Abstract
The virulence factor Mip (macrophage infectivity potentiator) contributes to the intracellular survival of Legionella pneumophila, the causative agent of Legionnaires' disease. The protein consists of two domains that are connected via a very long α-helix (A. Riboldi-Tunnicliffe et al., Nat. Struct. Biol. 8:779-783, 2001). The fold of the C-terminal domain (residues 100 to 213) is closely related to human FK506-binding protein (FKBP12), and like FKBP12, Mip exhibits peptidylprolyl cis/trans isomerase (PPIase) activity. The α-helical N-terminal domain is responsible for the formation of very stable Mip homodimers. In order to determine the importance of the homodimeric state of Mip for its biochemical activities and for infectivity of Legionella, a truncated, monomeric Mip variant [Mip(77-213)] was overexpressed in Escherichia coli and characterized biochemically. In vitro isomerase activity assays revealed that the altered protein exhibits full isomerase activity towards peptide substrates. However, the deletion resulted in a dramatic loss in the efficiency of refolding of reduced and carboxy-methylated RNase T1. By cis complementation of the Mip-negative mutant strain L. pneumophila JR32-2, we constructed the strain L. pneumophila JR32-2.4, which expresses an N-terminally truncated variant of Mip. Infection studies with these strains revealed that the N-terminal part and the dimerization of Mip but not its PPIase activity are necessary for full virulence in Acanthamoeba castellanii. Infection of guinea pigs showed that strains with dimerization-deficient Mip (JR32-2.4) or a very low PPIase activity (JR32-2.2) were significantly attenuated in the animal model. These results suggest a different role of the PPIase activity and the N-terminally mediated dimeric state of Mip in monocellular systems and during the infection of guinea pigs.
Legionella pneumophila, the etiological agent of Legionnaires' disease, is an environmental pathogen. In natural and man-made aquatic habitats legionellae multiply intracellularly within free-living protozoa (1, 2, 12). Human infection occurs by inhalation of aerosolized bacteria. During the colonization of the respiratory tract, Legionella enters and replicates within human alveolar macrophages and epithelial cells (6, 12, 33). In recent years, much progress has been made toward identifying specific virulence factors of Legionella. In addition to surface factors, secretion systems, iron acquisition determinants, and other factors, the Mip (macrophage infectivity potentiator) protein has been shown to contribute to survival of Legionella in different cell lines and to virulence in guinea pigs (4, 5, 48, 51).
The Legionella Mip is a basic 24-kDa protein (pI 9.8). It possesses an N-terminal signal sequence, which is cleaved off while the protein is transported through the cytoplasmic membrane (51), and finally the protein is found on the surface of Legionella cells (17). Mip exhibits a peptidylprolyl cis/trans isomerase (PPIase) activity and belongs to the enzyme family of FK506-binding proteins (FKBP) (10, 11, 42). Mip-like proteins or corresponding genes were also identified in other microorganisms, such as Chlamydia trachomatis (24), Chlamydia psittaci (40), Coxiella burnetii (30), Trypanosoma cruzi (31), and Escherichia coli (18). The addition of PPIase inhibitors such as FK506 or rapamycin to C. trachomatis and T. cruzi resulted in reduced infectivity of these organisms (25, 31). Mip-negative mutants of Legionella were approximately 10- to 100-fold less infective for Acanthamoeba castellanii and human mononuclear phagocytes than were their isogenic Mip-positive parental strains (51). Mip-negative mutants are also less infective for Hartmannella vermiformis and lung epithelial cells (5, 6). Site-specific Legionella mutants with strongly reduced PPIase activity exhibited wild-type growth rates in cell culture assays. However, it could not be determined whether the residual PPIase activity was sufficient for the virulent phenotype of the mutants (51).
Comparison of the primary structures showed that Mip-like proteins consist of a C-terminal FKBP-homologous domain and an N-terminal extension of 100 to 150 amino acid residues (43). Cross-linking experiments with dimethyl pimelimidate (DMP) revealed that Legionella Mip forms homodimers both in solution and on the bacterial surface (43, 44). In this respect the Legionella protein resembles FKBP22 and FkpA from E. coli, which were also shown to form dimers in solution (35, 36). More recently, the 2.4-Å crystal structure of the Mip protein from L. pneumophila Philadelphia 1 was determined (39). This study revealed that the Mip monomer consists of an N-terminal dimerization module, a very long connecting α-helix (α3), and a C-terminal PPIase domain. Formation of the nonglobular, V-shaped homodimer is mediated by an unusual four-helix bundle, involving two helices from the N-terminal domain of each monomer.
The exact function of Legionella Mip in vivo remains to be established. The hypothesis that the PPIase activity of Mip contributes to virulence is still justified since the residual PPIase activity of site-specifically altered Mip proteins may be sufficient to induce functional effects in eukaryotic cells (51). It has also to be considered that the enzymatic activities observed in vitro do not reflect the activity towards the putative in vivo substrate. Furthermore, it was recently demonstrated that specific monoclonal antibodies which block the PPIase active site of Mip in vitro reduced the infectivity of Legionella in A. castellanii and U937 cells (J. H. Helbig, unpublished data).
With the determination of the three-dimensional structure of Legionella Mip, a more rational starting point for functional analysis of this protein has been established. In this respect, two features of Mip appear to be prominent: (i) the N-terminal dimerization domain and (ii) the PPIase activity which is located in the C-terminal domain. Since the role of the homodimeric state of Legionella Mip is still unclear, we constructed and analyzed N-terminally truncated, monomeric Mip variants and tested their enzymatic properties. In addition, we studied the infection of A. castellanii and guinea pigs with these variants and previously established PPIase mutants.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The E. coli K-12 strain DH5α (14) and E. coli SY327 λpir (29) were used for transformation and propagation of recombinant plasmids. These strains were grown in Luria broth or on Luria broth agar plates (41). The L. pneumophila and the Mip-expressing recombinant E. coli strains used in this study are listed in Table 1. Solid medium for growth of L. pneumophila was ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered charcoal yeast extract medium (ABCYE) (pH 6.9), essentially as described previously (9). For broth culture, L. pneumophila was grown in ACES-buffered yeast extract broth (1% yeast), supplemented with 0.025% ferric pyrophosphate and 0.04% l-cysteine, at 37°C. For selection of recombinant bacteria, antibiotics were used at the indicated final concentrations: E. coli, 25 μg of chloramphenicol/ml and 50 to 100 μg of ampicillin/ml; L. pneumophila, 5 to 10 μg of chloramphenicol/ml and 80 μg of gentamicin/ml.
TABLE 1.
Mip expression of Legionella and recombinant E. coli strains
| Strain | Mip expression | Relative PPIase activitya (%) | Reference |
|---|---|---|---|
| L. pneumophila | |||
| Philadelphia 1 JR32 | Wild-type Mip (LpFKBP25) | 100 | 51 |
| Philadelphia 1 JR32-2 | Mip negative | 0.0 | 51 |
| Philadelphia 1 JR32-2.1 | Mutant complemented in cis with wild-type Mip (LpFKBP25) | 100 | 51 |
| Philadelphia 1 JR32-2.2 | Site-specifically altered Mip [FKBP25 (Y185A)] | 2.0 | 51 |
| Philadelphia 1 JR32-2.3 | Site-specifically altered Mip [FKBP25 (D142L)] | 6.2 | 51 |
| Philadelphia 1 JR32-2.4 | N-terminally truncated monomeric Mip [LpFKBP25(−20-3,80-213)] | 100 | This study |
| E. coli DH5α/pJMFe991 | N-terminally truncated monomeric Mip [Mip(77-213)] | 100 | This study |
Recombinant DNA techniques and plasmids.
