Abstract
Nonenterotoxigenic porcine Escherichia coli strains belonging to the serogroup O45 have been associated with postweaning diarrhea in swine and adhere to intestinal epithelial cells in a characteristic attaching and effacing (A/E) pattern. O45 porcine enteropathogenic E. coli (PEPEC) strain 86-1390 induces typical A/E lesions in a pig ileal explant model. Using TnphoA transposon insertion mutagenesis on strain 86-1390, we found a mutant that did not induce A/E lesions. The insertion was identified in a gene designated paa (porcine A/E-associated gene). Sequence analysis of paa revealed an open reading frame of 753 bp encoding a 27.6-kDa protein which displayed 100, 51.8, and 49% homology with Paa of enterohemorrhagic E. coli O157:H7 strains (EDL933 and Sakai), PEB3 of Campylobacter jejuni, and AcfC of Vibrio cholerae, respectively. Chromosomal localization studies indicated that the region containing paa was inserted between the yciD and yciE genes at about 28.3 min of the E. coli K-12 chromosome. The presence of paa and eae sequences in the porcine O45 strains is highly correlated with the A/E phenotype. However, the observation that three eae-positive but paa-negative PEPEC O45 strains were A/E negative provides further evidence for the importance of the paa gene in the A/E activity of O45 strains. As well, the complementation of the paa mutant restored the A/E activity of the 86-1390 strain, showing the involvement of Paa in PEPEC pathogenicity. These observations suggest that Paa contributes to the early stages of A/E E. coli virulence.
Attaching and effacing (A/E) Escherichia coli (AEEC) induces distinctive histopathological lesions on the intestinal mucosa, known as the A/E lesions. These lesions are characteristic of enteric pathogens such as enteropathogenic E. coli (EPEC), responsible for severe childhood diarrhea in developing countries (14, 38), enterohemorrhagic E. coli (EHEC), causing hemorrhagic colitis and hemolytic-uremic syndrome, a diarrheagenic E. coli strain of rabbits (RDEC-1), strains of Hafnia alvei isolated from children with diarrhea, and Citrobacter rodentium, causing transmissible colonic hyperplasia in mice (4, 16, 53). A/E lesions have also been associated with diarrhea in different animal species such as rabbits, calves, dogs, cats, lambs, pigs, and tamarins (8, 9, 22, 32, 37, 55).
A/E lesions result from intimate bacterial adherence to the apical surfaces of enterocytes and activation of several chromosomal gene products that interact with components of the host cell, leading to host cell protein phosphorylation, effacement of target brush borders, and disruption of the underlying actin cytoskeleton (11, 38). The genes are clustered in a chromosomal pathogenicity island called the locus of enterocyte effacement (LEE). Its location and size vary in different strains. In EPEC strain E2348/69 and EHEC O157:H7 strains, the LEE is inserted in the selC locus at about 82 min on the E. coli K-12 chromosome, but its size varies from 35 kb for EPEC to 43 kb for EHEC. In strains of serotype O26:H-, the LEE is about 35 kb and is inserted in the pheU gene (12, 34, 46). One of the LEE genes (eae) encodes intimin, a 94-kDa outer membrane protein involved in intimate attachment to host cells (24). Another encodes a translocated intimin receptor called Tir, which interacts with intimin and allows the intimate attachment of the bacteria to the epithelial cells (27). Other genes encode the secreted proteins EspA, EspB, EspD, and EspF, which are responsible for signal transduction in epithelial cells (15, 23, 28, 29, 31, 33, 35, 50) and which are secreted through a type III secretion apparatus, which is also encoded in the LEE (33). The recently identified EspC enterotoxin, whose gene is located within a pathogenicity island at 60 min on the chromosome of E. coli, may also play a role as an accessory virulence factor in some EPEC strains (36).
A/E lesions in naturally occurring swine postweaning diarrhea cases are often associated with E. coli of the O45 serogroup (19, 21, 55). This pig AEEC, termed porcine EPEC (PEPEC), possesses all the genes in the LEE. The A/E activity of PEPEC O45 isolates is highly correlated with the presence of the LEE (21, 55, 56). Although there is some heterogeneity in PEPEC strains with respect to the LEE insertion, all of these strains possess a β-intimin subtype. In PEPEC strain 86-1390, sequences of the eae, tir, and esp regions are closely related to those of other AEEC strains, particularly of rabbit EPEC (REPEC) strains (3). The presence of the eae β variant gene in the porcine O45 strain 86-1390 (57) is associated with the ability of this strain to produce A/E lesions in experimentally inoculated newborn gnotobiotic piglets (55) and in an homologous in vitro model using newborn piglet ileal explants (56). We have created a bank of PEPEC strain 86-1390 TnphoA mutants and screened for the loss of their capacity to induce the typical histopathological A/E lesions in pig intestinal ileal explants (2). One mutant, M155, did not induce A/E lesions, the TnphoA insertion occurring in a gene that was called paa, for porcine A/E associated. The distribution of paa in PEPEC O45 strains revealed that its presence was associated with that of the eae gene and its A/E phenotype in vivo. On examination of enteric E. coli isolates from humans and various animal species, a strong correlation between the presence of paa and eae in EHEC O157:H7 and O26 isolates and dog, rabbit, and pig isolates, and a lesser correlation in human EPEC isolates, was found (2). The aim of this study was to characterize the paa gene and to study the contribution of Paa to the development of A/E lesions due to PEPEC in a pig ileal explant model.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The wild-type pathogenic E. coli strain 86-1390 (serogroup O45, tetracycline [Tcr] and streptomycin [Smr] resistant) was isolated at the Faculté de Médecine Vétérinaire, Saint-Hyacinthe, Québec, Canada, from a 4-week-old pig with postweaning diarrhea. O45 strain 86-1390 induces typical A/E lesions both in vitro and in vivo and contains sequences homologous to the LEE (55, 56). A collection of 11 PEPEC strains was used for in vivo experiments. E. coli strain SM10λpir(pRT733) was used to introduce TnphoA into strain 86-1390 by conjugation (17). E. coli strain HB101 (supE44 hsdS20 (r− m−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1) (7) was used as host for recombinant plasmids in this study. The λZAPIIR system was used for construction of a genomic DNA library from strain 86-1390 (Stratagene, La Jolla, Calif.). The host strain E. coli XL1 Blue MRF′ {Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac[F′ proAB lacIq ZΔM15] Tn10(Tetr)} and E. coli strain SOLR {e14−(mcrA) Δ(mcrCB-hsdSMR-mrr)171 sbcC recB recJ umuC::Tn5(Kanr) uvrC lac gyrA96 relA1 λR[F′ proAB lacIq ZΔM15]Su− (nonsuppressing)}, as well as the helper phage M13 needed in the cloning procedure, were used according to the manufacturer's recommendations (Stratagene). E22 is an eae β-positive REPEC strain (40).
TnphoA mutagenesis.
