Abstract
A Tn551 insertional library of Staphylococcus aureus strain ISP479 was challenged with an antimicrobial peptide (CG 117-136) derived from human neutrophil cathepsin G (CG). After repeated selection and screening of surviving colonies, a mutant was identified that expressed increased resistance to CG 117-136. Southern hybridization analysis revealed that the Tn551 insert in this mutant (SK1) was carried on a 10.6-kb EcoRI chromosomal DNA fragment. Subsequent physical mapping of this Tn551 insert revealed that it was positioned between a putative promoter sequence and the translational start codon of the cspA gene, which encodes a protein (CspA) highly similar to the major cold shock proteins CspA and CspB of Escherichia coli and Bacillus subtilis, respectively. This Tn551 insertion as well as a separate deletion-insertion mutation in cspA decreased the capacity of S. aureus to respond to the stress of cold shock and increased resistance to CG 117-136. The results indicate for the first time that a physiologic link exists between bacterial susceptibility to an antimicrobial peptide and a stress response system.
Staphylococcus aureus is notorious for its capacity to both cause a wide range of infections, many of which can be life-threatening, and express resistance to multiple antibiotics. In recent years the problem of antibiotic resistance and S. aureus infections has gained significant attention because of the dearth of new effective antibiotics entering clinical practice and the emergence of strains expressing decreased susceptibility to vancomycin. Accordingly, new antimicrobials effective against antibiotic-resistant strains are needed. In this respect, there has been an increased interest in recent years in the development of antimicrobial peptides (APs) as therapeutic agents to treat bacterial infections that are not responsive to classical antibiotics (17).
Over the past 20 years, hundreds of APs have been isolated from plants, invertebrates, and vertebrates (6) and are thought to represent an integral arm of the host innate immune response to infection (3, 4, 7, 9, 10, 13, 42). In support of this hypothesis, the recent study of Nizet et al. (25) provided direct evidence for the role of an AP, termed LL-37, in host defense against group A streptococci. Levels of bacterial susceptibility to APs appear to be genetically controlled, since mutations in genes involved in cell wall or membrane structure or those encoding transcriptional regulatory proteins can alter levels of bacterial susceptibility to the lethal action of APs (12, 29). To date, the overwhelming amount of work dealing with the genetics of bacterial AP susceptibility has used gram-negative pathogens, notably Salmonella enterica serovar Typhimurium. Progress has been made, however, in recent years with respect to the identification of genes in gram-positive pathogens that are involved in determining AP susceptibility. As an example, Peschel et al. (29-32) have provided evidence for the role of certain covalent modifications of teichoic acid and phosphatidylglycerol in the capacity of S. aureus to express decreased susceptibility to APs.
APs and larger antimicrobial proteins that are stored within the cytoplasmic granules of human polymorphonuclear leukocytes (PMNs) have been implicated in the nonoxidative killing of phagocytosed bacteria (7, 42). Studies dealing with the identification of PMN-derived APs can provide insight as to how pathogens might survive killing by PMNs during infection. In this respect, Reeves et al. (33) suggested that the neutrophil-derived lysosomal proteases cathepsin G (CG) and elastase carry the burden of intraphagosomal killing of phagocytosed S. aureus. CG has been known for over 25 years for its capacity to exert in vitro bactericidal activity against staphylococci by a mechanism independent of its serine protease activity (27, 28). CG killing of S. aureus was reported (27) to be a consequence of depolarization of the cytoplasmic membrane.
We have described a cationic AP derived from full-length CG that, like CG, has broad-spectrum antibacterial action in vitro (37, 38). This peptide (CG 117-136) corresponds to residues 117 to 136 of mature CG (34). Of all the 20-mer peptides that span the full-length CG protein, it was the only one that possessed bactericidal activity against S. aureus (37, 38). CG 117-136 contains two domains (residues 117 to 123 and 129 to 136) of the full-length protein that, by X-ray crystallographic analysis, are predicted to be surface exposed (14). Our investigators recently reported that the bactericidal activity of CG 117-136 can be significantly enhanced by covalent attachment of the C-12 saturated fatty acid dodecanoyl to the N or C terminus (39). Circular dichroism measurements suggested that in phosphate buffer both the modified and unmodified peptides are in a random coil, but the C-12 attachment potentiates the formation of an α-helical structure of CG 117-136 in model membranes (22). The formation of this structure paralleled damage to artificial membranes, a finding that is consistent with earlier studies that linked CG killing of S. aureus to membrane depolarization (27).
In order to gain more information regarding the mechanism(s) by which CG 117-136 exerts bactericidal activity against S. aureus, we sought mutants that expressed decreased susceptibility to its lethal action. By screening an erm(C) insertional library of S. aureus (strain ISP479), we identified a mutant that expressed decreased susceptibility to CG 117-136 and its C-12-modified variant. The Tn551 insertion in this mutant was located in an untranslated sequence between a putative promoter sequence and the translational start codon for the major cold shock gene cspA (8). This mutation as well as an insertional-deletion mutation in the cspA coding sequence impacted both the susceptibility of S. aureus to the CG peptides and its capacity to respond to the stress imposed by cold shock (growth at 15°C). We propose that the physiologic events associated with cold shock response gene expression of S. aureus are linked to its susceptibility to an AP. To our knowledge, this is the first report that has associated a bacterial stress response gene to AP susceptibility.
MATERIALS AND METHODS
Bacterial strains and plasmids used.