Standard methods were used for recombinant DNA techniques (41). Transformation was performed by electroporation with a Bio-Rad Gene Pulser (Bio-Rad, Munich, Germany). E. coli cells were pulsed with the pulse controller set at 2.0 kV, 25 mF, and 200 Ω of resistance, and Legionella cells were pulsed with the pulse controller set at 2.3 kV, 25 mF, and 100 Ω of resistance (7).
The Legionella-specific mip promoter and the mip gene sequence were amplified in two fragments from the chromosome of L. pneumophila JR32. Standard PCRs were carried out using a Thermocycler apparatus (Progene Techne, Cambridge, United Kingdom). The mip promoter sequence including the first 3 amino acids of the mature Mip protein was amplified using the following forward and reverse primers: mip5′(XbaI), 5′ ATTTCTAGAACTCAGTTGCTG 3′, and mip3′(BglII), 5′ TGAGATCTGTTGCAGCCATTGCTG 3′ (8). For deletion of amino acid residues 4 to 79 of the mature Mip protein, the primers mip5′(BglII), 5′ TTAGATCTGAAAGCGGATGAA 3′, and mip3′(SacI), 5′ TTAGAGCTCCCGTCGCAAGCACTG 3′, were used. The obtained fragments were subcloned into pUC18 (Pharmacia, Freiburg, Germany), which resulted in the plasmids pJF1 and pJF2, respectively. Isolation of the fragments after digestion with XbaI, SacI, and BglII; subsequent ligation with T4 DNA ligase (GIBCO BRL, Eggenstein, Germany); and integration into the corresponding sites (XbaI and SacI) of the vector pBC KS(+) (Stratagene, La Jolla, Calif.) resulted in plasmid pJFM2. The correct reading frame of the mip fragment was confirmed by sequencing. By using the restriction enzymes XbaI and SacI, a 1.5-kb fragment of pJFM2 was subcloned into the pir-dependent shuttle vector pMSS704-1 (27), and the resulting plasmid pRK105 was used for chromosomal integration into L. pneumophila JR32-2. The mip promoter and the entire mip gene were amplified by using the primers Mip5′ 375, 5′ CTTGAGCTCATGGAGGCAGGATC 3′, and Mip3′ 1540, 5′ TAAGATCTTTTTTTCACTGAAATTAAGTG 3′. The obtained fragment was subcloned into pGEM-Teasy (Promega, Heidelberg, Germany), which resulted in plasmid pRK70.
In order to overproduce the truncated Mip(77-213) protein, the sequence coding for the polypeptide starting with Phe-77 was amplified by using the following forward and reverse primers: mip5′(NcoI), 5′ GCCATGGTCAATAAGAAAGCGGATGAA 3′, and mip3′(SacI), 5′ TTAGAGCTCCCGTCGCAAGCACTG 3′. The PCR product was purified using the High Pure PCR product purification kit (Boehringer, Mannheim, Germany) and subcloned into pUC18 with the SureClone kit (Pharmacia). The 720-bp fragment was isolated after digestion with NcoI and SacI and cloned into the high-level expression vector pTrc99 (Pharmacia), which resulted in plasmid pJMFe991. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis E. coli DH5α/pJMFe991 was grown to logarithmic phase (optical density at 600 nm of 0.5) and induced with IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) for 4 h.
Southern blot analysis.
The correct integration of shuttle vector pRK105 into the chromosome of the mip-negative strain L. pneumophila JR32-2 was confirmed by Southern blot analysis. Purification of chromosomal DNA was performed according to the manufacturer's protocol with Quick Prep columns (Qiagen, Hilden, Germany). The chromosomal DNA was digested with BamHI and ClaI and transferred onto nylon membranes after gel electrophoresis. Chromosomal DNA of strain L. pneumophila JR32 was used as template to amplify the mip-specific gene probe by using the PCR primers Mip2 92, 5′ ATTCTAGAAACTCAGTTGCTG 3′, and Mip3 912, 5′ GTGAGATCTGTTGCAGCCATTGCT 3′. The resulting 800-bp probe was labeled and detected by using the enhanced chemiluminescence detection kit (Amersham, Little Chalfont, Buckinghamshire, United Kingdom).
Purification and limited proteolysis of Mip.
Recombinant L. pneumophila Mip protein was purified as described previously (23). For purification of Mip(77-213), E. coli DH5α cells carrying the plasmid pJMFe991 were cultivated, harvested, ruptured with a French press, and centrifuged as described for the purification of wild-type Mip. The resulting supernatant was applied to a Fractogel EMD DEAE-650(M) column (2.5 by 20 cm), equilibrated with 5 mM Tricine buffer (pH 8.0). The Mip(77-213) protein passed the column unbound and was applied to a VivaSpin cell. The flowthrough of the VivaSpin cell was pooled and applied to a Fractogel EMD SO−3 650(M) column (1 by 6 cm) equilibrated with 5 mM Tricine buffer (pH 8.0). Fractions containing Mip(77-213) protein were obtained by running a linear gradient from 0 to 1 M NaCl in 100 ml of 5 mM Tricine buffer (pH 8.0). The identity of the purified protein was analyzed using electrospray ionization mass spectrometry (VG Bio-Q; Fisons Instruments, Manchester, United Kingdom) and automated N-terminal protein sequencing (476A sequencer; Applied Biosystems, Weiterstadt, Germany).
For limited proteolysis, 11 μM Mip was incubated with 30 μM α-chymotrypsin (Merck, Darmstadt, Germany) in 35 mM HEPES-NaOH (pH 7.8) at 25°C. After different incubation times, aliquots were removed and the proteolysis was stopped by the addition of 1 mM phenylmethylsulfonyl fluoride. Samples were separated on SDS-17.5% polyacrylamide gels and analyzed by Western blotting (22). The L. pneumophila Mip-specific monoclonal antibody 22/1 (16) was used for protein detection. Proteolysis samples were blotted onto a Selex-20 membrane (Schleicher & Schuell, Dassel, Germany) (47), and the N-terminal amino acid sequences of the fragments were determined by using an Applied Biosystems 476A sequencer. The molecular masses of the proteins were determined by using size-exclusion chromatography (Superdex 75 column; 1 by 30 cm) (Pharmacia) and reversed-phase high-pressure liquid chromatography (33). Matrix-assisted laser desorption ionization-time of flight mass spectrometry was performed as described previously (49).
Preparation of periplasmic proteins from E. coli and L. pneumophila.