Mutations were obtained from random insertion of the TnphoA sequence into the chromosomal DNA of E. coli strain 86-1390 (Smr Tcr). This was accomplished as described previously (17) by using the suicide vector pRT733, which carries the TnphoA insertion and the kanamycin resistance (Kmr) gene in E. coli strain SΜ10λpir (51). Exconjugants from the mating between E. coli strain SΜ10λpir(pRT733) and E. coli strain 86-1390 were selected on Luria-Bertani (LB) agar (Difco Laboratories, Detroit, Mich.) containing kanamycin and streptomycin (40 μg ml−1) and the alkaline phosphatase substrate XP (5-bromo-4-chloro-3-indolylphosphate) (Sigma Chemical Co., St. Louis, Mo.). Kanamycin- and streptomycin-resistant blue colonies resulting from the transposition of TnphoA into the genome of the recipient strain E. coli 86-1390 were stored in glycerol at −70°C. Of the Kmr and Smr transposon insertions, 1% were found to produce blue colonies on agar in the presence of alkaline phosphatase substrate XP.
Cloning and DNA sequencing phoA fusion regions.
Cloning the sequence flanking the 5′ end of phoA fusion regions was done with the kanamycin resistance gene as a selectable marker, and the appropriate DNA fragments were cloned into pBR322. One microgram of total DNA isolated from the mutant was digested by BamHI and ligated with 0.1 μg of similarly digested pBR322. The ligation mixture was electroporated into the strain HB101 with a Gene Pulser according to the manufacturer's instructions (Bio-Rad Laboratories Ltd., Mississauga, Ontario, Canada). Transformants carrying the desired recombinant plasmid were selected on LB agar plates supplemented with ampicillin (40 μg ml−1) and kanamycin (40 μg ml−1). To precisely identify the mutated gene, the double-stranded plasmid DNA at the junction of the site of TnphoA insertion was sequenced with a T7 sequencing kit (Pharmacia LKB Biotechnology Inc., Baie d'Urfé, Québec, Canada) according to the manufacturer's instructions. An oligonucleotide (5′AATAATCGCCTGAGC3′) corresponding to nucleotides 72 to 86 of TnphoA, synthesized on a Gene Assembler (Pharmacia LKB Biotechnology Inc.), was used as the primer. DNA sequence data were analyzed with the GeneWorks program (Intelligenetics, Inc., Mountain View, Calif.) and programs included in the GCG (Genetics Computer Group, Madison, Wis.) package (10). The deduced amino acid sequence was compared against the combined databases of the National Center for Biotechnology Information (Washington, D.C.) via the BLAST network service.
Cloning and sequencing paa.
To clone the full length of the paa gene, corresponding to the gene of the mutant M155 with the TnphoA insert, a genomic DNA library of PEPEC strain 86-1390 was constructed in the λZAPIIR vector. Chromosomal DNA was partially digested with EcoRI and pooled and fractionated through a 10 to 30% sucrose linear gradient. The desired fragments of 6 to 10 kb were isolated and ligated to dephosphorylated EcoRI-digested λZAPIIR vector arms and packaged with an in vitro packaging system (Stratagene). Bacteriophage particles were propagated in E. coli XL1 Blue and plated for plaque isolation. To screen the recombinant phages, a 350-bp PCR fragment derived from the 5′ end of the paa gene was generated by paa-specific primers M155-F1 (5′ATGAGGAAACATAATGGCAGG3′) and M155-R1 (5′TCTGGTCAGGTCGTCAATAC3′) annealed at nucleotides 91 to 110 and 428 to 447 of the paa gene, respectively. The 350-bp fragment was then radiolabeled with [α-32P]CTP as a probe by using an oligonucleotide random priming labeling kit (Pharmacia LKB) according to the manufacturer's instructions. Positive plaques were selected and excised with the ExAssist helper phage (M13) and E. coli strain SOLR system according to the Stratagene λZAPIIR instruction manual. Plasmid DNA was isolated by alkaline lysis, and the insert was sequenced by an automated DNA sequencer (AFL DNA sequencer; Pharmacia LKB) using the paa-specific oligonucleotides synthesized on a Gene Assembler (Pharmacia LKB).
Transcomplementation of the M155 paa::TnphoA mutant.
The paa gene was amplified with its promoter regions from strain 86-1390 DNA with the PaaHO/F (5′GGATCCTTAAAGGGCAGG3′) and PaaHO/R (5′GGATCCGATGTCAAGTGC3′) primers and cloned into the pGEM-T vector. The BamHI fragment was then inserted into the BamHI-linearized pACYC184 plasmid, resulting in the pACYC184-PaaHO construct, containing the wild-type paa gene. This construct was used as a complementation plasmid for paa in the M155 TnphoA mutant, leading to the M155c strain.
Quantification of A/E capacity of the mutants.
The A/E capacities of the TnphoA mutants generated in this study were examined by ileal explant culture as previously described (56). Briefly, overnight bacterial cultures were inoculated onto the villous surface of ileal explants from colostrum-deprived newborn piglets and incubated on a rocking platform at 37°C for 8 h in an atmosphere of 95% O2 and 5% CO2. RPMI 1640 culture medium (Gibco, BRL, Burlington, Ontario, Canada) was replaced with fresh medium at hourly intervals during the incubation to prevent acidic pH and overgrowth of bacteria. E. coli O45 strain 86-1390 and the porcine 862 strain, which does not possess the LEE, were used as positive and negative controls, respectively. Three or four ileal explants were used for each bacterial isolate, and the experiments were repeated three times. In some experiments, broth cultures were incubated at 37°C with an equal volume of lyophilized Paa antibody reconstituted with phosphate-buffered saline (PBS) for 30 min prior to the first explant inoculation. Tissues were processed for light microscopy examination as described previously (56); the intact villi with adherent bacteria were counted, and the number was expressed as a percentage of the total number of intact villi observed.
Southern analysis.
The number of TnphoA insertions was determined by Southern blot analysis as described previously (17). Briefly, total DNA was extracted from the strain by sodium dodecyl sulfate lysis, proteinase K treatment, phenol-chloroform extraction, and ethanol precipitation. Extracted DNA was digested with the restriction endonuclease SacI or EcoRV, neither of which cuts within TnphoA, under conditions described by the manufacturer (Pharmacia LKB). After separation by electrophoresis in a 0.7% agarose gel, DNA preparations were examined by Southern hybridization. An internal HindIII-BamHI fragment of TnphoA radiolabeled with [α-32P]CTP by using an oligonucleotide random priming labeling kit (Pharmacia LKB) according to the manufacturer's instructions was used as a probe.
Chromosomal localization of the paa gene.
First, the presence of paa between the rem and rel loci was investigated by PCR amplification. The remF (5′GATGCCTGCCACATCAGAGG3′) and relR (5′CCTAAGCCAGTACGTGTGAC3′) primers located at bp 2821 to 2840 and 3400 to 3420, respectively, were used to amplify a 580-bp fragment on the E. coli K-12 strain MG1655 chromosome. The PaaR primer (5′GCTACAAACCGATGAAGCGGC3′) was used in combination with remF to detect an insertion of the paa gene between the rem and rel loci, leading to a 605-bp amplicon. Second, the integrity of the yciD-yciE region was tested with the YciDF (5′AGTGGCGGCTTTGGCACTAA3′) and YciER (5′CGAATCTATGCTTGAATCCA3′) primers. They were used to amplify a 1,122-bp fragment on the MG1655 chromosome. The PCR mixture included 5 μl of template DNA, 5 μl of 2 mM deoxynucleoside triphosphate, 5 μl of 10× buffer (100 mM Tris-HCl, 15 mM MgCl2, 500 mM KCl; pH 8.3), 2.5 μl of each primer pair (25 pmol), 1 U of Taq DNA polymerase (Pharmacia), and sterile distilled water to 50 μl. The following cycles were used: 1 cycle of 94°C for 2 min and 30 cycles of 94°C for 30 s, 60°C for 45 s, and 72°C for 30 s, with a final extension of 72°C for 7 min. The PCR products were analyzed by agarose gel electrophoresis.
