivalent nature of the acquired immune response seen in resistant malaria-exposed humans. Mimicking this response has been proposed to be the best approach to produce an effective malaria vaccine (12). Our laboratory is now in the process of dividing a subpool of the original 30,000 plasmids contained within the VR1020/30K library into groups based on predicted functions (epitope types and homology with known malaria parasite sequences) and sizes, and these pools are being evaluated in vaccination experiments in an attempt to further define the protective epitopes in the vaccine.
Acknowledgments
This work was supported by Monash University, the Australia Indonesia Medical Research Initiative, the Cooperative Research Centre for Vaccine Technology, McGill University, the McGill Institute of Parasitology, and the Canada Research Chair program. A. Rainczuk is a recipient of an Australian Postgraduate Award scholarship and a scholarship from the Cooperative Research Centre for Vaccine Technology. T. Spithill holds a Canada Research Chair in Immunoparasitology.
We thank H. Nandurkar for providing the mouse cDNA and A. Lew for providing the CTLA4 expression vector.
Editor: J. M. Mansfield
REFERENCES
- 1.Alberti, E., A. Acosta, M. E. Sarmiento, C. Hidalgo, T. Vidal, A. Fachado, L. Fonte, L. Izquierdo, J. F. Infante, C. M. Finlay, and G. Sierra. 1998. Specific cellular and humoral immune response in Balb/c mice immunized with an expression genomic library of Trypanosoma cruzi. Vaccine 16:608-612. [DOI] [PubMed] [Google Scholar]
- 2.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. S. M. Matthew, B. Pollock, and D. Pye. 1998. Immunization with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. [DOI] [PubMed] [Google Scholar]
- 3.Barry, M. A., W. C. Lai, and S. A. Johnston. 1995. Protection against mycoplasma infection using expression-library immunization. Nature 377:632-635. [DOI] [PubMed] [Google Scholar]
- 4.Biragyn, A., I. M. Belyakov, Y. H. Chow, D. S. Dimitrov, J. A. Berzofsky, and L. W. Kwak. 2002. DNA vaccines encoding human immunodeficiency virus-1 glycoprotein 120 fusions with proinflammatory chemoattractants induce systemic and mucosal immune responses. Blood 100:1153-1159. [DOI] [PubMed] [Google Scholar]
- 5.Biragyn, A., K. Tani, M. C. Grimm, S. Weeks, and L. W. Kwak. 1999. Genetic fusion of chemokines to a self tumor antigen induces protective, T-cell dependent antitumor immunity. Nat. Biotechnol. 17:253-258. [DOI] [PubMed] [Google Scholar]
- 6.Boyle, J. S., J. L. Brady, and A. M. Lew. 1998. Enhanced responses to a DNA vaccine encoding a fusion antigen that is directed to sites of immune induction. Nature 392:408-411. [DOI] [PubMed] [Google Scholar]
- 7.Brayton, K. A., S. W. Vogel, and B. A. Allsopp. 1998. Expression library immunization to identify protective antigens from Cowdria ruminantium. Ann. N. Y. Acad. Sci. 849:369-371. [DOI] [PubMed] [Google Scholar]
- 8.Carlton, J. M., S. V. Angiuoli, B. B. Suh, T. W. Kooij, M. Pertea, J. C. Silva, M. D. Ermolaeva, J. E. Allen, J. D. Selengut, H. L. Koo, J. D. Peterson, M. Pop, D. S. Kosack, M. F. Shumway, S. L. Bidwell, S. J. Shallom, S. E. van Aken, S. B. Riedmuller, T. V. Feldblyum, J. K. Cho, J. Quackenbush, M. Sedegah, A. Shoaibi, L. M. Cummings, L. Florens, J. R. Yates, J. D. Raine, R. E. Sinden, M. A. Harris, D. A. Cunningham, P. R. Preiser, L. W. Bergman, A. B. Vaidya, L. H. van Lin, C. J. Janse, A. P. Waters, H. O. Smith, O. R. White, S. L. Salzberg, J. C. Venter, C. M. Fraser, S. L. Hoffman, M. J. Gardner, and D. J. Carucci. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419:512-519. [DOI] [PubMed] [Google Scholar]