E. coli and L. pneumophila cells grown to logarithmic phase were harvested by centrifugation (3,000 × g for 10 min) and washed once with ice-cold water. After centrifugation, the cells were resuspended in water and adjusted to an optical density at 600 nm of 1. The cell suspension was centrifuged (8,000 × g for 5 min) and resuspended in 1 ml of ice-cold phosphate-buffered saline buffer. After harvesting, the cells were resuspended in 150 μl of 10 mM Tris-HCl, pH 7.5, containing 20% sucrose and 5 μl of a 0.5 M EDTA stock solution. The sample was incubated on ice (10 min) and pelleted by centrifugation (20,000 × g for 2 min). The resulting pellet was resuspended in 100 μl of a 25 mM MgCl2 solution (10 min) and centrifuged again (20,000 × g for 2 min). After this step, the periplasmic proteins were present in the supernatant. The periplasmic fraction was not contaminated with cytoplasmic proteins as confirmed by Western blot analysis with antibodies (Pap Productions, Herbertshausen, Germany) against cytoplasmically localized cyclophilin. The same fraction showed a positive Western blot signal when an antibody to periplasmically localized cyclophilin was used. All preparation and incubation steps were carried out on ice, and centrifugations were performed at 4°C.
Analysis of the oligomeric state of Mip.
In order to analyze the oligomeric state of Mip, different concentrations of Mip(77-213) (0.5 to 40 μM) were incubated with a 20 mM concentration of the cross-linking reagent DMP in 0.1 M sodium phosphate (pH 8.0) for 1 h at 25°C (43). The reaction was stopped by the addition of SDS sample buffer and boiling. Samples were analyzed by SDS-PAGE and Coomassie blue staining. Wild-type Mip (3 μM) was used as control. Gel filtration was carried out by using a Superdex-75 column (Superdex-75 column; 0.32 by 30 cm) and the SMART system (Pharmacia) with 10 mM HEPES-NaOH, pH 7.8 (150 mM KCl, 1.5 mM MgCl2), and a flow rate of 40 μl/min. The column was calibrated using molecular mass markers in the range of 12.5 to 68 kDa (Boehringer).
Spectroscopic methods.
A Hitachi F4010 fluorescence spectrometer and an HP 8452A spectrophotometer were used for optical measurements. The concentration of reduced and carboxy-methylated RNase T1 (RCM-T1) was determined spectrophotometrically by using an absorption coefficient of ɛ280 = 21,060 M−1 cm−1 (13). For the Mip protein and the Mip(77-213) fragment, ɛ280 values of 17,780 and 16,500 M−1 cm−1 were calculated, respectively. Far-UV circular dichroism (CD) spectra were recorded using the J-710 spectropolarimeter (JASCO, Tokyo, Japan). The spectra were recorded 10 times. Data were analyzed by using software provided by the instrument manufacturer.
PPIase activity measurements.
PPIase activity of the proteins was measured using the protease-coupled (11, 35) or the protease-free (19) assay. Different tetrapeptides (Bachem, Weil am Rhein, Germany) with the amino acid sequence Suc-Ala-Xaa-Pro-Phe-4NA were used as substrates. Prior to the inhibition measurements, the enzyme was coincubated with rapamycin (Sigma, Taufkirchen, Germany) (dissolved in 50% ethanol) for 15 min. The final concentration of ethanol was kept constant and below 1%. To obtain the inhibition constant Ki (32), the data were analyzed according to the tight-binding model for competitive inhibition. All measurements were performed twice. The results did not differ more than 5%.
Protein folding experiments.
In order to measure the enzyme kinetics of the Mip protein and the Mip(77-213) fragment, RCM-T1 was used as substrate protein (28, 47). Unfolded RCM-T1 substrate was stored in 0.1 M Tris-HCl, pH 8.0, at −80°C. Refolding of RCM-T1 was initiated by dilution of the protein in buffer containing high salt concentrations (0.1 M Tris-HCl [pH 8.0], 2 M NaCl) and suitable concentrations of the Mip protein or the Mip(77-213) fragment at 15°C. The final concentration of RCM-T1 varied from 0.1 × 10−7 M up to 3.44 × 10−7 M. The folding reaction was analyzed by the increase of fluorescence of the protein at 320 nm after excitation at 268 nm. The small contribution of the Mip protein to the fluorescence was subtracted from the measured values in the individual experiments. Resulting progress curves of folding were analyzed by using the program DYNAFIT (21). A simple Michaelis-Menten kinetic in combination with a kinetic model for the uncatalyzed refolding of RCM-T1 was used to describe the obtained data. The rate constant of the uncatalyzed folding reaction was measured in the absence of the Mip protein. The determined value of k0 = 0.0016 s−1 which was independent from the concentration of RCM-T1 was used as a constant during the fitting process.
Infection of A. castellanii and intratracheal infection of guinea pigs.
The infection of A. castellanii was carried out as described previously (20, 51), except that L. pneumophila was added to the infection wells at a multiplicity of infection of 10. After 0, 3, 24, and 48 h, intracellular growth of L. pneumophila was examined by plating serial dilutions on ABCYE agar plates.
Guinea pigs were used as an animal model for experimental legionellosis as described previously (26). The 3- to 6-month-old male guinea pigs (600 to 800 g) were obtained from Charles River (Sulzfeld, Germany). Infection was initiated by intratracheal injection of 300 μl of a physiological sodium chloride suspension containing 107 bacteria. Forty-eight hours after infection, animals were killed and the lungs were removed aseptically. Lung tissue was homogenized in 0.9% saline, and aliquots of serial dilutions of the lung suspensions were plated on ABCYE agar. For determination of recovered Legionella, CFU were counted after 72 h of incubation. Mean values of bacterial concentrations recovered from guinea pig lungs or 48 h after infection of amoebae were analyzed by using one-way analysis of variance and the Minitab software. Differences of mean values from six different strains used for infection were compared by Fisher's multiple range test with a confidence interval set to 95%. The given P values were obtained from a nonpaired two-sample t test. In the case of large differences in the variance of means, data were transformed into their logarithms before analysis of variance. Differences were considered significant if P was <0.05.
RESULTS
Analysis of the chymotryptic digest of Mip.
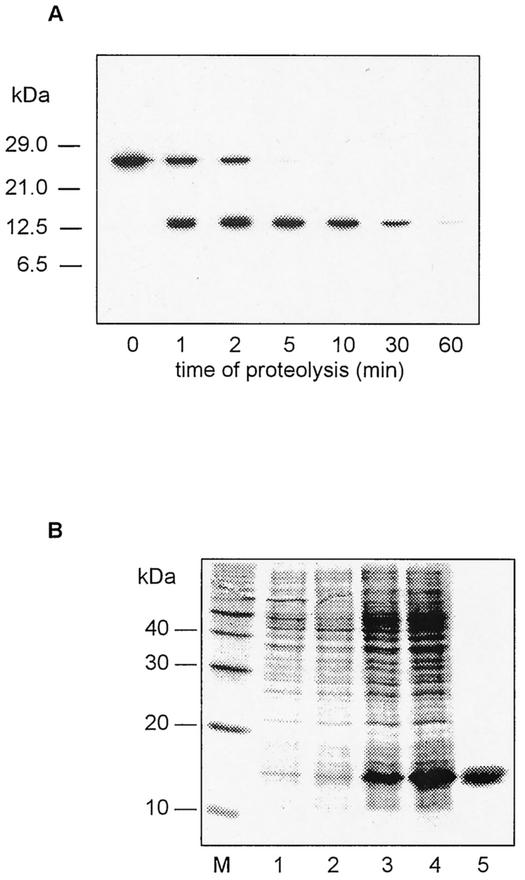
Limited proteolysis of the L. pneumophila Mip protein was one approach used to create stable fragments and characterize their enzymatic and biological activities. Analysis of the chymotryptic digest of Mip by Western blotting revealed a major fragment of about 14 kDa which disappeared after 60 min of proteolysis (Fig. 1A). The N-terminal sequence of this fragment was Asn-Lys-Lys-Ala-Asp. This indicates that the cleavage occurred after phenylalanine at position 77 of the mature Mip protein. Matrix-assisted laser desorption ionization-time of flight mass spectrometry confirmed the molecular mass of 14,526.8 Da, which is in good agreement with the theoretical value of 14,502.7 Da calculated for the Mip fragment (residues 78 to 213). These data indicate that the obtained fragment comprises the C-terminal FKBP-homologous domain and about half of the long α3 helix connecting the two domains. The obtained information was used for the construction of an N-terminal deletion variant of Mip.
