Abstract
Background
By screening a plasmid library for proteins that could cause silencing when targeted to the HMR locus in Saccharomyces cerevisiae, we previously reported the identification of Rtt107/Esc4 based on its ability to establish silent chromatin. In this study we aimed to determine the mechanism of Rtt107/Esc4 targeted silencing and also learn more about its biological functions.
Results
Targeted silencing by Rtt107/Esc4 was dependent on the SIR genes, which encode obligatory structural and enzymatic components of yeast silent chromatin. Based on its sequence, Rtt107/Esc4 was predicted to contain six BRCT motifs. This motif, originally identified in the human breast tumor suppressor gene BRCA1, is a protein interaction domain. The targeted silencing activity of Rtt107/Esc4 resided within the C-terminal two BRCT motifs, and this region of the protein bound to Sir3 in two-hybrid tests. Deletion of RTT107/ESC4 caused sensitivity to the DNA damaging agent MMS as well as to hydroxyurea. A two-hybrid screen showed that the N-terminal BRCT motifs of Rtt107/Esc4 bound to Slx4, a protein previously shown to be involved in DNA repair and required for viability in a strain lacking the DNA helicase Sgs1. Like SLX genes, RTT107ESC4 interacted genetically with SGS1; esc4Δ sgs1Δ mutants were viable, but exhibited a slow-growth phenotype and also a synergistic DNA repair defect.
Conclusion
Rtt107/Esc4 binds to the silencing protein Sir3 and the DNA repair protein Slx4 via different BRCT motifs, thus providing a bridge linking silent chromatin to DNA repair enzymes.
Background
Transcriptional silencing in the budding yeast Saccharomyces cerevisiae occurs at the silent mating-type loci HMR and HML, telomeres, and at the rDNA locus. At all of these silenced regions, DNA binding proteins recognize specific motifs and recruit a silencing protein complex (reviewed in [1]). HMR and HML are flanked by E and I "silencers." Each silencer has binding sites for ORC, and Rap1 or Abf1. The potent HMR-E silencer has a binding site for all three proteins. At telomeres, Rap1 also contributes a critical DNA-binding function, binding to the TG1–3 repeats.
At both the silent mating loci and at telomeres, the DNA-binding proteins recruit a Sir protein complex that can spread to silence genes at a distance (reviewed in [2]). At HMR-E, for example, this is achieved by ORC recruitment of Sir1 via a Sir1-Orc1 interaction [3], and Rap1 and Abf1 binding to Sir4 and Sir3 [4,5]. Sir4 and Sir3 multimerize, both with themselves and each other [4,6]. Sir4 also binds Sir2, and Sir2 plays a crucial role in the spreading of a Sir2, Sir3, Sir4 complex on chromatin by deacetylating histone H4 lysine 16. The deacetylation produces a novel product, 2' O-acetyl-ADP-ribose [7-9]. This compound has recently been shown to produce a conformational change in Sir3 that is likely to promote spreading of the Sir complex. Sir2, Sir3 and Sir4 are essential for silencing at the HM loci and at telomeres, while Sir1 plays a prominent role in silencing at the mating type loci but not at telomeres. The Yku70/Yku80 heterodimer that binds to DNA ends plays an important role in silencing at telomeres, while being dispensable for silencing at HM loci [10].
Deletion of the HMR-E silencer leads to loss of silencing of genes at the HMR locus [11,12]. However, if heterologous DNA binding sites (such as Gal4 sites) are integrated in place of HMR-E and the strain is transformed with certain Gal4 DNA binding domain (GBD) – silencing protein hybrids, silencing can be restored due to targeting of Sir proteins or proteins that bind to Sir proteins to HMR-E ([13]; see also Figure 1). This so-called "targeted silencing" has been a useful tool for investigating the process of Sir protein recruitment to silenced loci [3,13-17]. We previously described a screen for proteins capable of targeted silencing at HMR [16]. In this screen many known proteins were identified, as well as several proteins not characterized at the time, which we called Esc proteins because they establish silent chromatin. One of these was Esc4 whose characterization we describe in this report. RTT107/ESC4 was also identified in screens for mutants with increased Ty transposon mobility or DNA repair defects [18-20]. For simplicity we will only use the name ESC4 in the remainder of this paper. Esc4 has been shown to be phosphorylated by Mec1 kinase on SQ/TQ motifs in response to DNA damage during S phase [21]. The phosphorylation by Mec1 has quite recently been proposed to be regulated by Slx4, which was also shown to form a complex containing Esc4 [22].