Nonpolar mutation in paa.
A PCR fragment containing the gene and its promoter sequences was amplified with the PaaHO/F and PaaHO/R primers and cloned into the pGEM-T vector (Promega) according to the manufacturer's instructions. The construct was digested with KpnI, and an HincII fragment from pSB315 containing a kanamycin resistance cassette was ligated in the KpnI site, resulting in paa::kan. The construct was digested with BamHI, and the paa::kan fragment was ligated to the pKNG101 suicide vector cut with the same enzyme. The resulting construct was transferred to strain S17 λpir, from which it was mobilized into strain E22 by the membrane filter mating technique. Transconjugants were selected on M9 agar containing the appropriate antibiotic (kanamycin at 50 μg/ml). Selection for double-crossover allele replacement was obtained by sacB counterselection on LB agar plates without NaCl and containing 5% sucrose (25).
Pulsed-field gel electrophoresis.
Strains 86-1390, M155, STJ348 (O157:H7), and E2348/69 (EPEC) were inoculated 1/100 in 20 ml of LB medium and incubated at 37°C overnight with agitation. Bacteria were washed two times in SE (75 mM NaCl, 25 mM EDTA; pH 7.5) by centrifugation and resuspended in TE (10 mM Tris-HCl, 1 mM EDTA; pH 8). The optical density of the cells was adjusted to 1.5 to 1.8 at a wavelength of 600 nm. Low-melting-point agarose (Gibco, BRL) was prepared in distilled water to obtain a final concentration of 1.5%. Plugs were formed by mixing 500 μl of bacterial suspension with 500 μl of prewarmed (60°C) agarose. This mixture was then pipetted into plug molds (Bio-Rad Laboratories). After the plugs solidified, they were incubated at 50°C overnight in lysis buffer (1% [wt/vol] N-laurylsarcosine-0.5 M EDTA [pH 9.5] supplemented with 1 mg of proteinase K/ml). The lysis buffer was changed, and plugs were incubated for another 4 h. Plugs were washed three times for 1 h each time with 1 mM phenylmethylsulfonyl fluoride in 10 mM Tris-HCl, pH 8. Another set of three 30-min washes was done with 10 mM Tris-HCl, pH 8. The plugs were then preincubated for 30 min with 1 ml of the appropriate restriction enzyme buffer. The buffer was replaced by a fresh mixture containing 30 to 40 U of enzyme and incubated overnight at the appropriate temperature (37°C for XbaI and 50°C for SfiI). The next day, 10 U of enzyme was added to the plugs for a 2- to 3-h incubation period. Electrophoresis of the samples was performed on the CHEF-DRII system by using a 1% pulsed-field grade agarose gel (Sigma) with 2 liters of modified 0.5× TBE running buffer (10× TBE is 89 mM Tris-borate plus 25 mM EDTA, pH 8.3). The running conditions were as follows: switch of 5 to 35 s, 6 V/cm, and a run time of 20 h. Finally, the gel was stained in 10 mg of ethidium bromide/ml in distilled water for 30 min. Digested genomic DNA separated in agarose gels was transferred to positively charged nylon membranes (Immobilon-Ny+; Millipore Corporation, Bedford, Mass.) in accordance with the manufacturer's instructions and hybridized under stringent conditions as described by Sambrook et al. (45). The probe was labeled with biotin by PCR amplification using PaaF (5′GGATCCATGAGGAACATAA3′) and PaaR (5′CTCGAGAGTGCCTTTCCTGG3′).
Production of anti-Paa antibodies.
The paa gene of strain 86-1390 was amplified by PCR using primer pairs PaaF and PaaR. The amplicon was inserted into the pQE-30 expression vector (Qiagen) by using appropriate cloning sites, and fusion was confirmed by sequencing. E. coli M15(pREP4) (Qiagen) was used as the host strain for the expression of recombinant His-Paa. The His-Paa was purified from a Ni-nitrilotriacetic acid affinity column (Qiagen). Laying hens 25 weeks of age were immunized intramuscularly with 500 μl of incomplete Freund's adjuvant and an equal volume of purified His-Paa, corresponding to 50 μg of protein. Paa-specific immunoglobulin Y (IgY) was then extracted from egg yolks by the method described by Akita and Nakai (1), with some modifications. Briefly, egg yolks were separated from the albumin. An equal volume of PBS was added to the egg yolks, and the mixture was then homogenized by Vortex agitation. An equal volume of chloroform was added, and a solid homogenate was then obtained by mixing. The preparation was centrifuged for 5 min at 14,000 × g, and the supernatant containing IgY was recovered, lyophilized, and conserved until use. The anti-Paa IgY titer was determined by enzyme-linked immunosorbent assay using microtiter plates (Immulon 2HB; Dynec) precoated with 100 ng of purified protein in carbonate buffer (pH 9.6) per well.
Electron microscopy.
Electron microscopy and immunogold labeling were done as previously described (18) with modifications. Cultures of the wild-type strain 86-1390, complemented mutant strain M155c, and paa-negative control strain 862 were grown overnight at 37°C in tryptic soy broth (TSB) and washed three times in PBS. Bacterial pellets were obtained with an AirFuge air-driven ultracentrifuge (Beckman Instruments Inc., Palo Alto, Calif.) and were coated on 150-mesh Formvar-coated nickel grids (Marivac, St.-Laurent, Quebec, Canada). After a blocking step in 5% normal donkey serum, grids were labeled with appropriate dilutions of a chicken anti-Paa primary antibody, previously adsorbed against the paa-negative E. coli strain E2348/69, and a donkey anti-chicken IgY secondary antibody conjugated with 12-nm colloidal gold beads (Jackson ImmunoResearch Laboratories Inc.). Grids were negatively stained with 1% phosphotungstic acid at pH 6.0 and examined with a H-7100 transmission electron microscope at 75 kV (Hitachi High-Technologies, Rexdale, Ontario, Canada). The anti-Paa antibody adsorbed with the Paa-positive strain M155c was used as a negative control.
Ileal explant tissues were processed for transmission electron microscopy. Tissues were fixed for 2 h at room temperature in 2.5% glutaraldehyde and rinsed in 0.1 M cacodylate buffer at pH 7.3. Tissues were then postfixed in 2% osmium tetroxide, rinsed in water, dehydrated in a graded ethanol series, and finally embedded in Spurr resin (Marivac). Thin sections were mounted on copper grids, stained with uranyl acetate and lead citrate, and examined with a Philips 420 transmission electron microscope at 80 kV (Philips Electronics, Eindhover, The Netherlands).
Infection of gnotobiotic piglets.
Eleven porcine O45 strains were tested for A/E activity in experimentally inoculated newborn gnotobiotic piglets as previously described (55). Briefly, aseptically collected piglets were inoculated intragastrically with 10 ml of an overnight culture of E. coli (approximately 109 CFU) and 10 ml of 0.1% peptone-water. They were examined several times daily for development of clinical signs, and necropsy was performed when diarrhea occurred or at 120 h postinoculation (p.i.) if clinically normal.