The strains and origins of S. aureus and Escherichia coli and plasmids used in this investigation are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Genotype | Source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80 ΔlacZ ΔM15 Δ(lacZA-argF) U169 deoR recA1 endA1 hsdR17 (r−K m+K)phoA supE44 λ−thi-1 gyrA96 relA1 | Stratagene |
| TOP10 | Chemically competent E. coli strain F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZ ΔM15 ΔlacX74deoR recA1 araD139 Δ(ara-leu) 7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| S. aureus strains | ||
| RN4220 | rsbU res mutant strain used for electroporation with E. coli-replicated plasmids | J. Iandolo (18) |
| 8325-4 | rsbU mutant and plasmid free | J. Iandolo (26) |
| ISP479 | Library of rsbU mutant carrying Tn551 insertional sequence in the chromosome | J. Iandolo |
| SH1000 | rsbU+ variant of strain 8325-4 | K. Bayles (15) |
| SK1 | ISP479 mutant with Tn551 in 5′-UTR of cspA gene | This study |
| SK2 | ISP479 mutant with Tn551 insertion element upstream of the thioredoxin reductase gene | This study |
| SK10 | SK2 control with a Km cassette deletion-insertion of the cspA coding region | This study |
| SK12 | SH1000 with Tn551 in 5′-UTR of cspA gene | This study |
| SK29 | SH1000 with a Km cassette deletion-insertion of the cspA coding region | This study |
| Plasmids | ||
| pIM36 | Construct containing 1.2-kb probe for identification of Tn551 insertion sites by Southern hybridization | J. Iandolo |
| pUC18 | High-copy-number E. coli host Ampr | 45 |
| pUC18K | pUC18 with aphA-3 nonpolar Km cassette Ampr | 23 |
| pSPTI81(ts) | Low-copy-number shuttle vector with Ampr in E. coli and temperature-sensitive replication with Tetr in S. aureus | 16 |
| pBT2 | Low-copy-number shuttle vector with Ampr in E. coli and temperature-sensitive replication with Cmr in S. aureus | 5 |
| pCR2.1 | High-copy-number PCR cloning vector; Ampr and Kmr in E. coli | Invitrogen |
| pUCSA | pUC18 with 548bp cspA complementation sequence | This study |
| pCRCSKO | pCR2.1 vector with ligated 750-bp region upstream of the cspA start codon and 466-bp region downstream of the cspA stop codon | This study |
| pUCSKO | pUC18 vector with 750-bp region upstream of the cspA start codon and 466-bp region downstream of the cspA stop codon | This study |
| pUCSKOKm | pUCSKO with 850-bp nonpolar Km cassette ligated into SmaI site between cspA upstream and downstream regions | This study |
| pSPCSKOKm | pSPT181(ts) with the KOKm insert of pUCSKOKm | This study |
| pBCSA | pBT2 vector with 548-bp cspA complementation sequence | This study |
Growth conditions.
Bacteria were routinely grown on Trypticase soy agar (TSA) or in Trypticase soy broth (TSB), with or without antibiotics (see below), at 30 or 37°C. For cold shock experiments, overnight broth cultures were diluted 1:50 in TSB and grown at 30 or 37°C with shaking to an optical density at 600 nm (OD600) of 0.6, at which time the culture was split into two equal-volume samples. These split cultures were incubated at 15 or 37°C (or 30°C) for an additional 24 h, and growth was measured spectrophotometrically at 600 nm. For antimicrobial peptide assays (see below), the bacteria were grown at 37 or 30°C overnight in Mueller-Hinton Broth (MHB) with shaking and diluted 1:50 in MHB. The diluted cultures were then grown at the required temperature to mid-logarithmic phase.
APs used and bactericidal assays.
The preparation and purification of CG 117-136 and its modification by N-terminal covalent attachment of dodecanoyl have been described previously (37, 38). Prior to use in antimicrobial assays, all APs were dissolved in 0.01% (vol/vol) glacial acetic acid containing 0.2% (wt/vol) fatty acid-free bovine serum albumin (Sigma Chemical Co., St. Louis, Mo.) and stored at −30°C. The CFU reduction assay described previously (24, 37-41) was routinely employed. Results were calculated as average values ± standard deviations from at least three independent experiments, and the significance of values was determined by the paired Student t test.
Selection of Tn551-mediated mutants with increased resistance to CG 117-136.
The Tn551 insertional library of strain ISP479 was kindly provided by J. Iandolo (University of Oklahoma Health Sciences Center, Oklahoma City, Okla.). It was grown overnight in TSB containing 10 μg of erythromycin (Ery) per ml and diluted 1:50 in fresh broth lacking Ery. This culture was grown at 37°C with shaking to mid-logarithmic phase and diluted to 107 CFU/ml in 0.03×-strength TSB. This sample was then exposed to two separate additions of 200 μg of CG 117-136. The sample was incubated with each addition of peptide for 2 h at 37°C and then plated onto TSA with Ery. A separate culture not treated with CG 117-136 was similarly handled. The viability of both samples was determined by dilution plating onto TSA containing Ery, and all agar plates were incubated overnight at 37°C. From the untreated sample, a control colony (termed SK2) was recovered and saved for subsequent use as a control in bactericidal and growth assays. The colonies from the AP-treated culture were pooled and reexposed to CG 117-36 as described above. This procedure was repeated twice, and 10 colonies were then individually selected for testing against CG 117-136 in the CFU reduction assay.
Transduction.
Phage 80α was propagated on strain SK1 and used to transduce the Tn551/cspA sequence with the Eryr marker to strains 8325-4 and SH1000 essentially as described by Shafer and Iandolo (36). Transductants were selected on TSA containing 10 μg of Ery per ml. Insertion of Tn551 within the 5′-untranslated region of cspA was confirmed by PCR (see below). The same technique was used to transduce the cspA::Km sequence from strain SK10 (see below) to strain SH1000; these transductants were selected on TSA containing 25 μg of kanamycin (Km) per ml. The replacement of the cspA gene by the Km cassette was confirmed by PCR.
Isolation of chromosomal and plasmid DNA.
Chromosomal DNA from S. aureus strains was isolated from 1-ml samples containing the appropriate antibiotic from either mid-logarithmic or overnight cultures grown in TSB at 37°C with shaking. The DNeasy kit (Qiagen, Valencia, Calif.) was used with the following changes: lysostaphin at 50 μg/ml replaced the lysozyme in the required lysis buffer, incubation at 37°C was done for 1 h, and 4 μl of RNase A was added to each sample after lysis and incubated at room temperature for 10 min. The manufacturer's suggested protocol was then followed with elution by 200 μl of buffer AE applied to the spin column and the eluate reapplied to the column for a second elution. High-copy-number plasmids were extracted from 3 ml of Luria-Bertani (LB) overnight cultures of E. coli host cells using the standard Qiagen mini-preparation protocol. Medium- to low-copy-number plasmids [pBT2(ts), pSPTI81(ts), and their constructs] were extracted from 5 ml of TSB overnight cultures of S. aureus strains also by using the Qiagen mini-preparation technique (Qiagen). However, prior to the addition of the P2 lysis buffer, the cell pellet was suspended in 250 μl of P1 buffer plus 50 μg of lysostaphin/ml and incubated at 37°C for 1 h. High-copy-number plasmids, pCR2.1, pUC18, and their constructs, were eluted from the spin column with 50 μl of EB buffer. Medium- to low-copy-number plasmids [pSPT181(ts), pBT2(ts), and their constructs] were eluted from the spin column with 30 μl of EB buffer after a 1-min soak, and the eluate was reapplied to the column for another minute before the final spin and collection. Restriction digests were carried out with the appropriate enzymes and buffers (New England Biolabs, Beverly, Mass.).
Southern hybridization.
Five-microgram aliquots of chromosomal DNA samples were digested with EcoRI and subjected to agarose gel electrophoresis. After gel electrophoresis and transfer, the blot was probed with a digoxigenin-labeled (Roche, Indianapolis, Ind.) 1.2-kb HindIII fragment of the transposase gene of the Tn551 insertion element in pIM36 (kindly provided by J. Iandolo).