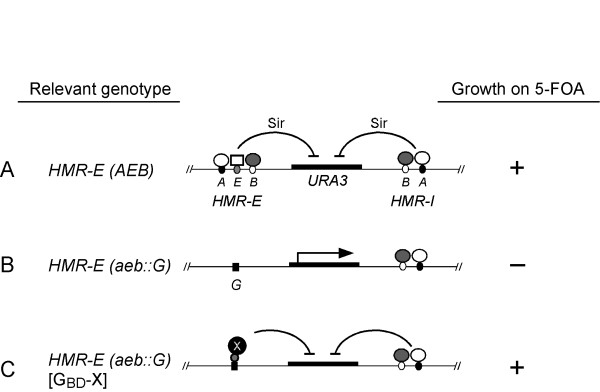
Figure 1.
Esc4 was identified in an HMR targeted silencing screen. (A) In the presence of both wild-type silencers, HMR-E and HMR-I, the URA3 reporter gene at HMR is completely silenced in a SIR-dependent manner leading to growth in the presence of the drug 5-FOA. (B) When the HMR-E silencer is deleted (and here replaced with a binding site for the Gal4 protein (G)), the HMR locus is no longer silenced and the URA3 reporter gene is expressed. This loss of silencing at hmr::URA3 leads to no growth on 5-FOA media. (C) When the strain in (B) is transformed with a GBD plasmid (GBD-X) capable of causing targeted silencing, this leads to restoration of silencing at hmr::URA3 and growth on 5-FOA. In this way, a GBD hybrid protein library was screened for factors capable of targeted silencing. A GBD-Esc4(1–1070) hybrid was identified.
In this report we show that Esc4 has 6 BRCT motifs and that they are important for its function. Many proteins that function in repair, and even a silencing protein, Rap1, contain BRCT (BRCA1 C-terminus) motifs. This motif was first identified by database searching using the C-terminus of the human breast cancer susceptibility protein, BRCA1 [23]. Since then, BRCT motifs from the human DNA repair protein XRCC1 and the two tandem BRCT motifs of BRCA1 protein have been crystallized [24,25] and crystal and/or solution structures have also been solved of BRCTs from 53BP1, DNA ligase III, and an NAD-dependent DNA ligase [26-29]. More recently, it was discovered that BRCT motifs could specifically bind to phosphoserine-containing proteins [30,31] and structures of such complexes were subsequently also determined [32-34]. Overall, BRCT motifs are thought to mediate a diverse array of protein-protein interactions, binding to proteins with different structures, as well as to other BRCTs, both inter- and intra-molecularly (reviewed in [35]).
DNA repair requires many DNA-modifying enzymes such as nucleases, ligases, topoisomerases, polymerases and helicases. Saccharomyces cerevisiae SGS1 is a member of the RecQ family of genes encoding DNA helicases. In addition to Escherichia coli recQ, this helicase gene family includes human BLM, WRN, RECQL, and Schizosaccharomyces pombe rqh1+. Members of the RecQ family of helicases have been implicated in genomic stability, aging and cancer. Yeast SGS1 has been shown by several labs to suppress DNA recombination, gross chromosomal rearrangements, and Ty1 transposition, and to exhibit 3' to 5' helicase activity [20,36-38]. Six SLX genes have been isolated in a synthetic lethal screen using an sgs1Δ mutant [39]. Two of these genes, SLX2 and SLX3, encode the Mus81/Mms4 nuclease that acts on branched DNA structures. SLX1 is also predicted to be a nuclease, based on its sequence [40]. Slx1 and Slx4 coimmunoprecipitate and slx1 and slx4 mutants display similar phenotypes and have been proposed to function together [39]. In addition, a genome-wide genetic interaction (synthetic genetic array, SGA) screen isolated a total of 24 genes that show a synthetic growth interaction with an sgs1Δ mutation [41]. This screen identified 4 of the 6 SLX genes previously identified in the sgs1Δ synthetic lethal screen described above [39], as well as three other genes known to show synthetic interactions [39,42-45]. The screen also identified 16 other genes that caused synthetic lethality or sickness when mutated with sgs1Δ. One of these genes was ESC4. Subsequently, there have been other reports confirming this genetic interaction between SGS1 and ESC4 [46,47].