Statistical analysis.
Results are presented as the means ± standard deviations of the means. A Kruskal-Wallis test was performed with commercially available software (SAS, version 8.1; SAS Institute, Cary, N.C.), and post hoc two-by-two comparisons were done to assess differences between the groups; a P value <0.0001 was taken to be significant.
Nucleotide sequence accession number.
The complete nucleotide sequence of paa was lodged with GenBank under accession number U82533 (paa was previously named anm).
RESULTS
Identification of a transposon mutant deficient in A/E activity.
After random insertion of TnphoA into the genome of porcine O45 E. coli strain 86-1390, mutants containing translational fusions between bacterial genes for extracytoplasmic proteins and phoA were screened on LB agar supplemented with kanamycin and streptomycin and with substrate XP. A total of 180 TnphoA mutants were then examined in a qualitative assay for adhesion to piglet ileal explants (56). Of these, 175 mutants were found to attach extensively to a similar extent as strain 86-1390, as observed by light microscopy (2) (Fig. 1A). In the five other mutants which attached less extensively to piglet ileal enterocytes, different insertion sites for TnphoA were observed. Three insertions were in genes found in E. coli K-12 (one in osmB, two in pstS), one was in IS100, and one was in an as yet uncharacterized gene (the mutant with this gene was named M155). This gene was called paa. Hence, only M155 was retained for further characterization. The presence of a single copy of TnphoA in the chromosomal DNA of the mutant M155 was demonstrated by Southern hybridization of genomic DNA digested by SacI or EcoRV and probed with an internal BamHI-HindIII fragment of TnphoA (data not shown).
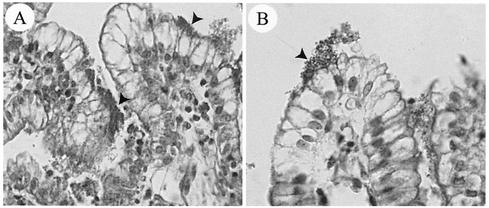
FIG. 1.
Light microscopy micrographs of ileal explants inoculated with the wild-type O45 strain 86-1390 (A) or with the LEE-negative strain 862 (B). Strain 86-1390 showed a typical intimate-adherence pattern (arrowheads) with irregularity of the associated epithelial cells, whereas a loose association of bacteria with the intestinal mucosa of some villi with no obvious change in associated epithelial cells (arrow) was observed for negative-control strain 862. Magnification, ×400.
Effect of the Paa mutation and complementation on the adherence phenotype.
The insertion of TnphoA in the paa gene (M155 mutant) resulted in a significantly reduced number of ileal villi showing bacterial adherence to epithelial intestinal cells, compared to that observed for the wild-type 86-1390, in our explant culture model (Fig. 2A). As observed for negative-control strain 862 (Fig. 1B), there was a patchy, loose association of mutant M155 with the mucosal surfaces of a low proportion of villi. Furthermore, the complementation of strain M155 with the pACYC184 plasmid carrying the paa gene and its promoter region (M155c strain) restored the adherence phenotype. On transmission electron microscopy, for the M155c and 86-1390 strains, bacteria demonstrated a tight attachment to epithelial cell surfaces, effacement of microvilli beneath the adherence site, and a dense region underneath the adherence site representing F-actin polymerization (Fig. 3A and 3B), whereas mutant M155 and control strain 862 showed no evidence of A/E lesion formation (Fig. 3C).
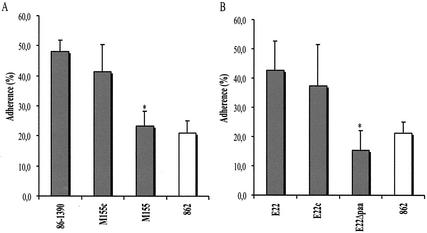
FIG. 2.
Adherence of wild-type strains and their paa mutant strains. (A) paa mutant strain M155 (n = 18) showed a decreased number of intact ileal villi with bacterial adherence to epithelial cells, compared to wild-type PEPEC strain 86-1390 (n = 12) and to the complemented mutant strain M155c (n = 20). The porcine strain 862 (n = 15), which does not possess the LEE, was used as a negative control. (B) paa mutant strain E22Δpaa (n = 19) showed a decreased number of intact ileal villi compared to wild-type REPEC strain E22 (n = 19) and to the complemented mutant strain E22c (n = 10). Error bars, standard deviations of the means. Asterisk, statistically significant difference (P < 0.0001, when compared by Kruskal-Wallis test) from wild-type strains 86-1390 (A) and E22 (B).
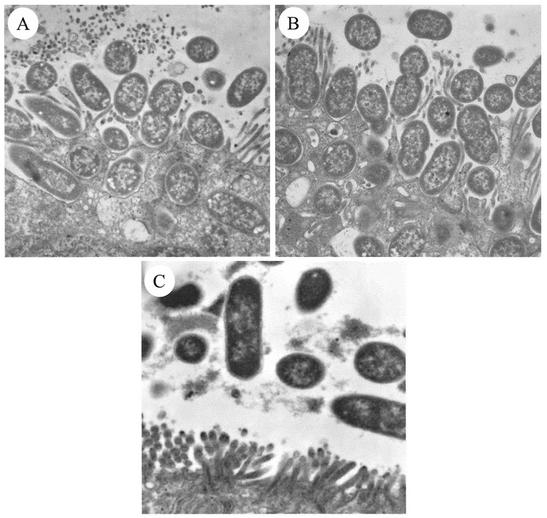
FIG. 3.
Transmission electron micrographs of ileal explants inoculated with the wild-type O45 strain 86-1390 (A; magnification, ×20,664), the complemented mutant strain M155c (B; magnification, ×20,702), or TnphoA mutant M155 (C; magnification, ×13,500). Typical A/E lesions were observed for both wild-type and complemented-mutant strains, whereas bacteria in the lumen without any direct contact with the epithelium were observed for the mutant M155.
Sequence analysis of paa and associated loci.
To further characterize the locus around the site of the TnphoA insertion of mutant M155, a genomic DNA library of PEPEC strain 86-1390 was screened. On primary screening, it was found that several of the plaques hybridized to the 350-bp PCR probe fragment which corresponded to the sequence adjacent to the TnphoA insertion of mutant M155 and which had been radiolabeled with [α-32P]CTP. One of these, with an insert of 6 kbp and designated λZAPIIR AN1, was chosen for further study. Using primers obtained from the sequence adjacent to the TnphoA insertion in M155, we determined the nucleotide sequence of the full length of the gene where TnphoA was inserted. It revealed an open reading frame of 753 bp. The region upstream of the first ATG was preceded by excellent matches to consensus sequences for −35 and −10 putative promoter regions and by a Shine-Dalgarno sequence (Fig. 4). Downstream of the TAG translational stop codon, a putative transcription terminator was evident (Fig. 4). The G+C content of paa was 44%, which is substantially lower than that of E. coli K-12 (50.8%) (6), suggesting that paa may have been acquired by 86-1390 through horizontal transfer. paa was predicted to encode a 251-amino-acid protein with an anticipated molecular mass of 27.6 kDa (Fig. 4). The prediction of the Paa protein localization site with the Expasy software suggested that the Paa peptide may be cleaved after the first 18 residues (54) (Fig. 4). The hydrophobicity profile indicated the presence of a potential transmembrane region (amino acids 1 to 18) corresponding to a Sec-dependent signal sequence and hydrophilic segments in mature Paa. However, the Expasy program also predicted that the entire Paa protein could be unstable due to its N-terminal end. The Domain Architecture Retrieval Tool (DART) from the National Center for Biotechnology Information (NCBI) identified a sulfate-binding motif in the C-terminal half of the protein. The comparison with the SWISS-PROT database showed that the amino acid sequence deduced from the paa gene displayed an identity of 100% with those encoded by the paa genes of the O157:H7 EDL933 and Sakai strains, 51.8% with PEB3, a major antigen of Campylobacter jejuni, and 49% with AcfC, a Vibrio cholerae accessory colonization factor (Fig. 5). AcfC and PEB3 also contain the same sulfate-binding motif.