Identification of the Tn551 insertion sites.
A combination of the techniques of restriction site PCR (RS-PCR) (35) and multiplex restriction site PCR (mRS-PCR) (43) was used to identify the insertion site of Tn551 at both the 5′ and 3′ ends of the element. This technique utilized a set of nested primers for the Tn551 element and three separate restriction site primers for EcoRI, HindIII, and BamHI (Table 2). The restriction site primers and oligomers were designed according to the pattern T7-phage promoter sequence-NNNNNNNNN-restriction site sequence. In brief, RS-PCRs utilized the restriction site primers for EcoRI, BamHI, and HindIII (Table 2) that were individually paired with the most distal Tn551 nested primer from its 5′ or 3′ junction for the initial PCR with chromosomal DNA from strain SK1. The thermal cycler program was 94°C for 5 min, 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min for 30 cycles, followed by 72°C for 10 min. The second RS-PCR was carried out with the same group of RS-PCR primers paired with the next Tn551 nested primer and the PCR template cDNA from the first RS-PCR. Distinct bands of PCR product were gel purified and subjected to automated DNA sequencing using a final Tn551 nested primer (Table 2). After the general location of the Tn551 insertion site was identified in strain SK1, a specific Tn551 primer, Tn515209, upstream of the insertion site and the specific cspA primer CSP3407 downstream of the insertion site were used to PCR amplify the junction of the Tn551 insertion element and the cspA sequence. This product was then subjected to bidirectional sequencing to confirm the exact insertion site. Automated DNA sequencing was performed by the DNA Sequencing Facility of Emory University.
TABLE 2.
Primers used for Tn551 insertion sites determination, complementation and insertion-deletion analysis, and RT-PCR
| Primer | Oligonucleotide sequence (5′ → 3′) |
|---|---|
| Tn55105 | GGGAGCATATCACTTTTCTTG |
| Tn55209 | TTTAGTGGGAATTTGTACCCC |
| Tn55242 | ATTCCCACTAAGCGCTCGGGA |
| Tn53184 | ACCGTTACCTGTTTGTGGCAA |
| Tn53097 | GACAGATGTCACCGTCAAGTT |
| Tn53027 | AAATTTCTCGTAGGCGCTCGG |
| Tn551BamHI | TCACACAGGAAACAGCTATGACC-aNNNNNNNNN-GGATCCb |
| Tn551EcoRI | TCACACAGGAAACAGCTATGACC-NNNNNNNNN-GAATTC |
| Tn551HindIII | TCACACAGGAAACAGCTATGACC-NNNNNNNNN-AAGCTT |
| Pstcsp5 | AACTGCAGAACCAATGCATTGACAAAATAATGAAGT |
| Bamcsp3 | CGGGATCCCGAATGTTGGTGATATAAAAAAAGAGTT |
| Scsp55KO | AACTGCAGAACCAATGCATTGGCATTTTGAACAGTACCCACAATG |
| Scsp53KO | TCCCCCGGGGAAATCTGAAACCTCCAAGACTA |
| Scsp35KO | TCCCCCGGGGGATTCTTAGATTTGAATCATTGA |
| Scsp33KO | CGGGATCCCGTTGCCATCATCTCTGCTTTAATAT |
| KO5371 | GATGACATGTTTAAGCACTATCAA |
| Km3471 | TCAGTAAGTAATCCAATTCGGCTA |
| KO3228 | TTAGCAACTAAACTAAGGATAAGA |
| Km5447 | TAGCCGAATTGGATTACTTACTGA |
| 516S547 | GGTGGCAAGCGTTATCCGGAATTA |
| 316S972 | CGAATTAAACCACATGCTCCACCG |
| Csp5237 | GGTTTAACGCTGAAAAAGGATTCG |
| Csp3407 | TAACAACGTTTGCAGCTTGTGGAC |
N is a randomly added base.
Underlined nucleotides represent restriction endonuclease site sequences.
cspA deletion-insertion construct.
A 750-bp DNA sequence positioned upstream of the cspA translational start codon and a 460-bp sequence downstream of the cspA termination codon were PCR amplified from SK2 chromosomal DNA using the primer pairs KO55/KO53 and KO35/KO33 (Table 2). Both PCR products were gel purified, digested with SmaI, and ligated together. The resulting 1.2-kb fragment was PCR amplified and cloned into pCR2.1 (pCRCSKO) (Invitrogen, Inc., Carlsbad, Calif.) and then subcloned into pUC18. The resulting construct (pUCSKO) was recovered from an E. coli DH5α transformant and digested with SmaI. The SmaI fragment from pUC18K containing the 850-bp nonpolar Km cassette aphA-3 (23) was gel purified and ligated into the SmaI-linearized pUCSKO, and transformants of E. coli DH5α were recovered by selection on LB agar plates containing 30 μg of Km per ml. A representative Km-resistant (Kmr) colony was grown, and its DNA plasmid (pUCSKOKm) was prepared as described above. The correct orientation of the Km cassette was verified by PCR using primers Km5447 and KO3228. In order to create the deletion-insertion in S. aureus control strain SK2, the CSKOKM insert of pUCSKOKm was removed by restriction digestion with PstI and BamHI, gel purified, and ligated into the linearized temperature-sensitive shuttle vector pSPT181(ts) (16). The ligated construct (pSPCSKOKM) was transformed into E. coli DH5α cells with selection on LB agar with 30 μg of Km per ml. The presence of both the pSPT181(ts) vector and the CSKOKm insert was verified from a selected Kmr colony by restriction digest analysis. The new construct (pSPCSKOKm) was then electroporated (1) into S. aureus RN4220, and recovered plasmid DNA from an electroporant was introduced into strain SK2. The protocol of Janzon and Arvidson (16) was followed to generate a double crossover deletion-insertion of the Km cassette for the cspA coding sequence of the S. aureus control strain SK2. In this case, tetracycline (Tet) (5 μg/ml) in TSA and Km (25 μg/ml) in TSA were used to select clones at the nonpermissive temperature, which carried the single crossover of the construct pSPCSKOKm into the chromosome of SK2. The Tet-sensitive (Tets) and Kmr phenotype was used to select clones that had lost the vector pSPT181(ts) and retained the Km cassette at the nonpermissive temperature. The presence of the Km cassette and loss of the cspA coding region in the new construct (SK10) were confirmed by PCR.
Complementation analysis.