Here we demonstrate that the C-terminal two BRCT motifs of Esc4 bind to Sir3 and are sufficient for SIR-dependent targeted silencing at HMR. Furthermore, the N-terminal four BRCT motifs in Esc4 bind to Slx4, thus linking this DNA repair protein to silent chromatin.
Results
Esc4 establishes targeted silencing when targeted to HMR
In a screen described previously, we identified Esc4 as a protein that could restore silencing when targeted to an HMR locus harboring a deletion of the HMR-E silencer [16]. Targeting of proteins to HMR was mediated by the binding of a Gal4 DNA binding domain (GBD)-hybrid protein to a Gal4 DNA binding site (G) that replaced the HMR-E silencer (Figure 1). Silencing was assessed using a URA3 reporter gene integrated at the HMR locus.
Targeted silencing by Esc4 is SIR-dependent
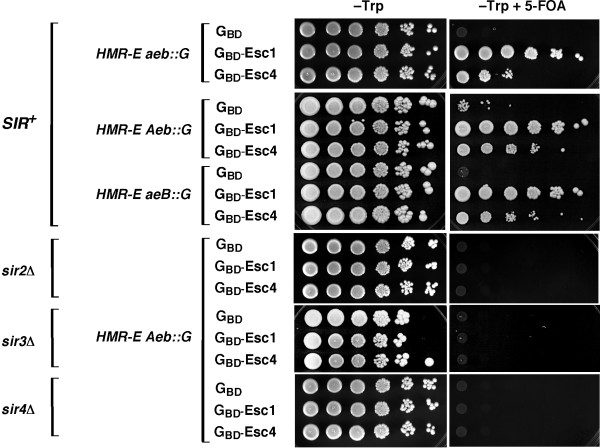
In order to understand how Esc4 promoted silencing when targeted to the HMR locus, we tested silencing by GBD-Esc4 in various strains. First, targeted silencing by GBD-Esc4 was compared to GBD alone or to the potent targeted silencing factor GBD-Esc1 in the strain in which the screen was performed, which has the entire HMR-E silencer deleted and replaced with a Gal4 DNA binding site (HMR-E aeb::G) [17]. The fraction of cells silenced by GBD-Esc4 was not as great as by GBD-Esc1 but, nevertheless, significant targeted silencing at HMR was observed despite the complete absence of the HMR-E silencer (Figure 2, HMR-E aeb::G). When targeted silencing by GBD-Esc4 was assessed in strains harboring deletions of just the E and B sites (HMR-E Aeb::G) or the A and E sites (HMR-E aeB::G) of the HMR-E silencer, silencing was increased roughly 50 to 100-fold, as expected for strains with at least one element of the natural silencer (Figure 2).
Figure 2.
Targeted silencing by Esc4 is SIR-dependent. Targeted silencing by GBD-Esc4 (1–1070) was assessed in hmr::URA3 silencing reporter strains containing different HMR-E elements replaced by a Gal4 DNA binding site (G), similar to the situation shown in Figure 1B. A TRP1-marked plasmid expressing GBD alone served as a negative control and targeted silencing by the potent targeted silencing factor GBD-Esc1(1124–1658) served as a positive control [17]. Targeted silencing by GBD-Esc4 in strains with wild-type SIR genes (SIR+) or sir2Δ, sir3Δ, or sir4Δ mutations was examined.
To test whether targeted silencing by GBD-Esc4 was SIR-dependent, it was tested in strains deleted for the SIR2, SIR3 or SIR4 genes. As seen in Figure 2, silencing by Esc4 required each of these SIR genes. This was expected since targeted silencing by other proteins has been shown to be SIR-dependent in every case examined previously [14,16]. It thus seemed likely that Esc4 caused silencing by recruiting the Sir protein complex.