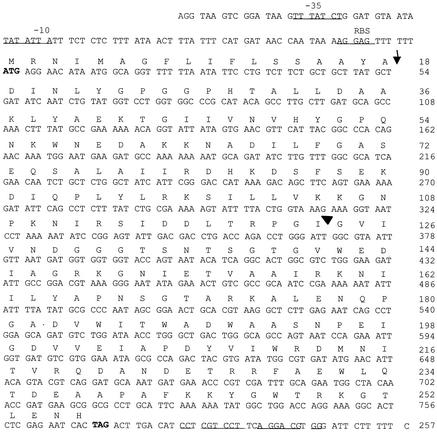
FIG. 4.
The nucleotide sequence of paa and its flanking sequences and the deduced amino acid sequences. The putative −10 and −35 promoter sites, the ribosome binding site (RBS), and the putative transcription terminator are underlined. The translation initiation codon and the TAG translation termination codon are in boldface. Vertical arrow, potential peptide signal cleavage site; arrowhead, insertion site of TnphoA.
FIG. 5.
Alignment of the deduced amino acid sequences of the Paa of the 86-1390 strain and the Paa proteins of the O157:H7 EDL933 and Sakai E. coli strains, the AcfC protein of V. cholerae, and the PEB3 protein of C. jejuni. ∗, identical or conserved residues in all sequences in the alignment; colons, conserved substitutions; periods, semiconserved substitutions. The amino acid sequence alignment was performed with the Clustal W program.
Localization of paa on the chromosome.
The 3.5-kb region containing paa was also 100% identical to the same region in the O157:H7 EDL933 and Sakai strains. Upstream of paa, there was homology with the prpH gene, encoding a fimbrial protein precursor of Pap-related pilus H. Downstream of paa was a sequence displaying identity to the rem gene from E. coli K-12. This may indicate that paa has interrupted the relB-rem region. Indeed the amplification by PCR of relB-rem showed that this region is disrupted in many paa-positive strains (data not shown). On the other hand, amplification was successful when a set of paa-rem primers was used. Localization of paa in the genome of O157:H7 strains EDL933 and Sakai demonstrated that paa was inserted at 28.3 min within the yciD-yciE locus of the K-12 chromosome. PCR studies confirmed this result: 9 out of 14 (64%) EPEC strains isolated from pigs, 2 of 7 (29%) REPEC strains, and 3 of 4 EHEC strains from humans (75%) were interrupted in the region between the yciD and yciE genes from E. coli K-12 (Table 1). Thus, in the O157:H7 strains EDL933 and Sakai, paa belongs to O island 57 and the Sp9 region, respectively, inserted within yciD and yciE. These islands contain incomplete lambda-like phage sequences (phage CP-933O for EDL933, phage Sp9 for Sakai). In PEPEC strains, the paa region is also within yciD-yciE. Moreover, in these genomes the relB-rem region is disturbed and a copy of rem is found near the 3′ end of paa. Genome analysis of the two studied O157:H7 strains indicates that paa is in a region specific to these pathogenic strains which harbors the sequence of a lambda phage.
TABLE 1.
Integrity of the yciDE region in paa-positive strains
| Strain type | No. of strainsa
|
||
|---|---|---|---|
| Total | yciDE negativeb | yciDE positive | |
| EHEC | 4 | 3 | 1 |
| REPEC | 7 | 2 (E22, RDEC-1) | 5 |
| PEPEC | 14 | 9 (86-1390) | 5 |
| Total | 25 | 14 (56%) | 11 (44%) |
Strains considered positive amplified a 1,122-bp fragment, whereas no amplification was seen in the negative strains.
Strains 86-1390 and E22 were used for testing the role of Paa; RDEC-1 is an REPEC strain.
The chromosomes of different AEEC strains were digested with SfiI or XbaI and examined by pulsed-field electrophoresis and Southern blotting. For both digests, a paa biotinylated probe hybridized with only one fragment of about 210 (XbaI) or 120 kbp (SfiI) for strains 86-1390 and M155 and about 290 (XbaI) or 210 kbp (SfiI) for the EHEC O157:H7 STJ348 strain, indicating that the chromosomal arrangement in the last strain was different from that in the other two. There was no hybridization for the paa-negative EPEC strain E2348/69. In paa-positive strains, paa was present in only one copy in the chromosome.
Creation of a paa mutant by allelic exchange and complementation.
Creation of a paa mutant by allelic exchange on the chromosome of PEPEC strain 86-1390 was unsuccessful. However, a paa mutant (E22Δpaa) was obtained from strain E22, a paa-positive REPEC strain that induces A/E lesions in our porcine ex vivo model. Strain E22Δpaa showed a reduced-adherence phenotype (Fig. 2B) and was not able to induce A/E lesions in the ex vivo model. The complementation of E22Δpaa with the wild-type paa restored this phenotype (Fig. 2B).
Development of A/E lesions in vivo by paa-positive and paa-negative PEPEC strains.
Most tested eae- and paa-positive porcine O45 isolates induced severe A/E lesions leading to diarrhea between 24 and 70 h after infection (Table 2). The severity and extent of the A/E lesions appeared to be related to the time of onset and severity of diarrhea in the inoculated piglets. On the other hand, eae-positive, paa-negative isolates induced less-severe or no A/E lesions and piglets developed no diarrhea or mild diarrhea after 83 h p.i.
TABLE 2.
Clinical and histopathological findings in piglets inoculated with porcine E. coli O45 isolates
| Straine | No. of pigs with diarrhea/no. inoculated | Onset of diarrhea (h)b | Extent of A/E lesionsd | Presence of paa gene |
|---|---|---|---|---|
| 81-4420 | 2/2 | 24 | ++++ | + |
| 86-1390 | 2/2 | 30 | ++++ | + |
| 91-19-172 | 2/2 | 35 | ++++ | + |
| 90-2061 | 2/2 | 39 | +++ | + |
| 90-1513 | 2/2 | 41 | +++ | + |
| 88-4299 | 1/1a | 44 | ++ | + |
| 86-4733 | 2/2 | 70 | + | + |
| 83-2315 | 2/2 | 83 | ++ | − |
| 88-1861 | 1/1a | 91 | − | − |
| 89-56-196 | 1/2 | 96 | − | − |
| 82-4378 | 0/2 | NMc | + | − |
| 81-1786 | 1/2 | 85 | − | − |
One piglet died of causes unrelated to the infection within 20 h after birth.
Mean time of onset of diarrhea p.i.
NM, nonmeasurable data.