A 548-bp segment of the cspA gene of S. aureus strain SK2 was generated by PCR using the primers PSTCSP5 and BAMCSP3 (Table 2). The sequence of the PCR product was confirmed, and the product was treated with PstI and BamHI and ligated into identically digested pUC18 to form the construct pUCSA. Because the copy number of the construct pUCSA was very low in an E. coli DH5α background, it was necessary to carry out a CsCl plasmid purification from a 500-ml culture grown in LB with 50 μg of ampicillin per ml in order to obtain enough cspA insert to ligate the 548-bp PstI- and BamHI-restricted fragment into the temperature-sensitive shuttle vector pBT2(ts) (5). This construct was termed pBCSA. After sequence confirmation of the insert in pBCSA, the construct was transformed into S. aureus strain RN4220 and subsequently into SK10 for complementation of the deletion-insertion of cspA in strain SK10.
RT-PCR.
In order to determine the specific difference in the quantity of cspA RNA that may be responsible for the variation in susceptibility of the SK1 mutant to the APs and to cold shock, samples of both SK1 and SK2 were collected during a cold shock experiment. A 20-ml sample of each culture at 37°C was collected at the split point of the cold shock experiment (OD600 of 0.550 to 0.650), followed by collection of 20-ml samples at 2 h from both the 37°C and 15°C cultures. The samples were centrifuged at 6,000 ×g for 10 min, decanted, and frozen at −70°C. The CFU per milliliter for the initial split sample was determined by serial and plate dilutions on TSA. RNA was extracted using the RNeasy Midi kit from Qiagen, Inc. The manufacturer's protocol was followed, with the suspension of cells at 5.5 × 108 to 1.0 ×1010 CFU in Tris-EDTA buffer, the addition of lysostaphin at 50 μg/ml, and a 37°C incubation for 1 h. SuperScript II RNase H reverse transcriptase (Invitrogen, Inc.) was used with 200 ng of RNA for each sample according to the manufacturer's instructions for first-strand cDNA synthesis, while AmpliTaq (Applied Biosystems) was used to generate the amplified PCR transcriptional products. Primers for the 16S rRNA control transcript were 516S547 and 316S972. Primers for the cspA transcriptional product were CSP5237 and CSP3407 (Table 2). The relative amounts of transcript were determined from equal volumes (10 μl of a 50-μl reaction volume) of the reverse transcription-PCR (RT-PCR) product. Samples were electrophoresed into a 1.2% agarose gel containing ethidium bromide. The bands of product were photographed using a Stratagene transilluminator 4000 and a Kodak DC 290 digital camera. The Kodak 1D analysis program was used to estimate pixel intensities of equal areas of each of the 12 bands.
Nucleotide sequence accession number.
The nucleotide sequence of the S. aureus cspA gene was assigned accession number AF259960 in GenBank.
RESULTS AND DISCUSSION
Identification of aTn551 insertional mutant with increased resistance to CG 117-136.
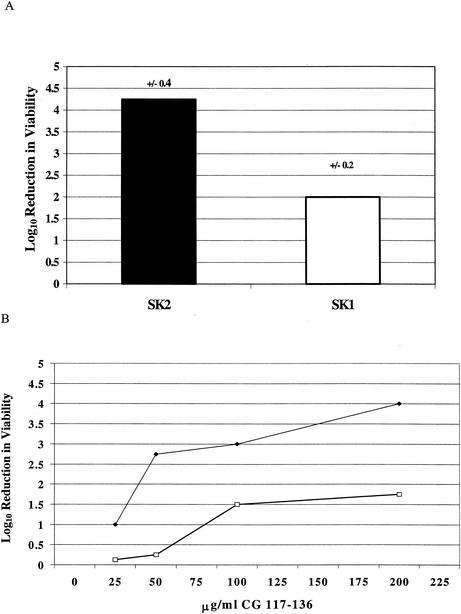
Using the selection process described in Materials and Methods, 4 of the 10 colonies recovered after repeated exposure to CG 117-136 expressed increased resistance, compared to control strain SK2, to this AP (Fig. 1). Southern blot hybridization analysis of EcoRI-digested chromosomal DNA from the four colonies revealed that Tn551 was carried on a 10.6-kb EcoRI fragment (data not presented) compared to the 8.1-kb Tn551-containing fragment of control strain SK2. As all four resistant colonies displayed the same Southern blot hybridization profile, we viewed them as being siblings and concentrated our efforts on a single colony, which we termed strain SK1. Phage 80α transduction of the Ery resistance marker of Tn551 from strain SK1 to strain 8325-4 confirmed linkage of the insert with increased resistance to the peptide (data not presented). This result indicated that the peptide resistance property of strain SK1 was most likely due to the Tn551 insertion and not that of an unlinked, secondary mutation.
FIG. 1.
Susceptibility of strains SK1 and SK2 to CG 117-136. (A) The log10 reduction in viability (CFU per milliliter) when strains SK1 and SK2 were incubated with CG 117-136 (200 μg/ml). Values are averages from three independent experiments, with the standard deviation shown above each bar. The difference in susceptibility was significant (P = 0.012). (B) Susceptibility of strains SK1 (□) and SK2 (♦) to different concentrations of CG 117-136.
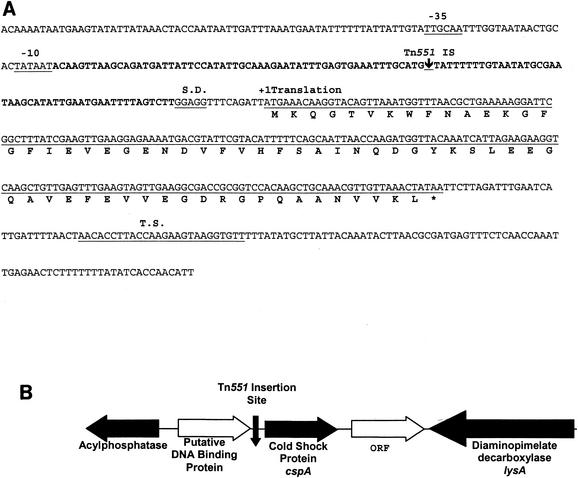
Characterization of the Tn551 insertional site and surrounding DNA sequence in strain SK1.
The site of the Tn551 insertion in strain SK1 was determined as described in Materials and Methods. As is shown in Fig. 2, the insertion site in strain SK1 was located within a T-rich untranslated region (UTR) located 61 bp upstream of the putative translational start codon of an open reading frame previously identified as the cspA gene (8); the location and orientation of surrounding open reading frames are also shown in Fig. 2. PCR and DNA sequencing analysis confirmed that the transductant of strain 8325-4 bearing the Eryr determinant from strain SK1 (see above) also contained Tn551 inserted at this site (data not presented).
FIG. 2.