Saccharomyces Esc4 proteins contain six BRCT motifs and are homologous to S. pombe Brc1
Esc4 contains several copies of the BRCT motif, originally identified in the human breast cancer susceptibility gene, BRCA1 [23]. The presence of these motifs in Esc4 (encoded by yeast ORF YHR154w) has been observed previously by database searches [48,49], although there has been disagreement as to how many BRCTs exist in this protein, with some reports suggesting six BRCTs [49,50] and others only four [21,22,48]. Our own analyses including use of Pfam [51,52] confirms the first five putative BRCTs in Esc4, and it seems likely that residues 935–1049, which are BRCT-like with a conserved W or Y residue near the C-terminus, also form a domain that folds into a BRCT type structure as previously reported [49] (Figure 3). Furthermore, analysis of a homolog of Esc4 from S. pombe, Brc1, using Pfam shows six BRCTs in this homologous protein, with the last two again being the most alternative (E-values of 4.1 × 10-5 and 5.8 × 10-2 for the fifth and sixth, respectively, compared to values ranging from 2.8 × 10-6 to 1.7 × 10-14 for the first four BRCTs), further supporting a conserved total of six motifs in these homologs (Figure 3).
Figure 3.
Saccharomyces cerevisiae Esc4 has six BRCT motifs that are conserved in both sequence and domain architecture among yeast homologs. The six BRCT motifs in Esc4 are indicated with brackets and yellow color and were described previously [49]. The motifs in Esc4 are shown as they align with a subset of Esc4 homologs: three from other budding yeasts Ashbya gossipii, Kluyveromyces lactis and Candida glabrata and one from the evolutionarily distant fission yeast Schizosaccharomyces pombe Brc1. The highly conserved aromatic residue, a hallmark of BRCT motifs, is marked above the alignment with a red asterisk. Blue color indicates similarity with the predominant residue at the position in the alignment, with darkness correlated with greater similarity as calculated by ClustalW software and displayed with Jalview alignment editor's BLOSUM62 score viewing option [67, 68].
The six BRCT motifs of Esc4 protein exist as a set of four tandem motifs at the N-terminus of the protein and two more at the C-terminus (Figure 3, yellow lines). These two sets of BRCT motifs are separated by a 375 amino acid linker region. Although there are proteins from various budding yeasts with compelling sequence similarity to S. cerevisiae Esc4 (e.g. Ashbya gossipii, Kluyveromyces lactis, and Candida glabrata, as shown in Figure 3), currently the only obvious non-budding yeast homolog candidate is Brc1 from the evolutionarily distant fission yeast Schizosaccharomyces pombe (Figure 3, bottom line). S. pombe Brc1 is 878 amino acids in length and also contains four BRCTs at the N-terminus and two at the C-terminus, separated by a linker region. Esc4 and Brc1 are 52% similar and 21% identical overall. Human and mouse PTIP proteins have been reported to be quite similar in sequence [21,53], but do not contain the classically-conserved C-terminal aromatic residue present in all of the six BRCTs highlighted with a red asterisk in the alignment shown in Figure 3. Therefore, it is not yet clear if these proteins are bona fide Esc4/Brc1 homologs.
The C-terminal two BRCT motifs of Esc4 are sufficient for targeted silencing
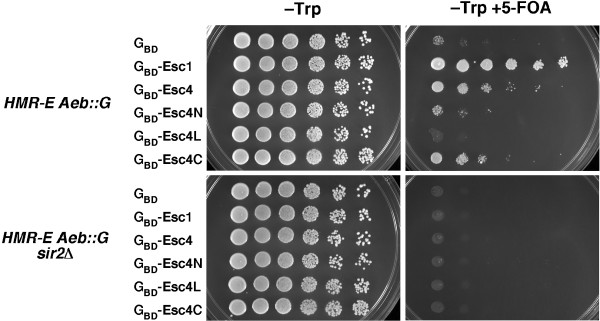
The Esc4 hybrid protein isolated in our targeted silencing screen was full-length [16] and therefore contained all six predicted BRCT motifs. To determine which part of Esc4 and which BRCT motifs were responsible for the SIR-dependent targeted silencing, we constructed three GBD hybrids: one to the N-terminal four BRCT motifs (GBD-Esc4N, aa 1–480), one to the linker between the N- and C-terminal sets of motifs (GBD-Esc4L, aa 480–836), and one to the C-terminal two BRCT motifs (GBD-Esc4C, aa 818–1070). These constructs were tested for targeted silencing in a strain harboring deletions of the E and B sites (HMR-E Aeb::G) and in a sir2Δ derivative of that strain (Figure 4; see Table 1 for strain information). Significant targeted silencing was observed by GBD-Esc4C, although it was not as much as with full-length Esc4. The observed silencing by GBD-Esc4C was SIR-dependent, as observed for the full-length protein (Figure 2). No significant silencing was seen with Esc4N or with the linker region, Esc4L (Figure 4).