FIG. 1.
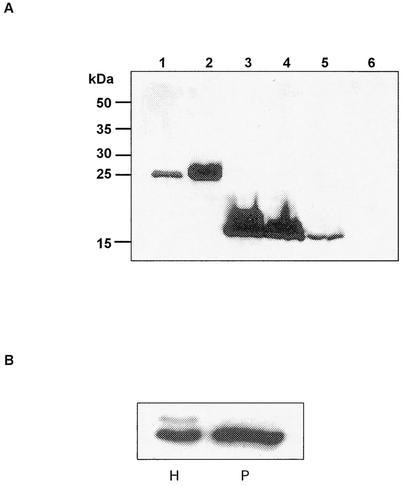
(A) Limited proteolysis of L. pneumophila Mip. The chymotryptic digestion of Mip was stopped after different incubation times. Western blot analysis shows bands of the intact Mip (24 kDa) and a chymotryptic Mip fragment (14 kDa). (B) Coomassie blue-stained SDS-PAGE of crude extracts of E. coli DH5α expressing the Mip(77-213) fragment. Lanes 1 to 4, E. coli DH5α/pJMFe991 before (lanes 1 and 2) and after (lanes 3 and 4) induction with IPTG; lane 5, homogeneous Mip(77-213) fragment after the last chromatographic purification step; lane M, molecular mass markers.
Overexpression, purification, and biochemical characterization of the Mip(77-213) fragment.
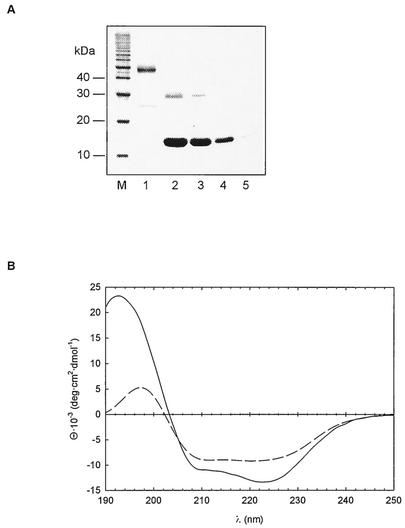
In order to further characterize the C-terminal fragment obtained by limited proteolysis, we expressed an N-terminal deletion variant of Mip in E. coli DH5α. The expressed protein Mip(77-213), which lacked amino acid residues 1 to 76, was purified to homogeneity (Fig. 1B). The structure of Mip(77-213) was analyzed by gel filtration, cross-linking experiments, and far-UV CD spectroscopy. In gel filtration experiments, the dimeric wild-type Mip exhibited a molecular mass of 62 kDa (41). For Mip(77-213), a value of 17 kDa was obtained. This value corresponds well to the molecular mass (14,650 Da) calculated from the deduced amino acid sequence (data not shown).
As reported previously, wild-type Mip dimers can be cross-linked by DMP (43). Comparable concentrations of Mip(77-213) (5 μM) and DMP did not result in a dimeric protein (Fig. 2A). This indicates that Mip(77-213) is monomeric in solution. At higher concentrations (25 to 40 μM), Mip(77-213) was partly cross-linked and exhibited weak bands by SDS-PAGE. This result agrees with observations that we made in similar cross-linking experiments with monomeric human FKBP12 (data not shown) and suggests an unspecific interaction of the FKBP domain at high protein concentrations.
FIG. 2.
(A) Analysis of the oligomeric state of Mip. A 3 μM concentration of wild-type Mip (lane 1) and 40 (lane 2), 25 (lane 3), 5 (lane 4), and 0.5 (lane 5) μM concentrations of Mip(77-213) were cross-linked by DMP and analyzed by SDS-PAGE and Coomassie blue staining. (B) Far-UV CD spectra of wild-type Mip (solid line) and Mip(77-213) (dashed line). The data are shown as mean residue ellipticities versus wavelengths. Spectra were recorded at 20°C in 10 mM sodium phosphate (pH 7.5).
The positive band at 192 nm and the negative bands at 206 and 222 nm in the far-UV CD spectrum of wild-type Mip protein indicated the presence of α-helices. The contribution of β-structures to the CD spectrum can be estimated from a positive band in the range of 196 nm and a negative band at 218 nm (50) (Fig. 2B). The CD spectrum of Mip(77-213) was in agreement with a secondary structure content different from that of wild-type Mip. From the weaker intensity of the positive band at 192 nm a reduced α-helical content of Mip(77-213) can be deduced. The spectrum also differed significantly from that observed for human FKBP12 (hFKBP12) (49). These data suggest that the residues (77 to 98) remaining of the long α3 helix linking the two domains in the native Mip protein retained their conformation in the fragment.
The PPIase activity of Mip(77-213) was measured using the protease-coupled and protease-free assays. No significant differences between wild-type Mip and Mip(77-213) were detected (Table 1). Similarly, the determination of the inhibition constants Ki for the PPIase inhibitor rapamycin exhibited only slight differences for wild-type Mip (Ki = 15 ± 3 nM) and Mip(77-213) (Ki = 34 ± 5 nM).
Refolding of RCM-T1 by Mip and Mip(77-213).