The major cold shock gene, cspA, of S. aureus. (A) The putative −10 and −35 sites for the promoter and the Shine-Dalgarno (S.D.) site of cspA are labeled and underlined. The putative 5′ UTR is shown in bold and covers a region from the −10 site to the Shine-Dalgarno site. A down-pointing arrow designates the Tn551 transposon insertion site, located within the 5′ UTR. The start site of translation is labeled and marked with a +1. The putative coding sequence is underlined. The amino acid sequence is identified by the single-letter designation under the coding sequence, with an asterisk for the stop codon. After the coding sequence, a putative hairpin termination site (T.S.) is labeled above the sequence and underlined. (B) The chromosomal arrangement of identified and unidentified genes and their protein products located upstream and downstream of the major cold shock gene, cspA. A down-pointing arrow designates the Tn551 transposon insertion site.
The cspA gene of S. aureus would encode a 66-amino-acid polypeptide (Mr of 7.32 kDa) with a predicted pI equal to 4.54. It has 60% and 76% identity, over the entire amino acid sequence, with the major cold shock proteins (CspA and CspB) of E. coli (44) and Bacillus subtilis (11), respectively (Fig. 3).
FIG. 3.
Alignment of the major cold shock proteins of S. aureus (CspA), B. subtilis (CspB), and E. coli (CspA). The amino acid sequence of S. aureus CspA is shaded along with identical amino acids in the B. subtilis and E. coli protein sequences. A missing amino acid is noted by a dash. A consensus sequence is located over each section of the alignment and is underlined.
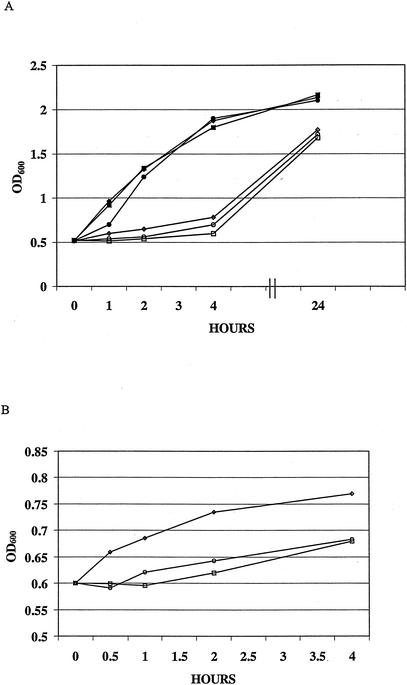
Growth of strains SK1 and SK2 after cold shock.
In order to determine whether the Tn551 insertion in strain SK1 impacted its ability to respond to cold shock, we monitored the growth of strains of SK1 and SK2 in TSB at 15 and 37°C after initially growing the strains to mid-logarithmic phase at 37°C. In several experiments we observed that strain SK1 was less able than strain SK2 to respond to the shift in temperature (37 to 15°C) during the first 4 h of incubation at 15°C (Fig. 4), but ultimately it reached the same OD as strain SK2.
FIG. 4.
Growth of S. aureus strains SK1 (cspA::Tn551), SK2 (Control), and SK10 (cspA::Km) at 37 and 15°C. (A) The growth curves began when the cultures were split at mid-logarithmic phase (OD600 = 0.55 to 0.65) at 37°C and continued for 24 h at both 37 and 15°C. The ODs of samples were measured at the split and at 1, 2, 4, and 24 h after the initiation of cold shock. (B) The expanded cold shock growth curves from panel A of strains SK1, SK2, and SK10 at 15°C after the split at mid-logarithmic phase. Samples were measured at the split and at 1, 2, and 4 h. All ODs for 15°C growth are reported at an initial OD600 of 0.600. (▪), SK1 at 37°C; □, SK1 at 15°C; ♦, SK2 at 37°C; ⋄, SK2 at 15°C; •, SK10 at 37°C; ○, SK10 at 15°C.
Transcriptional analysis of cspA.
In order to determine whether the Tn551 insert in strain SK1 impacts transcription of cspA, we used RT-PCR to compare transcript levels in strains SK1 and SK2. Cultures were grown in TSB to mid-logarithmic phase at 37°C. Twenty-milliliter samples were processed for RNA, and the remaining culture volume was divided into two equal samples. These cultures were grown for 2 h at 37 or 15°C, and total RNA was then isolated. Equal quantities (0.2 μg) of RNA were used in RT-PCRs to detect the cspA transcript and 16S rRNA. The results, shown in Fig. 5, showed that the level of cspA in strain SK2 was higher than that in strain SK1 in all three RNA samples. A comparison of the ratio of band intensities (cspA/16S rRNA), also shown in Fig. 5, supported this conclusion. It should be noted, however, that the degree of induction of cspA expression in the 15°C culture compared to that in the 37°C culture was approximately the same (1.84 for SK1 versus 1.82 for SK2). Taken together, we concluded that while the Tn551 insertion in SK1 down-regulated expression of cspA, it did not eliminate its induction during growth at 15°C.
FIG. 5.
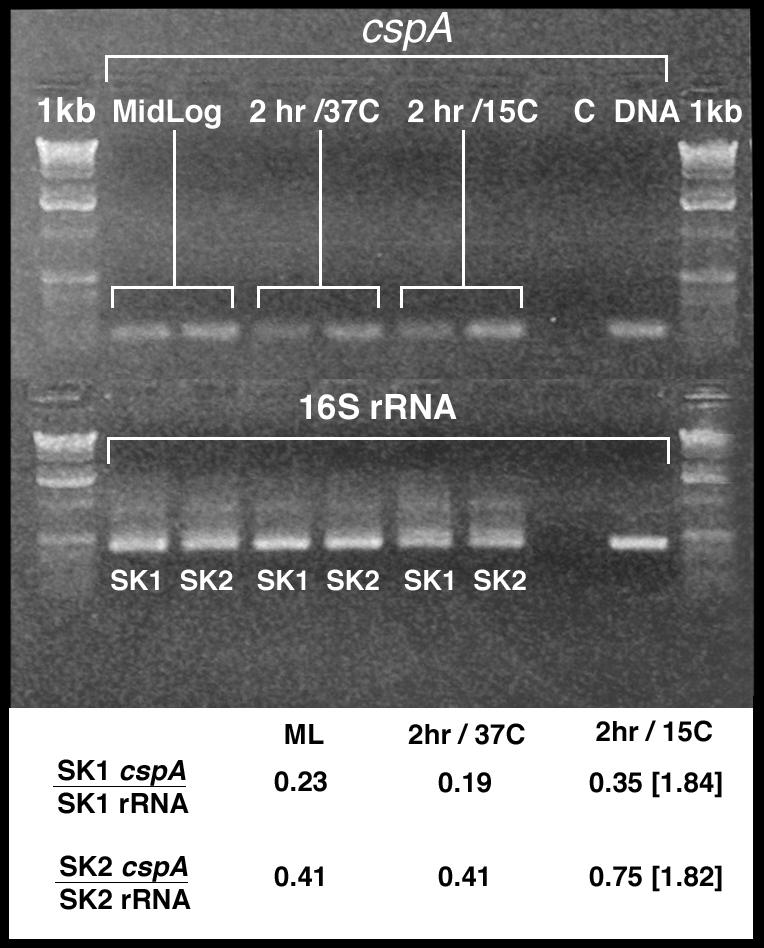
RT-PCR analysis of cspA and 16S rRNA transcripts in the strains SK1 and SK2. (Upper panel) Culture samples for RNA extraction were taken at mid-logarithmic phase, 2 h post-mid-log at 37°C, and 2 h post-mid-log at 15°C. RT-PCR was performed with 200 ng of RNA for both cspA and 16S rRNA reactions. (Lower panel) Changes in cspA transcription were estimated from the ratio of cspA band intensity to that of the 16S rRNA band intensity. The brackets represent induction of cspA, calculated as the ratios obtained from the 15°C versus 37°C samples at the 2-h time point.