Figure 4.
Targeted silencing activity is achieved by the C-terminus of Esc4. Three different regions of Esc4, containing either the first four BRCT motifs (1–480), linker region (480–836), or last two BRCTs (818–1070) were tested for targeted silencing in a strain with an HMR-E Aeb::G silencer and URA3 reporter gene at HMR. GBD-Esc1 served as a potent positive control and GBD as a negative control. SIR2-dependence of targeted silencing was also determined.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or Source |
| W303-1a | MATaleu2–3,112 ura3-1 his3–11,15 trp1–1 ade2-1 can1–100 | R. Rothstein |
| W303-1b | MATα version of W303-1a | R. Rothstein |
| W303 | Diploid from W303-1a × W303-1b | This study |
| YDZ5 | W303 MATa/MATα esc4Δ::his5+/ESC4 | This study |
| YDZ174 | W303 MATa/MATα esc4Δ::his5+/esc4Δ::his5+ | This study |
| YDZ3 | W303-1a esc4Δ::his5+ | This study |
| YDZ4 | W303-1b esc4Δ::his5+ | This study |
| YEA78 | W303-1b hmr::URA3 HMR-E (aeb::G) gal4::LEU2 | This study |
| YEA76 | YEA78 but HMR-E (Aeb::G) | This study |
| YEA77 | YEA78 but HMR-E (aeB::G) | Thi study |
| YAM7 | YEA76 sir2Δ::kanMX6 | This study |
| YEA118 | YEA76 but GAL4 sir3::LEU2 | This study |
| YKAB17 | YEA76 sir4Δ::kanMX6 | This laboratory |
| YDS631 | W303-1b adh4::URA3-(C1–3A)n | [13] |
| YDZ22 | YDS631 esc4Δ::his5+ | This study |
| YAM1 | W303-1a slx4Δ::his5+ | This study |
| YAM2 | W303-1b slx4Δ::his5+ | This study |
| YAM3 | W303-1a slx4Δ::kanMX6 | This study |
| YAM4 | W303-1b slx4Δ::kanMX6 | This study |
| YAM13 | W303-1b sgs1Δ::kanMX6 | This study |
| YDZ243 | W303-1_ sgs1Δ::kanMX6 esc4Δ::his5+ | This study |
| YAM6 | W303-1a esc4Δ::his5+slx4Δ::kanMX6 | This study |
| YAS501 | W303-1a asf1Δ::his5+ | This laboratory |
| L40 | MATahis3Δ200 trp1–901 leu2–3,112 ade2 URA3::(lexAop)8-lacZ LYS2::(lexAop)4-HIS3 | [54] |
| YRJ3 | L40 sir3Δ::kanMX6 | This study |
| YJL103 | L40 sir4Δ::kanMX6 | This study |
The C-terminal BRCT motifs of Esc4 interact with Sir3
Because the C-terminal two BRCT motifs of Esc4 gave targeted silencing, we suspected that this region of the protein was binding to a silencing protein to recruit the Sir complex. Using a LexA-Esc4C hybrid and the two-hybrid reporter strain L40 [54], we tested for two-hybrid interactions with several Gal4 activation domain (GAD)-silencing protein constructs, including Sir1, Sir2, Sir3, Sir4 and Rap1 [3,17,55]. A strong interaction (i.e., the lacZ reporter gene generated blue color from X-gal visible in 15 minutes) with GAD-Sir3 (aa 252–978) was identified, as well as a weaker interaction with GAD-Sir4 (aa 839–1358) (Table 2). None was detected with Sir1, Sir2 or Rap1. Because the GAD-Sir4 (aa 839–1358) hybrid contained the region known to bind to Sir3 [7], we hypothesized that LexA-Esc4 was binding to GAD-Sir4 via a bridge of endogenous Sir3. To test this, we used a derivative of strain L40 harboring a sir3Δ mutation and examined the LexA-Esc4 interaction with GAD-Sir4. In this case, the interaction with GAD-Sir4 was no longer observed, whereas the interaction with GAD-Sir3 and an unrelated two-hybrid control interaction were unaffected (Table 2). When a sir4Δ derivative of L40 was used, no change in the LexA-Esc4 interactions with GAD-Sir3 or GAD-Sir4 was observed, further supporting the idea that Esc4 requires Sir3 to interact with Sir4, and not vice versa. Taken together, the targeted silencing data strongly suggest an interaction (probably a direct one) between the C-terminal BRCT motifs and the silencing protein Sir3.