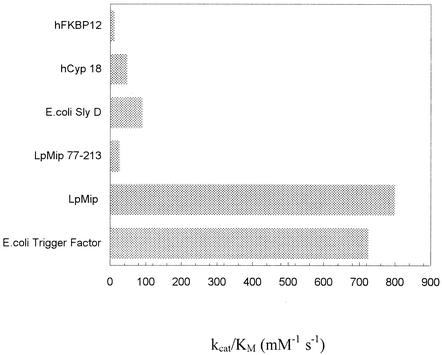
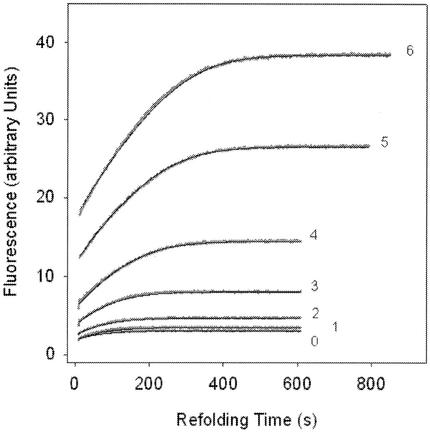
We applied the RCM-T1 refolding assay to elucidate whether the N-terminal half influences the catalytic properties of the Mip protein. By using different Mip concentrations (3.2 to 39 nM) and 0.02 μM RCM-T1, we were able to estimate a catalytic efficiency kcat/Km of 0.8 × 106 M−1 s−1 for wild-type Mip. This value is similar to the catalytic efficiency of the E. coli trigger factor and remarkably high compared to those of other previously analyzed PPIases such as hFKBP12 or hCyp18 (52) (Fig. 3). In contrast, Mip(77-213) exhibited a low kcat/Km value of 0.028 × 106 M−1 s−1. In order to analyze the folding efficiency of the enzyme, we determined kcat and Km separately by using a constant Mip concentration of 13 nM and RCM-T1 concentrations ranging from 0.1 × 10−7 to 3.44 × 10−7 M (Fig. 4). To dissect catalyzed and uncatalyzed folding, we also measured the refolding in the absence of Mip. The resulting values of the kinetic constants for the Mip-catalyzed folding, 0.095 s−1 for kcat and 75 nM for Km, are about 10-fold lower than those for the E. coli trigger factor (Table 2). The ratio of these two values (1.2 × 106 M−1 s−1) is consistent with the composite estimate for kcat/Km of 0.8 × 106 M−1 s−1. Since the substrate binding of Mip(77-213) is very weak, it was not possible to determine the corresponding kinetic constants. The data summarized in Table 2 confirm that the kinetics of Mip-catalyzed folding are adequately described by the Michaelis-Menten equation.
FIG. 3.
Specific activities of L. pneumophila Mip, Mip(77-213), and other previously analyzed PPIases (52) for the protein substrate RCM-T1. Measurements were performed in 0.1 M Tris-HCl-2 M NaCl (pH 8.0) at 15°C in the protein folding assay. All kcat/Km values were determined directly (52).
FIG. 4.
RCM-T1 refolding by Mip. The progress curves of refolding are shown for seven different concentrations of RCM-T1, 0.1 × 10−7 M (line 0), 0.21 × 10−7 M (line 1), 0.36 × 10−7 M (line 2), 0.43 × 10−7 M (line 3), 0.86 × 10−7 M (line 4), 1.72 × 10−7 M (line 5), and 3.44 × 10−7 M (line 6), in the presence of 13 nM L. pneumophila Mip protein. Measurements were performed in 0.1 M Tris-HCl-2 M NaCl (pH 8.0) at 15°C.
TABLE 2.
Kinetic constants for refolding of RCM-T1 by different PPIases
Expression of an N-terminally truncated Mip protein in L. pneumophila JR32-2.
In order to analyze the N-terminally truncated monomeric Mip in vivo, the Mip-negative strain JR32-2 was complemented with a Mip variant that lacks the amino acid residues 4 to 79 [designated LpFKBP25(−20-3,80-213)].
pJFM2 and pRK105 were introduced into E. coli DH5α and E coli SY327 λpir, respectively. pRK105 was used to complement the Mip-negative strain L. pneumophila JR32-2 in cis. The proper integration of the plasmid was confirmed by Southern blot analysis (data not shown). The expression of the truncated monomeric Mip protein in E. coli pJFM2, E. coli pRK105, and L. pneumophila JR32-2.4 was confirmed by Western blot analysis (Fig. 5A). The observed 15-kDa bands in the Western blot correspond with the predicted mass of the truncated Mip protein. E. coli carrying pRK70 (wild-type Mip), L. pneumophila JR32 (wild-type Mip), and L. pneumophila JR32-2 (Mip negative) were used as controls.
FIG. 5.
(A) Western blot analysis of E. coli and L. pneumophila strains expressing the 24-kDa wild-type Mip (LpFKBP25) and the 15-kDa truncated Mip [LpFKBP25(−20-3,80-213)]. The expression of the proteins from whole-cell lysate was detected by using the monoclonal antibody 22/1. Lane 1, L. pneumophila JR32 (Mip+); lane 2, E. coli pRK70 (Mip+); lane 3, E. coli pJFM2 (Mip monomer); lane 4, E. coli pRK105 (Mip monomer); lane 5, L. pneumophila JR32-2.4 (Mip monomer); lane 6, L. pneumophila JR32-2 (Mip−). (B) Western blot analysis of whole-cell lysate (H) and the periplasmic compartment (P) of L. pneumophila strains expressing the truncated Mip protein [LpFKBP25(−20-3,80-213)]. The bands detected in the whole-cell lysate correspond to the unprocessed [LpFKBP25(−20-3,80-213)] and processed [LpFKBP25(80-213)] truncated Mip protein. Only the correctly processed [LpFKBP25(80-213)] truncated Mip protein was detectable in the periplasmic compartment.
To confirm the correct localization of the truncated Mip protein [LpFKBP25(−20-3,80-213)] within the bacterial cell, we analyzed periplasmic preparations and cell lysates of the complemented mutant strain L. pneumophila JR32-2.4. Western blot analysis with the monoclonal antibody 22/1 revealed two proteins in the whole-cell lysate (Fig. 5B). The molecular weight of the upper band corresponds to that of the unprocessed truncated Mip protein which contains a signal peptide. The lower band represents the processed truncated Mip protein. In the periplasmic preparation, only the processed truncated Mip protein was present. The same expression pattern was obtained for the expression of the truncated Mip protein in E. coli DH5α (data not shown). This indicates that a similar processing of the protein occurred. N-terminal automated protein sequencing from the processed and unprocessed protein exhibited the expected amino acid sequence (data not shown). Taken together, these results suggest that the processing and localization of LpFKBP25 and LpFKBP25(−20-3,80-213) are basically the same.
In vivo influence of the truncated Mip protein on intracellular multiplication in A. castellanii.
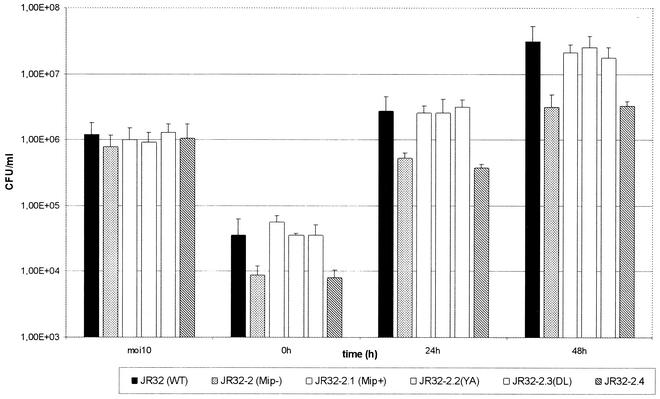
To examine the influence of the truncation of Mip on intracellular multiplication, cells of the natural host A. castellanii cells were infected with the L. pneumophila strains JR32 (wild-type Mip), JR32-2 (Mip-negative), JR32-2.1 (Mip-negative mutant complemented with wild-type Mip), JR32-2.2 (site-specifically altered Mip with 2% PPIase activity), JR32-2.3 (site-specifically altered Mip with 6.2% PPIase activity), and JR32-2.4 [Mip-negative mutant complemented with LpFKBP25(−20-3,80-213)], respectively. The results of the gentamicin infection assays (Fig. 6) revealed similarly reduced growth rates of strain JR32-2 and JR32-2.4. Both strains exhibited a 10-fold-lower CFU than that of the wild-type strain L. pneumophila JR32 (P < 0.02) and strain JR32-2.1 (P < 0.05). For JR32-2.2 and JR32-2.3, we observed wild-type growth rates which confirmed previous results (51). Infections of macrophage-like U937 cells additionally confirmed our findings (data not shown). Thus, the infection experiments suggest that the N-terminal part but not the PPIase activity is necessary for wild-type growth in monocellular host systems.