- 9.Crewther, P. E., M. L. S. M. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 64:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly, T. M., and C. A. Long. 1995. Humoral response to a carboxyl-terminal region of the merozoite surface protein-1 plays a predominant role in controlling blood-stage infection in rodent malaria. J. Immunol. 155:236-243. [PubMed] [Google Scholar]
- 11.Deliyannis, G., J. S. Boyle, J. L. Brady, L. E. Brown, and A. M. Lew. 2000. A fusion DNA vaccine that targets antigen-presenting cells increases protection from viral challenge. Proc. Natl. Acad. Sci. USA 97:6676-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doolan, D. L., and S. L. Hoffman. 2002. Nucleic acid vaccines against malaria. Chem. Immunol. 80:308-321. [DOI] [PubMed] [Google Scholar]
- 13.Drew, D. R., J. S. Boyle, A. M. Lew, M. W. Lightowlers, P. J. Chaplin, and R. A. Strugnell. 2001. The comparative efficacy of CTLA-4 and L-selectin targeted DNA vaccines in mice and sheep. Vaccine 19:4417-4428. [DOI] [PubMed] [Google Scholar]
- 14.Good, M. F. 2001. Towards a blood-stage vaccine for malaria: are we following all the leads? Nat. Rev. Immunol. 1:117-125. [DOI] [PubMed] [Google Scholar]
- 15.Groux, H., and J. Gysin. 1990. Opsonization as an effector mechanism in human protection against asexual blood stages of Plasmodium falciparum: functional role of IgG subclasses. Res. Immunol. 141:529-542. [DOI] [PubMed] [Google Scholar]
- 16.Hirunpetcharat, C., P. Vukovic, X. QinLiu, D. C. Kaslow, L. H. Miller, and M. F. Good. 1999. Absolute requirement for an active immune response involving B cells and Th cells in immunity to Plasmodium yoelii passively acquired with antibodies to the 19-kDa carboxyl-terminal fragment of merozoite surface protein-1. J. Immunol. 162:7309-7314. [PubMed] [Google Scholar]
- 17.Ho, A. S., S. H. Wei, A. L. Mui, A. Miyajima, and K. W. Moore. 1995. Functional regions of the mouse interleukin-10 receptor cytoplasmic domain. Mol. Cell. Biol. 15:5043-5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodder, A. N., P. E. Crewther, and R. F. Anders. 2001. Specificity of the protective antibody response to apical membrane antigen 1. Infect. Immun. 69:3286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman, S. L., and D. L. Doolan. 2000. Can malaria DNA vaccines on their own be as immunogenic and protective as prime-boost approaches to immunization? Dev. Biol. (Basel) 104:121-132. [PubMed] [Google Scholar]
- 20.Kumar, S., J. E. Epstein, T. L. Richie, F. K. Nkrumah, L. Soisson, D. J. Carucci, and S. L. Hoffman. 2002. A multilateral effort to develop DNA vaccines against falciparum malaria. Trends Parasitol. 18:129-135. [DOI] [PubMed] [Google Scholar]
- 21.Langhorne, J., S. J. Quin, and L. A. Sanni. 2002. Mouse models of blood-stage malaria infections: immune responses and cytokines involved in protection and pathology. Chem. Immunol. 80:204-228. [DOI] [PubMed] [Google Scholar]
- 22.Lew, A. M., J. L. Brady, and J. S. Boyle. 2000. Site-directed immune responses in DNA vaccines encoding ligand-antigen fusions. Vaccine 18:1681-1685. [DOI] [PubMed] [Google Scholar]
- 23.Linsley, P. S., P. M. Wallace, J. Johnson, M. G. Gibson, J. L. Greene, J. A. Ledbetter, C. Singh, and M. A. Tepper. 1992. Immunosuppression in vivo by a soluble form of the CTLA-4 T cell activation molecule. Science 257:792-795. [DOI] [PubMed] [Google Scholar]