++++, extensive bacterial colonization and severe effacement of microvilli; +++, large areas of bacterial colonization and heavy effacement; ++, focal lesions; +, small scattered focal lesions; −, no lesions observed.
Strains were eae and LEE positive, except for 81-1786, which was a control eae-negative strain.
Capacity of anti-Paa antibodies to block adhesion.
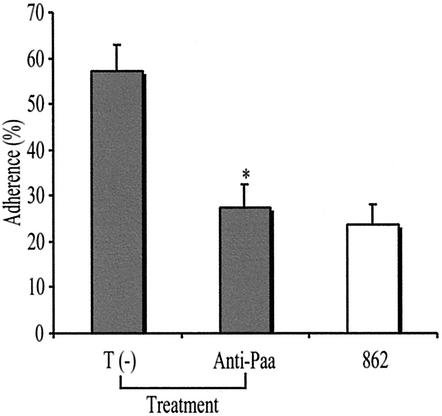
Treatment with chicken egg yolk anti-Paa antibodies significantly reduced, by up to 53%, the proportion of intact villi showing intimate adherence, following inoculation of pig ileal explants with PEPEC strain 86-1390 ex vivo (Fig. 6), compared to treatment with egg yolk antibodies from hens immunized with a sonicated preparation from the Paa-negative host strain M15(pREP4).
FIG. 6.
Reduction of the percentage of intact villi showing intimate adherence when pig ileal explants are inoculated with PEPEC strain 86-1390 following treatment with anti-Paa antibodies, compared to percentages for explants inoculated with strain 86-1390 following treatment with antibodies from hens immunized with a sonicate preparation from host strain M15(pREP4) (T −). The porcine strain 862, which does not have the LEE, was used as a negative control. Asterisk, statistically significant difference (P < 0.0001, when compared by Kruskal-Wallis tests) from the T (−) treatment.
The Paa protein is located at the bacterial surface.
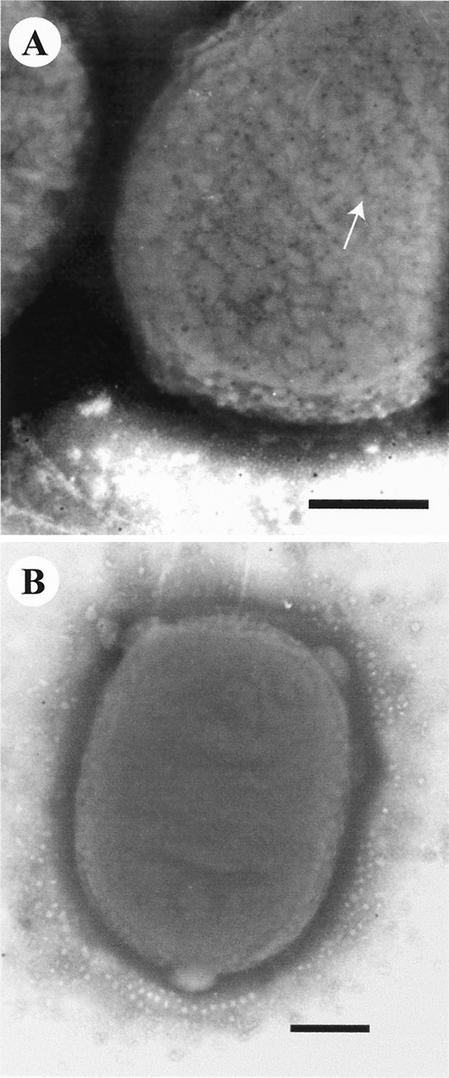
Immunogold labeling was performed in order to locate the Paa protein in the wild-type strain 86-1390 and the complemented mutant strain M155c labeled with the anti-Paa polyclonal antiserum. The Paa protein was uniformly distributed over the bacterial surface of the trans-complemented mutant strain M155c (Fig. 7A) and, to a lesser extent, on the bacterial surface of strain 86-1390 (data not shown). Moreover, the expression of the Paa protein was sevenfold higher in M155c than in the wild-type strain 86-1390 (data not shown). Low expression of Paa was confirmed by testing the PhoA activity of the fusion protein Paa-PhoA of the TnphoA mutant M155 in similar conditions (data not shown). When strains were labeled with the Paa-adsorbed antibody preparation, only a few gold beads were present, mostly in the background, confirming the specificity of the labeled antibody (Fig. 7B).
FIG. 7.
Transmission electron micrographs showing the uniform distribution of immunogold-labeled Paa protein (arrow) over the bacterial surface of the complemented strain M155c (A) following overnight growth at 37°C in TSB. When anti-Paa serum was adsorbed against the Paa protein, only a few gold beads were observed for strain M155c, mostly in the background (B). Bars = 300 nm.
DISCUSSION
In this study, we have identified by transposon mutagenesis using a PEPEC strain a locus important for development of A/E lesions, which we named paa. The paa::TnphoA mutant was no longer able to adhere to microvilli of intestinal epithelial cells and to create A/E lesions. paa sequences are often present in A/E strains, especially O157:H7 strains. The predicted amino acid sequence of Paa is identical to those of the Paa proteins of O157:H7 strains EDL933 and Sakai (20, 41) and very similar to those of the PEB3 and AcfC proteins. PEB3 is a major surface antigen involved in the pathogenicity of C. jejuni and was shown to be very unstable at ambient temperature. This feature is also shared with Paa. The acfC gene is part of the toxin-coregulated pilus (tcp) acf gene cluster, which has previously been shown to be required for efficient intestinal colonization and biogenesis of the toxin-associated pilus of V. cholerae (42). Genomic analysis of numerous V. cholerae strains (O1, non-O1, and O139) revealed that only strains capable of causing epidemic Asiatic cholera possessed the TCP-accessory colonization factor colonization gene cluster (30). AcfC is possibly secreted by V. cholerae cells into the culture supernatant (13). It is not known if Paa plays a role similar to that of AcfC. Paa, in contrast to AcfC, was shown to be associated mostly with the bacterial pellet and was best expressed in LB broth at 30°C (data not shown). Furthermore, immunogold labeling indicated that the Paa protein is distributed on the bacterial surface in strains 86-1390 and M155c. Nevertheless, the paa gene encodes a protein involved in the mechanism of pathogenesis of infection due to strain 86-1390. This gene is absent in nonpathogenic E. coli, and the G+C content of paa (44%) differs from that of E. coli K-12 (50.8%).
The regions flanking paa in PEPEC 86-1390 were sequenced for a total of 3.5 kb. Upstream of paa, there is a truncated gene homologous to prpH, coding for a subunit of the H pilus, a member of the Pap family, and downstream of paa are two genes homologous to gef and rem. The gef gene encodes a putative toxic protein similar to the Hok/Gef family, and rem has no known function. The region containing paa in the 86-1390 strain is 100% identical to the region containing paa in the O157:H7 strains EDL933 and Sakai. In the Sakai strain, this region is enclosed in a region of 58.2 kb, specific to the pathogen, localized between yciD and yciE of E. coli K-12 MG1655. This 58.2-kb region contains a lambda prophage that harbors virulence-related genes encoding proteins such as Lom and TrcA homologues. Lom is a member of a family of outer membrane proteins associated with virulence in two enterobacterial species. Expressed in lysogens, this protein confers the ability to survive in macrophages (5). TrcA is reported to be a chaperone molecule in EPEC strains (52). The prophage contains insertions of insertion sequence elements and deletions and thus is presumably defective (39). In strain EDL933, the region containing paa is inserted in a larger region of 103.1 kb, also localized between yciD and yciE. We suppose that these flanking sequences are also found in the PEPEC 86-1390 strain. These data suggest that paa could be part of a new putative pathogenicity islet.