FIG. 6.
Intracellular replication of L. pneumophila JR32 (wild type, Mip+), JR32-2 (Mip−), JR32-2.1 (Mip+, complementation in cis), JR32-2.2 YA (Mip+; PPIase activity, 2%), JR32-2.3 DL (Mip+; PPIase activity, 6.2%), and JR32-2.4 (truncated monomer) in A. castellanii. The cells were infected at a multiplicity of infection of 10. CFU were determined in duplicate. Error bars indicate the standard deviations obtained from four independent experiments.
Survival of different Legionella Mip mutant strains in the guinea pig model.
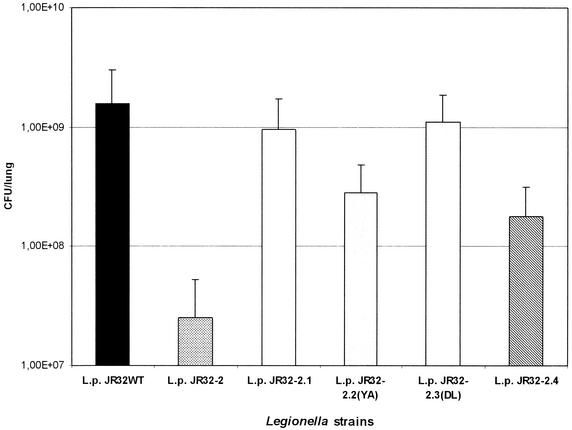
From the analysis of virulence factors of Legionella, it is known that the results with infections of different model systems may vary (15). Since only infected guinea pigs exhibit symptoms which closely resemble human disease, we analyzed the survival of the strains JR32, JR32-2, JR32-2.1, JR32-2.2, JR32-2.3, and JR32-2.4 in this experimental model for legionellosis (Fig. 7). The number of bacteria which could be recovered from lungs infected with the Mip-negative mutant strain JR32-2 was more than 60-fold reduced compared to that for the wild-type strain JR32 (P < 0.02). This indicates that the Mip-negative phenotype has a more dramatic effect in the animal model than in the monocellular system A. castellanii. The CFU determined for strain JR32-2.4 were found to be 10-fold reduced (P < 0.03). However, compared to the Mip-negative strain JR32-2, the truncated monomeric Mip-expressing strain showed a sevenfold-higher recovery rate (P < 0.03). This indicates that the N-terminal half, probably the N-terminally mediated dimerization, as well as the PPIase activity of Mip, is necessary to establish full virulence. This conclusion is further confirmed by the fact that strain JR32-2.2 exhibited a fivefold reduction in the animal model compared to the growth rate of the wild-type strain JR32 (P < 0.02). The site-specific mutant strain JR32-2.2 exhibits only a residual in vitro PPIase activity of 2.0% (51). Interestingly, the strain JR32-2.3 with 6.2% PPIase activity (51) was able to exhibit a higher growth than was strain JR32-2.2 (P < 0.04). Thus, the infection experiments with guinea pigs reveal that Legionella strains with N-terminally truncated Mip or very low PPIase activity were significantly attenuated.
FIG. 7.
Investigation of growth of L. pneumophila JR32 (wild type, Mip+), JR32-2 (Mip−), JR32-2.1 (Mip+, complementation in cis), JR32-2.2 YA (Mip+; isomerase activity, 2%), JR32-2.3 DL (Mip+; isomerase activity, 6.2%), and JR32-2.4 (truncated monomer) in the guinea pig animal model. The animals were infected intratracheally with 107 bacteria. The number of recovered bacteria (CFU) was determined 48 h after infection. CFU were determined in duplicate for three independent experiments for each strain.
DISCUSSION
Pathogenic bacteria that are able to invade and multiply in eukaryotic host cells often produce Mip-like proteins exhibiting an FK506-sensitive PPIase activity. However, the exact function of Mip proteins in vivo remains to be established. In order to approach this question, the structure of the L. pneumophila Mip dimer, both as the free enzyme and in complex with FK506, has recently been determined by X-ray crystallography (39). The crystal structure revealed that dimerization of the protein is mediated by an unusual four-helix bundle formed by two helices at the N terminus of each Mip monomer. The N-terminal domain is connected to the C-terminal, FKBP-like domain by the very long α3 helix (44 residues). FK506 was shown to bind to a hydrophobic site in the FKBP domain. The overall structure of FK506-bound Mip is similar to that of the free enzyme.
In the present study it was our aim to elucidate the functional organization of Mip. Characterization of the stable chymotryptic fragment of Mip showed that the long interdomain helix α3 of Mip had been cleaved between amino acid residues 77 and 78. CD-spectroscopic characterization of the respective Mip(77-213) fragment as expressed in E. coli indicated that the α-helical structure between residues 77 and 94 had been retained. Furthermore, it could be shown that the loss of the N-terminal dimerization domain resulted in a monomeric protein fragment. A similar monomeric fragment of the Mip-like protein FkpA from E. coli was previously obtained by tryptic digestion (3).
By using the protease-coupled and the protease-free assay, it was demonstrated that the PPIase activity of the Mip protein and the inhibition by rapamycin were not impaired by the N-terminal truncation. This suggests an independent action of the two PPIase active sites of the dimer. However, it cannot be excluded that both catalytic centers are necessary for a putative natural binding partner.
Differences between wild-type Mip and Mip(77-213) were observed in the RCM-T1 refolding assay. The catalytic efficiency of wild-type Mip was comparable to values obtained for the trigger factor and the Mip-like FkpA from E. coli (Table 2) (36, 52). The kcat/Km values of these enzymes are extremely high compared to those for other PPIases such as hCyp18 and hFKBP12. It has been shown that the high enzymatic folding activity of E. coli trigger factor in the RCM-T1 assay originates from a low Michaelis constant (Km) (45, 52). Similarly, our determination of the kinetic parameters kcat and Km revealed a tight binding of the protein substrate to L. pneumophila wild-type Mip.
The deletion of 76 N-terminal residues of Mip resulted in a dramatic loss of efficiency in refolding RCM-T1. This is indicated by a 28-fold-reduced kcat/Km value of Mip(77-213) compared to that for the wild-type Mip. The kcat/Km of Mip(77-213) corresponds to the value found for the hFKBP12 (kcat/Km = 1.2 × 10−4 M−1 s−1) (52), and it is likely that the reduced folding efficiency of Mip(77-213) originates in the loss of the dimeric state as well as of residues involved in substrate binding. In analogy, it has been shown that the high affinity of the E. coli trigger factor for unfolded protein substrates can be attributed to the N- and C-terminal domains which encompass the FKBP-like domain (45, 46, 52). Another example is the chaperone-like activity of E. coli FkpA, which requires the PPIase active site and the full-length structure of the protein during the refolding of the maltose-binding protein MalE31 (3, 37). We suggest that the protein binding site of L. pneumophila Mip during the catalysis of RCM-T1 refolding extends beyond the PPIase active site and involves much of the linker helix α3 and perhaps the N-terminal dimerization domain of the protein. In addition, the dimeric state of Mip may be necessary to keep two substrate-binding sites at the right distance.