The cspA-coding region is required for CG 117-136 susceptibility.
In order to verify that the cspA gene is important in determining both levels of bacterial susceptibility to CG 117-136 and its capacity to respond to the stress of cold shock, we created an insertional-deletion mutant of cspA in strain SK2, as described in Materials and Methods. A colony was obtained (SK10) that contained the cspA::Km sequence from pSPCSK0Km in the chromosome replacing the wild-type cspA sequence; this replacement was verified by PCR (data not presented). Similar to the delayed cold shock response of strain SK1, strain SK10 displayed a reduced capacity to respond to the cold shock stress when grown in TSB at 15°C after initially growing at 37°C (Fig. 4). Strain SK10 was found to be nearly threefold more resistant than parental strain SK2 to the bactericidal action of CG 117-136 and the C-12-modified peptide variant (data not presented).
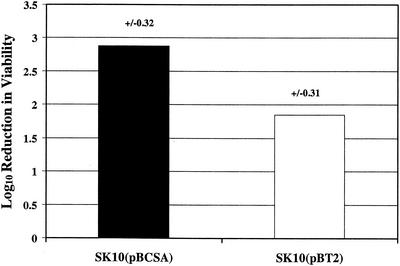
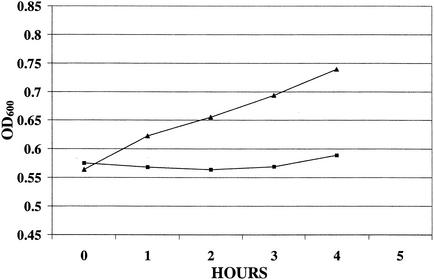
We expressed wild-type cspA in trans in strain SK10 to determine if it would complement its cspA insertional-deletion mutation and to ascertain what impact cspA copy number might have on AP susceptibility levels and the ability of S. aureus to respond to the stress imposed by cold shock. A plasmid construct (pBSCA) bearing the cspA-coding region and 214 bp of upstream and 134 bp of downstream noncoding DNA was introduced into strain SK10. In a separate electroporation reaction, the vector plasmid (pBT2) was first introduced into strain RN4220, and from this host it was electroporated into strain SK10. Because pBT2 is temperature sensitive for replication (5), bactericidal and temperature-shift growth assays were performed at 30°C. The results revealed that SK10(pBSCA) was less resistant to the C-12-modified variant of CG 117-136 (CG 117-136C12) than strain SK10(pBT2) (Fig. 6). As is shown in Fig. 7, strain SK10(pBSCA) grew more rapidly at 15°C than strain SK10(pBT2), indicating that the presence of cspA was needed for a rapid acclimation to cold shock.
FIG. 6.
Susceptibility of strains SK10(pBCSA) and SK10(pBT2) to CG 117-136C12. Shown are the log10 values for the reduction in viability (in CFU per milliliter) of strains incubated with CG 117-136C12 (25 μg/ml). Values are averages from three independent experiments, with the standard deviation shown above each bar. The difference in susceptibility was significant (P = 0.002).
FIG. 7.
Cold shock growth curves of strains SK10(pBCSA) and SK10(pBT2) at 15°C after the split at mid-logarithmic phase (OD600 = 0.55 to 0.65). The ODs of samples were measured at the split and 1, 2, 3, and 4 h after the initiation of cold shock. ▴, SK10(pBCSA); ▪, SK10(pBT2).
CspA-mediated susceptibility to CG 117-136C12 is independent of the secondary sigma factor SigB.
SigB is critically involved in the response of S. aureus to certain stress conditions (15, 19) and in the capacity of the pathogen to express genes encoding virulence factors (15, 20). The strains ISP479 and 8325-4 used in the experiment described above produce an inactive SigB because of an 11-bp deletion in their rsbU gene (15), which encodes a phosphatase that controls the activity of RsbV. Dephosphorylated RsbV binds the anti-SigB competitor RsbW to activate SigB. In strains ISP479 and 8325-4, RsbV remains phosphorylated and SigB is inactive because it is constitutively bound by RsbW. In order to determine whether SigB might regulate the capacity of CspA to modulate staphylococcal susceptibility to CG 117-136C12, the Tn551 insertion upstream of cspA of strain SK1 and the cspA::Km sequence of strain SK10 were introduced into strain SH1000, which is a derivative of strain 8325-4 with a wild-type rsbU gene (15). The presence of these mutations was confirmed by PCR (data not presented). We tested strains SH1000, 8325-4 (parent of strain SH1000) (15), and the genetic derivatives of strain SH1000 having Tn551 upstream of cspA (strain SK12) or cspA::Km (strain SK29) for their susceptibility to CG 117-136C12. There was no significant difference in the susceptibility of strain 8325-4 (SigB inactive) and SH1000 (SigB active) to the lethal action of CG 117-136C12, suggesting that SigB does not control S. aureus susceptibility to this AP (data not presented). However, as is shown in Table 3, strains SK12 and SK29 were significantly less susceptible than the parental strain SH1000 to this AP. Interestingly, strain SK29 was significantly (P = 0.045) less susceptible to CG 117-136C12 than strain SK12. This observation is consistent with cspA being deleted in strain SK29, as opposed to changes in gene expression due to the presence of Tn551 in strain SK12. Hence, taken together with the data obtained with strains SK1 and SK10, the susceptibility of S. aureus to CG 117-136C12 mediated by cspA expression is not dependent on the presence of an active SigB.
TABLE 3.
SigB-active strains of S. aureus have decreased susceptibility to CG 117-136C12 due to cspA mutations
| Strain | CspA | Log10 reduction in viabilitya
|
|
|---|---|---|---|
| +CG 117-136C12b | Controlc | ||
| SH1000 | Wild-type | 2.61 (±0.44) | −0.51 (±0.47) |
| SK12 | Tn551 insertion upstream of cspA | 1.45 (±0.56) | −0.41 (±0.35) |
| SK29 | cspA::Km | 0.83 (±0.48) | −0.32 (±0.19) |
Results are from four assays that employed 25 μg of CG 117-136C12 per ml.