The distribution of paa in PEPEC O45 strains revealed that it was associated with the presence of eae and the A/E phenotype in vivo and in vitro (2). The correlation between the presence of paa and eae among the isolates from humans and animals suggests that paa may be more frequently required for the A/E activity of EHEC and dog isolates than for the A/E activity of rabbit, pig O45, and human EPEC isolates (2). The presence of the paa gene could reflect some differences in the mechanisms of A/E activity and/or the development of diarrhea for isolates from different animal species and categories such as EHEC and EPEC. The explant culture technique has proved to be an efficient way to study the A/E phenotype of PEPEC strains ex vivo (56). Moreover, use of ileal explants from the same animal species as those from which the isolates originated eliminates problems due to lack of species-specific recognition of receptors by bacterial adhesins. The observation that three eae-positive but paa-negative porcine O45 strains were A/E negative provides further evidence for the importance of the paa gene in the A/E activity of porcine O45 strains. These results are confirmed by demonstration in the ex vivo model of a clear decrease in the number of ileal villi showing bacterial intimate adherence for paa mutants compared to the numbers for wild-type PEPEC and REPEC strains. Complementation of the mutants with the paa gene restored adherence capacity to a level similar as that for the wild-type strain (Fig. 1), confirming the importance of paa in PEPEC O45 strain 86-1390 and REPEC strain E22. However, we observed that the growth rate of strain M155 complemented with the high-copy-number plasmid pCRII carrying the paa gene and its promoter sequence was lower than that of M155 complemented with paa carried by the low-copy-number plasmid pACYC184. This suggests that overexpression of Paa may be toxic in the wild-type E. coli strain. In experimental infection of newborn piglets, the paa-negative isogenic strain was less adherent than the wild-type strain in the ileum but as adherent as the wild-type strain in the cecum and colon (data not shown). This reflects the results obtained with the ileal ex vivo model, in which the paa-negative strain is less adherent than the wild type. This also could indicate that paa has a more important role in early colonization of the ileum. Moreover, the localization of the Paa protein at the bacterial surface and the ability of Paa-specific antibodies to reduce the adherence level of the PEPEC strain 86-1390 clearly demonstrate the involvement of the Paa protein in A/E lesion formation, possibly in the initial-adherence process. These results also indicate that the Paa protein could be a potential candidate for a vaccine, together with Eae and Tir.
Interestingly, Paa contains a sulfate-binding domain; such motifs are also associated with microbial adherence. For instance, numerous pathogens such as Neisseria gonorrhoeae, Helicobacter pylori, and Pseudomonas aeruginosa bind to the host cell surface via heparan sulfate (HS). Gram-positive bacteria, viruses, and parasites also bind HS on host cells (43, 44). Furthermore, infection studies of gnotobiotic piglets also suggested that the Paa protein is involved in the first step of PEPEC pathogenicity, particularly in initial bacterial adherence, since paa-defective strains showed a reduced adherence and infected piglets had no, or delayed-onset, diarrhea. All these data indicate that Paa contributes to the intimate-adherence phenotype and might be a new adhesin. Its receptor could be HS as with other pathogens. Paa may have a role similar to that of other adherence-conferring molecules of E. coli such as Efa1, Iha, ToxB, and Afa. Efa1 influences colonization of the bovine intestine by Shiga toxin-producing E. coli (47), while Iha facilitates the adherence of E. coli O157:H7 to epithelial cells (48). ToxB is important for full expression of adherence by affecting the production and secretion of some virulence factors required for the development of A/E lesions with O157:H7 strains (49), and it was suggested that EPEC Afa functions as an initial adhesin (26). The more precise role of paa, which is associated not only with AEEC but also with some pig enterotoxigenic E. coli strains (2), is under investigation.
Acknowledgments
This work was supported in part by the Fonds pour la Formation des Chercheurs et l'Aide à la Recherche du Québec, grant 0214, by the Natural Sciences Engineering Research Council of Canada, grant 215841-98, and by EU-Community Quality of Life QLK2 2000-0060.
We thank Bernadette Foiry for her technical assistance, Guy Beauchamp for statistical analysis, Diane Montpetit and Robert Alain for electron microscopy, Hojabr Dezfulian for pulsed-field gel electrophoresis, and John Leong for critical reading of the manuscript.
Editor: B. B. Finlay
REFERENCES
- 1.Akita, E. M., and S. Nakai. 1993. Comparison of four purification methods for the production of immunoglobulins from eggs laid by hens immunized with an enterotoxigenic E. coli strain. J. Immunol. Methods 160:207-214. [DOI] [PubMed] [Google Scholar]
- 2.An, H., J. M. Fairbrother, C. Desautels, and J. Harel. 1999. Distribution of a novel locus called Paa (porcine attaching and effacing associated) among enteric Escherichia coli. Adv. Exp. Med. Biol. 473:179-184. [DOI] [PubMed] [Google Scholar]
- 3.An, H., J. M. Fairbrother, C. Desautels, T. Mabrouk, D. Dugourd, H. Dezfulian, and J. Harel. 2000. Presence of the LEE (locus of enterocyte effacement) in pig attaching and effacing Escherichia coli and characterization of eae, espA, espB and espD genes of PEPEC (pig EPEC) strain 1390. Microb. Pathog. 28:291-300. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin, T. J., W. Ward, A. Aitken, S. Knutton, and P. H. Williams. 1991. Elevation of intracellular free calcium levels in HEp-2 cells infected with enteropathogenic Escherichia coli. Infect. Immun. 59:1599-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barondess, J. J., and J. Beckwith. 1990. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature 346:871-874. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 8.Broes, A., R. Drolet, M. Jacques, J. M. Fairbrother, and W. M. Johnson. 1988. Natural infection with an attaching and effacing Escherichia coli in a diarrheic puppy. Can. J. Vet. Res. 52:280-282. [PMC free article] [PubMed] [Google Scholar]
- 9.Cantey, J. R., and R. K. Blake. 1977. Diarrhea due to Escherichia coli in the rabbit: a novel mechanism. J. Infect. Dis. 135:454-462. [DOI] [PubMed] [Google Scholar]
- 10.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1992. Enteropathogenic Escherichia coli. Infect. Immun. 60:3953-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 13.Everiss, K. D., K. J. Hughes, and K. M. Peterson. 1994. The accessory colonization factor and toxin-coregulated pilus gene clusters are physically linked on the Vibrio cholerae 0395 chromosome. DNA Seq. 5:51-55. [DOI] [PubMed] [Google Scholar]
- 14.Fagundes-Neto, U. 1996. Enteropathogenic Escherichia coli infection in infants: clinical aspects and small bowel morphological alterations. Rev. Microbiol. 27:117-119. [Google Scholar]