In order to distinguish the in vivo influence of the PPIase activity from the dimerization of Mip, we analyzed the virulence of different Legionella mutants in Acanthamoeba cells and guinea pigs. In earlier studies, we had demonstrated that site-specifically altered Mip proteins with strongly reduced PPIase activity were still able to support the intracellular survival of Legionella in monocellular systems like Acanthamoeba, U937 cells, and blood monocytes (51). Our results presented here clearly demonstrated that the dimeric state or certain parts of the N-terminal half of Mip are essential for Legionella virulence in Acanthamoeba. Although the truncated Mip protein retained its PPIase activity towards peptide substrates, we found that the corresponding Legionella mutant JR32-2.4 exhibited intracellular growth rates similar to those of the Mip-negative strain JR32-2. This suggests either that the binding of the natural binding partner is insufficient or that the PPIase activity is dispensable. Since it was demonstrated recently that Legionella Mip can effectively substitute for the monomeric macrophage infectivity potentiator protein from T. cruzi (TcMIP) in invasion assays (34), a detailed structural comparison of the two proteins may provide clues to the identification of a common cellular target.
In guinea pig infections, it could be demonstrated for the first time that the PPIase activity and the full-length structure of Mip are crucial for full virulence of Legionella. The finding that the reduced PPIase activity of strain JR32-2.2 results in an attenuated phenotype in guinea pig infections additionally indicates the importance of the PPIase activity. Further support for the importance of the enzymatic activity comes from amino acid sequence comparisons. The Mip sequences from 35 Legionella species were 82 to 99% conserved at the amino acid level. A total conservation was found for amino acids which are known to be involved in PPIase activity (38).
Taken together, this work provides evidence for a different role of the PPIase activity of Mip in monocellular systems and during the infection of higher organisms. Based on the data observed in vitro and in guinea pigs, we propose that interaction of Mip with its putative substrate requires both the dimeric state of the protein and its PPIase activity. However, the exact function of the PPIase activity and the reason for the different outcomes in different host model systems remain a future challenge.
Acknowledgments
We thank Silke Killinger for technical assistance.
R.K. and J.F. contributed equally to the work.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG STE/3-2, SFB 604-B3), the Studienstiftung des Deutschen Volkes, and the Fonds der Chemischen Industrie.
Editor: S. H. E. Kaufmann
REFERENCES
- 1.Abu Kwaik, Y. 1998. Fatal attraction of mammalian cells to Legionella pneumophila. Mol. Microbiol. 30:689-696. [DOI] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y., L.-Y. Gao, B. J. Stone, C. Venkataraman, and O. S. Harb. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64:3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arie, J. P., N. Sassoon, and J. M. Betton. 2001. Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol. Microbiol. 39:199-210. [DOI] [PubMed] [Google Scholar]
- 4.Cianciotto, N. P., B. I. Eisenstein, C. H. Mody, and N. C. Engleberg. 1990. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J. Infect. Dis. 162:121-126. [DOI] [PubMed] [Google Scholar]
- 5.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cianciotto, N. P., J. K. Stamos, and D. W. Kamp. 1995. Infectivity of Legionella pneumophila mip mutant for alveolar epithelial cells. Curr. Microbiol. 30:247-250. [DOI] [PubMed] [Google Scholar]
- 7.Dietrich, C., K. Heuner, B. C. Brand, J. Hacker, and M. Steinert. 2001. Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 69:2116-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engleberg, N. C., C. Carter, D. R. Weber, N. P. Cianciotto, and B. I. Eisenstein. 1989. DNA sequence of mip, a Legionella pneumophila gene associated with macrophage infectivity. Infect. Immun. 57:1263-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10:437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer, G., H. Bang, B. Ludwig, K. Mann, and J. Hacker. 1992. Mip protein of Legionella pneumophila exhibits peptidyl-prolyl-cis/trans isomerase (PPIase) activity. Mol. Microbiol. 6:1375-1383. [DOI] [PubMed] [Google Scholar]
- 11.Fischer, G., H. Bang, and C. Mech. 1984. Determination of enzymatic catalysis for the cis-trans-isomerization of peptide binding in proline-containing peptides. Biomed. Biochim. Acta 43:1101-1111. (In German.) [PubMed]
- 12.Gao, L.-Y., B. J. Stone, J. K. Brieland, and Y. Abu Kwaik. 1998. Different fates of Legionella pneumophila pmi and mil mutants within human-derived macrophages and alveolar epithelial cells. Microb. Pathog. 25:291-306. [DOI] [PubMed] [Google Scholar]
- 13.Gill, S. C., and P. H. Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Harb, O. S., and Y. Abu Kwaik. 2000. Characterization of a macrophage-specific infectivity locus (milA) of Legionella pneumophila. Infect. Immun. 68:368-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helbig, J. H., B. Ludwig, P. C. Lück, A. Groh, W. Witzleb, and J. Hacker. 1995. Monoclonal antibodies to Legionella Mip proteins recognize genus- and species-specific epitopes. Clin. Diagn. Lab. Immunol. 2:160-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helbig, J. H., P. C. Lück, M. Steinert, E. Jacobs, and M. Witt. 2001. Immunolocalization of Mip protein of extracellularly and intracellularly grown Legionella pneumophila. Lett. Appl. Microbiol. 32:83-88. [DOI] [PubMed] [Google Scholar]
- 18.Horne, S. M., and K. D. Young. 1995. Escherichia coli and other species of the Enterobacteriaceae encode a protein similar to the family of Mip-like FK506-binding proteins. Arch. Microbiol. 163:357-365. [DOI] [PubMed] [Google Scholar]
- 19.Janowski, B., S. Wollner, M. Schutkowski, and G. Fischer. 1997. A protease-free assay for peptidyl prolyl cis/trans isomerases using standard peptide substrates. Anal. Biochem. 252:299-307. [DOI] [PubMed] [Google Scholar]
- 20.Köhler, R., A. Bubert, W. Goebel, M. Steinert, J. Hacker, and B. Bubert. 2000. Expression and use of the green fluorescent protein as a reporter system in Legionella pneumophila. Mol. Gen. Genet. 262:1060-1069. [DOI] [PubMed] [Google Scholar]
- 21.Kuzmic, P. 1996. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 237:260-273. [DOI] [PubMed] [Google Scholar]
- 22.Kyhse-Anderson, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10:203-209. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig, B., J. Rahfeld, B. Schmidt, K. Mann, E. Wintermeyer, G. Fischer, and J. Hacker. 1994. Characterization of Mip proteins of Legionella pneumophila. FEMS Microbiol. Lett. 118:23-30. [DOI] [PubMed] [Google Scholar]