Results from CFU reduction assay are reported as average values (standard deviation). The differences between strains SH1000 and SK12 and between SH1000 and SK29 were significant (P values of 0.044 and 0.008, respectively).
Negative numbers reflect growth of the control culture over the 2-h incubation.
Bacteria can acclimate themselves to the stress imposed by suboptimal growth temperatures through the enhanced production of a group of low-molecular-weight, cationic proteins. In general, these cold shock proteins (Csp's) have been divided into two classes (44). Class I Csp's are synthesized by bacteria at low levels when grown at 37°C and then are significantly overproduced when the culture is shifted to 15°C. Class II Csp's are produced to some extent at 37°C and only moderately increased during cold shock. The major Csp of E. coli (CspA) is a member of the class I category. Based on our RT-PCR results with the cspA gene of S. aureus, it would appear that it is also up-regulated after a temperature shift from 37 to 15°C (Fig. 5). The production of CspA at 37°C may be of significance in vivo because it appears to be a major antigen recognized by the humoral immune system during systemic S. aureus infection (21). The functional activity of such anti-CspA antibodies is unknown. Nevertheless, the presence of such antibodies predicts that CspA is produced by S. aureus during human infections. Accordingly, it is important to determine the function of CspA with respect to the physiology of S. aureus and its pathogenic mechanisms.
CspA of E. coli and CspB of B. subtilis have been studied in detail with respect to their capacities to regulate gene expression and their production at 37 and 15°C; these topics have been extensively reviewed (44). CspA has the capacity to regulate genes at the level of transcription by behaving as a single-strand DNA-binding protein. It also regulates protein synthesis by its RNA-binding ability, which prevents formation of stable and unfavorable secondary structures in mRNA. Moreover, the capacity of CspA to regulate gene expression is likely to be temperature independent because, although it is overproduced during cold shock, a considerable amount is present in bacteria at 37°C. We have recently examined the impact of loss of CspA in S. aureus strain COL and found that loss of CspA decreased the susceptibility of strain COL to CG 117-136 (data not presented). Interestingly, both strains were equally sensitive to the bactericidal action of two unrelated APs, LL-37 and protegrin-1 (MICs of 50 and 3.9 μg/ml, respectively), suggesting that the mechanisms of staphylococcalcidal action of these APs may differ.
Loss of CspA in strain COL resulted in a change in the levels of at least 14 bacterial proteins, as judged by comparison of two-dimensional isoelectric focusing-sodium dodecyl sulfate-polyacrylamide gel electrophoresis profiles, regardless of whether the CspA+ or CspA− derivatives of strain COL were grown at 37 or 15°C (data not presented). A significant finding with these strains (and that of strains Newman and SH1000) is that loss of CspA resulted in a concomitant loss of pigment production, which was restored by complementation of the cspA gene in trans (S. Katzif and W. M. Shafer, unpublished data). Because there are numerous changes in protein content due to loss of CspA in S. aureus strains grown at 37 or 15°C, it is not yet possible to determine how many are linked directly to enhanced resistance to CG 117-136 or its C-12-modified variant. How might loss or decreased production of CspA impact levels of S. aureus susceptibility to CG 117-136? It is clear that CspA homologs in other bacteria (44) can impact transcription of certain genes or translation of their respective transcripts. It is possible that the products of these CspA-regulated genes might impact expression of other genes involved in susceptibility to this AP. Alternatively, these proteins could either modify its binding to the bacterial surface or alter its ability to insert into the bacterial cytoplasmic membrane due to changes in membrane structure and/or fluidity. Testing of these hypotheses will require the identification and characterization of the CspA-regulated genes and confirmation of their linkage with susceptibility to CG 117-136.
Recent studies by Nizet et al. (25), Reeves et al. (33), and Belaaouaj et al. (2) have emphasized the importance of APs and the lysosomal serine proteases CG and elastase in neutrophil-killing bacteria. These agents exert bactericidal activity by nonoxidative mechanisms, possibly through depolarizing the cytoplasmic membrane. While the significance of this killing process has been underappreciated due to the presumed potency of the oxidative killing systems, these more recent studies suggest that nonoxidative killing systems carry the burden of intraleukocytic killing of bacteria. With respect to S. aureus, the importance of PMNs can be surmised because neutropenic patients are at increased risk for infection. Accordingly, continued studies that determine the mechanism by which APs and serine proteases exert staphylococcalcidal activity are warranted. Based on our results with CspA, we suggest that genes subject to its regulation encode proteins that modulate CG killing of S. aureus by human PMNs.
Acknowledgments
We thank K. Bayles, J. Iandolo, C. Lee, and A. Peschel for providing bacterial strains and plasmids and L. Pucko for help in preparing the manuscript. We thank P. Dunman for the RNA protocol, and A. T. Abdelal, C. D. Lu, and D. Walthall for their assistance with the CsCl plasmid preparation.
This work was supported by National Institutes of Health grant AI-43316 (W.M.S.) as well as funds from the VA Medical Research Service. W.M.S. is the recipient of a Senior Research Career Scientist Award from the VA Medical Research Service.
Editor: F. C. Fang
REFERENCES
- 1.Augustin, J., and F. Götz. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 66:203-208. [DOI] [PubMed] [Google Scholar]
- 2.Belaaouaj, A., K. S. Kim, and S. D. Shapiro. 2000. Degradation of outer membrane protein A in Escherichia coli killing by neutrophil elastase. Science 289:1185-1188. [DOI] [PubMed] [Google Scholar]
- 3.Bevins, C. L. 1994. Antimicrobial peptides as agents of mucosal immunity. CIBA Found. Symp. 186:250-269. [DOI] [PubMed] [Google Scholar]
- 4.Boman, H. G. 1991. Antibacterial peptides: key components needed in immunity. Cell 65:205-207. [DOI] [PubMed] [Google Scholar]
- 5.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 6.Devine, D. A., and R. E. W. Hancock. 2002. Cationic peptides: distribution and mechanisms of resistance. Curr. Pharm. Des. 8:703-714. [DOI] [PubMed] [Google Scholar]
- 7.Elsbach, P., and J. Weiss. 1988. Phagocytic cells: oxygen-independent systems, p. 445-470. In J. Gallin, I. M. Goldstein, and R. Snyderman (ed.), Inflammation: basic principles and clinical correlations. Raven Press, New York, N.Y.