- 24.Melby, P. C., G. B. Ogden, H. A. Flores, W. Zhao, C. Geldmacher, N. M. Biediger, S. K. Ahuja, J. Uranga, and M. Melendez. 2000. Identification of vaccine candidates for experimental visceral leishmaniasis by immunization with sequential fractions of a cDNA expression library. Infect. Immun. 68:5595-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mota, M. M., K. N. Brown, V. E. Do Rosario, A. A. Holder, and W. Jarra. 2001. Antibody recognition of rodent malaria parasite antigens exposed at the infected erythrocyte surface: specificity of immunity generated in hyperimmune mice. Infect. Immun. 69:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mota, M. M., K. N. Brown, A. A. Holder, and W. Jarra. 1998. Acute Plasmodium chabaudi chabaudi malaria infection induces antibodies which bind to the surfaces of parasitized erythrocytes and promote their phagocytosis by macrophages in vitro. Infect. Immun. 66:4080-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mumper, R. J., and H. C. Ledebur, Jr. 2001. Dendritic cell delivery of plasmid DNA. Mol. Biotechnol. 19:79-95. [DOI] [PubMed] [Google Scholar]
- 28.Piedrafita, D., D. Xu, D. Hunter, R. A. Harrison, and F. Y. Liew. 1999. Protective immune responses induced by vaccination with an expression genomic library of Leishmania major. J. Immunol. 163:1467-1472. [PubMed] [Google Scholar]
- 29.Rainczuk, A., P. M. Smooker, L. Kedzierski, C. G. Black, R. L. Coppel, and T. W. Spithill. 2003. The protective efficacy of MSP4/5 against lethal P. chabaudi adami challenge is dependent on the type of DNA vaccine vector and vaccination protocol. Vaccine 21:3030-3042. [DOI] [PubMed]
- 30.Sansom, D. M. 2000. CD28, CTLA-4 and their ligands: who does what to whom? Immunology 101:169-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smooker, P. M., Y. Y. Setiady, A. Rainczuk, and T. W. Spithill. 2000. Expression library immunization protects mice against a challenge with virulent rodent malaria. Vaccine 18:2533-2540. [DOI] [PubMed] [Google Scholar]
- 32.Sykes, K. F., M. G. Lewis, B. Squires, and S. A. Johnston. 2002. Evaluation of SIV library vaccines with genetic cytokines in a macaque challenge. Vaccine 20:2382-2395. [DOI] [PubMed] [Google Scholar]
- 33.Taylor-Robinson, A. W., and R. S. Phillips. 1994. B cells are required for the switch from Th1- to Th2-regulated immune responses to Plasmodium chabaudi chabaudi infection. Infect. Immun. 62:2490-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unkeless, J. C., Z. Shen, C. W. Lin, and E. DeBeus. 1995. Function of human Fc gamma RIIA and Fc gamma RIIIB. Semin. Immunol. 7:37-44. [DOI] [PubMed] [Google Scholar]
- 35.Warmerdam, P. A., P. W. Parren, A. Vlug, L. A. Aarden, J. G. van de Winkel, and P. J. Capel. 1992. Polymorphism of the human Fc gamma receptor II (CD32): molecular basis and functional aspects. Immunobiology 185:175-182. [DOI] [PubMed] [Google Scholar]
- 36.Weidanz, W. P., J. R. Kemp, J. M. Batchelder, F. K. Cigel, M. Sandor, and H. C. Heyde. 1999. Plasticity of immune responses suppressing parasitemia during acute Plasmodium chabaudi malaria. J. Immunol. 162:7383-7388. [PubMed] [Google Scholar]
- 37.Wipasa, J., S. Elliott, H. Xu, and M. F. Good. 2002. Immunity to asexual blood stage malaria and vaccine approaches. Immunol. Cell Biol. 80:401-414. [DOI] [PubMed] [Google Scholar]
- 38.Wipasa, J., C. Hirunpetcharat, Y. Mahakunkijcharoen, H. Xu, S. Elliott, and M. F. Good. 2002. Identification of T cell epitopes on the 33-kDa fragment of Plasmodium yoelii merozoite surface protein 1 and their antibody-independent protective role in immunity to blood stage malaria. J. Immunol. 169:944-951. [DOI] [PubMed] [Google Scholar]
- 39.Wipasa, J., H. Xu, M. Makobongo, M. Gatton, A. Stowers, and M. F. Good. 2002. Nature and specificity of the required protective immune response that develops postchallenge in mice vaccinated with the 19-kilodalton fragment of Plasmodium yoelii merozoite surface protein 1. Infect. Immun. 70:6013-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, H., A. N. Hodder, H. Yan, P. E. Crewther, R. F. Anders, and M. F. Good. 2000. CD4+ T cells acting independently of antibody contribute to protective immunity to Plasmodium chabaudi infection after apical membrane antigen 1 immunization. J. Immunol. 165:389-396. [DOI] [PubMed] [Google Scholar]