- 24.Lundemose, A. G., S. Birkelund, S. J. Fey, P. M. Larsen, and G. Christiansen. 1991. Chlamydia trachomatis contains a protein similar to the Legionella pneumophila mip gene product. Mol. Microbiol. 5:109-115. [DOI] [PubMed] [Google Scholar]
- 25.Lundemose, A. G., J. E. Kay, and J. H. Pearce. 1993. Chlamydia trachomatis Mip-like protein has peptidyl-prolyl cis/trans isomerase activity that is inhibited by FK506 and rapamycin and is implicated in initiation of chlamydial infection. Mol. Microbiol. 7:777-783. [DOI] [PubMed] [Google Scholar]
- 26.Lüneberg, E., U. Zähringer, Y. A. Knirel, D. Steinmann, M. Hartmann, I. Steinmetz, M. Rohde, J. Kohl, and M. Frosch. 1998. Phase-variable expression of lipopolysaccharide contributes to the virulence of Legionella pneumophila. J. Exp. Med. 188:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Bacteriophage P22 transduction of integrated plasmids: single-step cloning of Salmonella typhimurium gene fusions. J. Bacteriol. 175:7086-7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayr, L. M., C. Odefey, M. Schutkowski, and F. X. Schmid. 1996. Kinetic analysis of the unfolding and refolding of ribonuclease T1 by a stopped-flow double-mixing technique. Biochemistry 35:5550-5561. [DOI] [PubMed] [Google Scholar]
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mo, Y. Y., N. P. Cianciotto, and L. P. Mallavia. 1995. Molecular cloning of a Coxiella burnetii gene encoding a macrophage infectivity potentiator (Mip) analogue. Microbiology 141:2861-2871. [DOI] [PubMed] [Google Scholar]
- 31.Moro, A., F. Ruiz-Cabello, A. Fernandez-Cano, R. P. Stock, and A. Gonzalez. 1995. Secretion by Trypanosoma cruzi of a peptidyl-prolyl cis-trans isomerase involved in cell infection. EMBO J. 14:2483-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison, J. F. 1969. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim. Biophys. Acta 185:269-286. [DOI] [PubMed] [Google Scholar]
- 33.Payne, N. R., and M. A. Horwitz. 1987. Phagocytosis of Legionella pneumophila is mediated by human monocyte complement receptors. J. Exp. Med. 166:1377-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira, P. J., M. C. Vega, E. Gonzales-Rey, R. Fernandez-Carazo, S. Macedo-Ribeiro, F. X. Gomis-Ruth, A. Gonzalez, and M. Coll. 2002. Trypanosoma cruzi macrophage infectivity potentiator has a rotamase core and a highly exposed α-helix. EMBO Rep. 3:88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rahfeld, J. U., K. P. Rucknagel, G. Stoller, S. M. Horne, A. Schierhorn, K. D. Young, and G. Fischer. 1996. Isolation and amino acid sequence of a new 22-kDa FKBP-like peptidyl-prolyl cis/trans-isomerase of Escherichia coli. Similarity to Mip-like proteins of pathogenic bacteria. J. Biol. Chem. 271:22130-22138. [DOI] [PubMed] [Google Scholar]
- 36.Ramm, K., and A. Plückthun. 2000. The periplasmic Escherichia coli peptidyl prolyl cis, trans-isomerase FkpA II. Isomerase-independent chaperone activity in vitro. J. Biol. Chem. 275:17106-17113. [DOI] [PubMed] [Google Scholar]
- 37.Ramm, K., and A. Plückthun. 2001. High enzymatic activity and chaperone function are mechanistically related features of the dimeric E. coli peptidyl-prolyl-isomerase FkpA. J. Mol. Biol. 310:485-498. [DOI] [PubMed] [Google Scholar]
- 38.Ratcliff, R. M., S. C. Donnellan, J. A. Lanser, P. A. Manning, and M. W. Heuzenroed. 1997. Interspecies sequence differences in the Mip protein from the genus Legionella: implications for function and evolutionary relatedness. Mol. Microbiol. 25:1149-1158. [DOI] [PubMed] [Google Scholar]
- 39.Riboldi-Tunnicliffe, A., B. König, S. Jessen, M. S. Weiss, J. Rahfeld, J. Hacker, G. Fischer, and R. Hilgenfeld. 2001. Crystal structure of Mip, a prolylisomerase from Legionella pneumophila. Nat. Struct. Biol. 8:779-783. [DOI] [PubMed] [Google Scholar]
- 40.Rockey, D. D., B. B. Chesebro, R. A. Heinzen, and T. Hackstadt. 1996. A 28 kDa major immunogen of Chlamydia psittaci shares identity with Mip proteins of Legionella spp. and Chlamydia trachomatis—cloning and characterization of the C. psittaci mip-like gene. Microbiology 142:945-953. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schmid, F. X. 1993. Prolyl isomerase: enzymatic catalysis of slow protein-folding reactions. Annu. Rev. Biophys. Biomol. Struct. 22:123-142. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt, B., J. Rahfeld, A. Schierhorn, B. Ludwig, J. Hacker, and G. Fischer. 1994. A homodimer represents an active species of the peptidyl-prolyl cis/trans isomerase FKBP25mem from Legionella pneumophila. FEBS Lett. 352:185-190. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt, B., S. König, D. Svergun, V. Volkov, G. Fischer, and M. H. Koch. 1995. Small-angle X-ray solution scattering study on the dimerization of the FKBP25mem from Legionella pneumophila. FEBS Lett. 372:169-172. [DOI] [PubMed] [Google Scholar]
- 45.Scholz, C., G. Stoller, T. Zarnt, G. Fischer, and F. X. Schmid. 1997. Cooperation of enzymatic and chaperone functions of trigger factor in the catalysis of protein folding. EMBO J. 16:54-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scholz, C., M. Mucke, M. Rape, A. Pecht, A. Pahl, H. Bang, and F. Schmid. 1998. Recognition of protein substrates by the prolyl isomerase trigger factor is independent of proline residues. J. Mol. Biol. 277:723-732. [DOI] [PubMed] [Google Scholar]
- 47.Stoller, G., K. P. Rucknagel, K. H. Nierhaus, F. X. Schmid, G. Fischer, and J. U. Rahfeld. 1995. A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as trigger factor. EMBO J. 14:4939-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 49.Tradler, T., G. Stoller, K. P. Rucknagel, A. Schierhorn, J. U. Rahfeld, and G. Fischer. 1997. Comparative mutational analysis of peptidyl prolyl cis/trans isomerase: active sites of Escherichia coli trigger factor and human FKBP12. FEBS Lett. 407:184-190. [DOI] [PubMed] [Google Scholar]
- 50.Venyaminov, S. Y., and K. S. Vassilenko. 1994. Determination of protein tertiary structure class from circular dichroism spectra. Anal. Biochem. 222:176-184. [DOI] [PubMed] [Google Scholar]
- 51.Wintermeyer, E., B. Ludwig, M. Steinert, B. Schmidt, G. Fischer, and J. Hacker. 1995. Influence of site specifically altered Mip proteins on intracellular survival of Legionella pneumophila in eukaryotic cells. Infect. Immun. 63:4576-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zarnt, T., T. Tradler, G. Stoller, C. Scholz, F. X. Schmid, and G. Fischer. 1997. Modular structure of the trigger factor required for high activity in protein folding. J. Mol. Biol. 271:827-837. [DOI] [PubMed] [Google Scholar]