- 8.Francis, K. P., and G. S. A. B. Stewart. 1997. Detection and speciation of bacteria through PCR using universal major cold-shock protein primer oligomers. J. Ind. Microbiol. Biotechnol. 19:286-293. [DOI] [PubMed] [Google Scholar]
- 9.Gallo, R. L., and K. M. Huttner. 1998. Antimicrobial peptides: an emerging concept in cutaneous biology. J. Investig. Dermatol. 111:739-743. [DOI] [PubMed] [Google Scholar]
- 10.Ganz, T. 1999. Defensins and host defense. Science 286:420-421. [DOI] [PubMed] [Google Scholar]
- 11.Graumann, P., K. S. Schroder, R. Schmid, and M. A. Maraheil. 1996. Cold-shock stress proteins in Bacillus subtilis. J. Bacteriol. 128:4611-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groisman, E. A. 1996. Bacterial responses to host-defense peptides. Trends Microbiol. 4:127-128. [DOI] [PubMed] [Google Scholar]
- 13.Hancock, R. E. W., and G. Diamond. 2000. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 8:402-410. [DOI] [PubMed] [Google Scholar]
- 14.Hof, P., I. Mayr, R. Huber, E. Korzus, J. Potempa, J. Travis, J. Powers, and W. Bode. 1996. The 1.8 Å crystal structure of human cathepsin G in complex with Suc-Val-Pro-PheP-(Oph)2: a janus-faced proteinase with two opposite specificities. EMBO J. 15:5481-5491. [PMC free article] [PubMed] [Google Scholar]
- 15.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. Sigma B modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janzon, L., and S. Arvidson. 1990. The role of the δ-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9:1391-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley, K. J. 1996. Using host defenses to fight infectious diseases. Nat. Bio/Technol. 14:587-590. [DOI] [PubMed] [Google Scholar]
- 18.Kreiswirth, B., S. Lofdahl, M. J. Betley, M. O'Reilly, M. P. Shlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 35:709-712. [DOI] [PubMed] [Google Scholar]
- 19.Kullik, I., and P. Giachino. 1997. The alternative sigma factor sigma B in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167:151-159. [DOI] [PubMed] [Google Scholar]
- 20.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenz, U., K. Ohlsen, H. Karch, M. Hecker, A. Thiede, and J. Hacker. 2000. Human antibody response during sepsis against targets expressed by methicillin-resistant Staphylococcus aureus. FEMS. Immunol. Med. Microbiol. 29:145-153. [DOI] [PubMed] [Google Scholar]
- 22.Mak, P., J. Pohl, A. Dubin, M. S. Reed, S. E. Bowers, M. T. Fallon, and W. M. Shafer. 2003. The increased bactericidal activity of a fatty acid-modified synthetic antimicrobial peptide of human cathepsin G correlates with its enhanced capacity to interact with model membranes. Int. J. Antimicrob. Agents 21:13-19. [DOI] [PubMed] [Google Scholar]
- 23.Ménard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyasaki, K. T., and A. L. Bodeau. 1991. In vitro killing of oral Capnocytophaga by granule fractions of human neutrophils is associated with cathepsin G activity. J. Clin. Investig. 87:1585-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nizet, V., T. Ohtake, X. Lauth, J. Trowbridge, J. Rudisill, R. A. Dorschner, V. Pestonjamasp, J. Piraino, K. Huttner, and R. L. Gallo. 2001. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 414:454-457. [DOI] [PubMed] [Google Scholar]
- 26.Novick, R. P. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33:155-166. [DOI] [PubMed] [Google Scholar]
- 27.Odeberg, H., and I. Olsson. 1975. Mechanisms for the microbicidal activity of cationic proteins of human granulocytes. Infect. Immun. 14:1269-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odeberg, H., and I. Olsson. 1976. Antibacterial activity of cationic proteins from human granulocytes. J. Clin. Investig. 56:1118-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 30.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Götz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 31.Peschel, A., C. Vuong, M. Otto, and F. Götz. 2000. The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolysins. Antimicrob. Agents Chemother. 44:2845-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. M. van Kellsl, and J. A. G. van Strijk. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reeves, E. P., H. Lu, H. L. Jacobs, C. G. M. Messina, S. Bolsover, G. Gabella, E. O. Potma, A. Warley, J. Roes, and A. W. Segal. 2002. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416:291-297. [DOI] [PubMed] [Google Scholar]
- 34.Salvesen, G., D. Farley, J. Shuman, A. Przybla, C. Reiley, and J. Travis. 1987. Molecular cloning of human cathepsin G: structural similarity to mast cell and cytotoxic T lymphocyte proteinases. Biochemistry 26:2289-2293. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar, G., R. T. Turner, and M. E. Bolander. 1993. Restriction-site PCR: a direct method of unknown sequence retrieval adjacent to a known locus by using universal primers. PCR Methods Appl. 2:318-322. [DOI] [PubMed] [Google Scholar]
- 36.Shafer, W. M., and J. J. Iandolo. 1979. Genetics of staphylococcal enterotoxin B in methicillin-resistant isolates of Staphylococcus aureus. Infect. Immun. 25:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shafer, W. M., M. E. Shepherd, B. Boltin, L. Wells, and J. Pohl. 1993. Synthetic peptides of human lysosomal cathepsin G having potent antipseudomonal activity. Infect. Immun. 61:1900-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shafer, W. M., F. Hubalek, M. Huang, and J. Pohl. 1996. Bactericidal activity of a synthetic peptide (CG 117-136) of human cathepsin G is dependent on arginine content. Infect. Immun. 64:4842-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shafer, W. M., S. Katzif, S. Bowers, M. Fallon, M. Hubalek, M. S. Reed, P. Veprek, and J. Pohl. 2002. Tailoring an antibacterial peptide of human lysosomal cathepsin G to enhance its broad-spectrum action against antibiotic-resistant bacterial pathogens. Curr. Pharm. Des. 8:695-702. [DOI] [PubMed] [Google Scholar]
- 40.Shafer, W. M., V. C. Onunka, and L. E. Martin. 1986. Antigonococcal activity of human neutrophil cathepsin G. Infect. Immun. 54:184-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shafer, W. M., and V. C. Onunka. 1989. Mechanisms of staphylococcal resistance to non-oxidative antimicrobial action of human neutrophils: importance of pH and ionic strength in determining the bactericidal activity of cathepsin G. J. Gen. Microbiol. 135:825-830. [DOI] [PubMed] [Google Scholar]
- 42.Spitznagel, J. K. 1990. Antibiotic proteins of human neutrophils. J. Clin. Investig. 86:1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber, K. L., M. E. Bolander, and G. Sarkar. 1998. Rapid acquisition of unknown DNA sequence adjacent to a known segment by multiplex restriction site PCR. BioTechniques 25:415-419. [DOI] [PubMed] [Google Scholar]
- 44.Yamanaka, K. 1999. Cold shock response in Escherichia coli. J. Mol. Microbiol. Biotechnol. 1:193-202. [PubMed] [Google Scholar]
- 45.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. J. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]