- 15.Foubister, V., I. Rosenshine, M. S. Donnenberg, and B. B. Finlay. 1994. The eaeB gene of enteropathogenic Escherichia coli is necessary for signal transduction in epithelial cells. Infect. Immun. 62:3038-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis, D. H., J. E. Collins, and J. R. Duimstra. 1986. Infection of gnotobiotic pigs with an Escherichia coli O157:H7 strain associated with an outbreak of hemorrhagic colitis. Infect Immun. 51:953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harel, J., C. Forget, M. Ngeleka, M. Jacques, and J. M. Fairbrother. 1992. Isolation and characterization of adhesin-defective TnphoA mutants of septicaemic porcine Escherichia coli of serotype O115:K-:F165. J. Gen. Microbiol. 138:2337-2345. [DOI] [PubMed] [Google Scholar]
- 18.Harel, J., M. Jacques, J. M. Fairbrother, M. Bosse, and C. Forget. 1995. Cloning of determinants encoding F165(2) fimbriae from porcine septicaemic Escherichia coli confirms their identity as F1C fimbriae. Microbiology 141:221-228. [DOI] [PubMed] [Google Scholar]
- 19.Harel, J., H. Lapointe, A. Fallara, L. A. Lortie, M. Bigras-Poulin, S. Lariviere, and J. M. Fairbrother. 1991. Detection of genes for fimbrial antigens and enterotoxins associated with Escherichia coli serogroups isolated from pigs with diarrhea. J. Clin. Microbiol. 29:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 21.Helie, P., M. Morin, M. Jacques, and J. M. Fairbrother. 1991. Experimental infection of newborn pigs with an attaching and effacing Escherichia coli O45:K“E65” strain. Infect. Immun. 59:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janke, B. H., D. H. Francis, J. E. Collins, M. C. Libal, D. H. Zeman, and D. D. Johnson. 1989. Attaching and effacing Escherichia coli infections in calves, pigs, lambs, and dogs. J. Vet. Diagn. Investig. 1:6-11. [DOI] [PubMed] [Google Scholar]
- 23.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaniga, K., I. Delor, and G. R. Cornelis. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137-141. [DOI] [PubMed] [Google Scholar]
- 26.Keller, R., J. G. Ordonez, R. R. de Oliveira, L. R. Trabulsi, T. J. Baldwin, and S. Knutton. 2002. Afa, a diffuse adherence fibrillar adhesin associated with enteropathogenic Escherichia coli. Infect. Immun. 70:2681-2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 28.Kenny, B., L. C. Lai, B. B. Finlay, and M. S. Donnenberg. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20:313-323. [DOI] [PubMed] [Google Scholar]
- 29.Knutton, S., M. M. Baldini, J. B. Kaper, and A. S. McNeish. 1987. Role of plasmid-encoded adherence factors in adhesion of enteropathogenic Escherichia coli to HEp-2 cells. Infect. Immun. 55:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovach, M. E., M. D. Shaffer, and K. M. Peterson. 1996. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology 142:2165-2174. [DOI] [PubMed] [Google Scholar]
- 31.Lai, L. C., L. A. Wainwright, K. D. Stone, and M. S. Donnenberg. 1997. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect. Immun. 65:2211-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansfield, K. G., K. C. Lin, D. Xia, J. V. Newman, D. B. Schauer, J. MacKey, A. A. Lackner, and A. Carville. 2001. Enteropathogenic Escherichia coli and ulcerative colitis in cotton-top tamarins (Saguinus oedipus). J. Infect. Dis. 184:803-807. [DOI] [PubMed] [Google Scholar]
- 33.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 35.McNamara, B. P., A. Koutsouris, C. B. O'Connell, J. P. Nougayrede, M. S. Donnenberg, and G. Hecht. 2001. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Investig. 107:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mellies, J. L., F. Navarro-Garcia, I. Okeke, J. Frederickson, J. P. Nataro, and J. B. Kaper. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect. Immun. 69:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moxley, R. A., and D. H. Francis. 1986. Natural and experimental infection with an attaching and effacing strain of Escherichia coli in calves. Infect Immun. 53:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohnishi, M., K. Kurokawa, and T. Hayashi. 2001. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 9:481-485. [DOI] [PubMed] [Google Scholar]
- 40.Oswald, E., H. Schmidt, S. Morabito, H. Karch, O. Marches, and A. Caprioli. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect. Immun. 68:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 42.Peterson, K. M., and J. J. Mekalanos. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect. Immun. 56:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plotkowski, M. C., A. O. Costa, V. Morandi, H. S. Barbosa, H. B. Nader, S. de Bentzmann, and E. Puchelle. 2001. Role of heparan sulphate proteoglycans as potential receptors for non-piliated Pseudomonas aeruginosa adherence to non-polarised airway epithelial cells. J. Med. Microbiol. 50:183-190. [DOI] [PubMed] [Google Scholar]
- 44.Rostand, K. S., and J. D. Esko. 1997. Microbial adherence to and invasion through proteoglycans. Infect. Immun. 65:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 46.Sperandio, V., J. B. Kaper, M. R. Bortolini, B. C. Neves, R. Keller, and L. R. Trabulsi. 1998. Characterization of the locus of enterocyte effacement (LEE) in different enteropathogenic Escherichia coli (EPEC) and Shiga-toxin producing Escherichia coli (STEC) serotypes. FEMS Microbiol. Lett. 164:133-139. [DOI] [PubMed] [Google Scholar]
- 47.Stevens, M. P., P. M. van Diemen, G. Frankel, A. D. Phillips, and T. S. Wallis. 2002. Efa1 influences colonization of the bovine intestine by Shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect. Immun. 70:5158-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tatsuno, I., M. Horie, H. Abe, T. Miki, K. Makino, H. Shinagawa, H. Taguchi, S. Kamiya, T. Hayashi, and C. Sasakawa. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, K. A., C. B. O'Connell, P. W. Luther, and M. S. Donnenberg. 1998. The EspB protein of enteropathogenic Escherichia coli is targeted to the cytoplasm of infected HeLa cells. Infect. Immun. 66:5501-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tobe, T., I. Tatsuno, E. Katayama, C. Y. Wu, G. K. Schoolnik, and C. Sasakawa. 1999. A novel chromosomal locus of enteropathogenic Escherichia coli (EPEC), which encodes a bfpT-regulated chaperone-like protein, TrcA, involved in microcolony formation by EPEC. Mol. Microbiol. 33:741-752. [DOI] [PubMed] [Google Scholar]
- 53.Tzipori, S., R. Gibson, and J. Montanaro. 1989. Nature and distribution of mucosal lesions associated with enteropathogenic and enterohemorrhagic Escherichia coli in piglets and the role of plasmid-mediated factors. Infect. Immun. 57:1142-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Heijne, G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, C., J. Harel, M. Jacques, C. Desautels, M. S. Donnenberg, M. Beaudry, and J. M. Fairbrother. 1994. Virulence properties and attaching-effacing activity of Escherichia coli O45 from swine postweaning diarrhea. Infect. Immun. 62:4153-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, C., J. Harel, M. Jacques, and J. M. Fairbrother. 1995. Interaction with pig ileal explants of Escherichia coli O45 isolates from swine with postweaning diarrhea. Can. J. Vet. Res. 59:118-123. [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, C., S. Menard, J. D. Dubreuil, and J. M. Fairbrother. 1996. Detection and localization of the EaeA protein of attaching and effacing Escherichia coli O45 from pigs using a monoclonal antibody. Microb. Pathog. 21:205-213. [DOI] [PubMed] [Google Scholar]