Table 2.
Esc4 binds to Sir3 in the two-hybrid system
| Interaction in two-hybrid strainsa | ||||
| LexA hybrid | GAD hybrid | L40 | L40 Δ sir3 | L40 Δ sir4 |
| LexA-Esc4 (818–1070) | GAD-Sir3 (252–978) | ++ | ++ | ++ |
| LexA-Esc4 (818–1070) | GAD-Sir4 (839–1358) | + | - | + |
| LexA-Esc4 (818–1070) | GAD vector | - | - | - |
| LexA-Esc4 (1–480) | GAD-Slx4 (383–748) | +++ | +++ | +++ |
a Three +++ signs means an easily visible blue color from the lacZ reporter gene was visible within 15 minutes, whereas two ++ signs means it was visible within 30 minutes and one + sign means it was visible between 40 minutes and two hours.
The N-terminal four BRCT motifs of Esc4 bind to Slx4
The above results indicated that Esc4 caused SIR-dependent targeted silencing primarily by binding to Sir3 through its C-terminal two BRCT motifs. In addition, Esc4 also contains four BRCT motifs at its N-terminus and these BRCT motifs are more similar to those found in various proteins from diverse eukaryotes (Figure 3, and ref. [49]). In order to identify proteins that bind to these N-terminal BRCT motifs, a two-hybrid screen was performed using a LexA-Esc4N (aa 1–480) fusion protein. Strikingly, from screening ~2 × 107 library plasmids expressing GAD protein hybrids, thirteen clones containing in-frame fusions of GAD to Slx4 were identified (Table 3). In-frame GAD fusions to the six different positions in Slx4 were isolated (see Table 3) and all contained at least the C-terminal half of the protein (residues 383–748). The Esc4N-Slx4C two-hybrid interaction was a very strong one and was specific. Also, Slx4 did not show an interaction with Esc4C. Thus, the N-terminal BRCT motifs of Esc4 are not only necessary for binding Slx4, as reported while this manuscript was in preparation [22], but are indeed sufficient for this interaction in vivo. Furthermore, our two-hybrid data show that the region of Slx4 sufficient for Esc4 binding resides in its C-terminus.
Table 3.
The amino-terminal BRCT motifs of Esc4 bind to Slx4
| LexA hybrid | GAD hybrid | Number of isolates |
| LexA-Esc4 (1–480) | GAD-Slx4 (166–748) | 1 |
| LexA-Esc4 (1–480) | GAD-Slx4 (194–748) | 2 |
| LexA-Esc4 (1–480) | GAD-Slx4 (260–748) | 1 |
| LexA-Esc4 (1–480) | GAD-Slx4 (280–748) | 4 |
| LexA-Esc4 (1–480) | GAD-Slx4 (368–748) | 3 |
| LexA-Esc4 (1–480) | GAD-Slx4 (383–748) | 2 |
Genetic and phenotypic analysis of ESC4
A heterozygous diploid strain with a complete deletion of ESC4 was constructed and dissected to generate a null mutant haploid. This esc4Δ mutant grew normally and also mated with normal efficiency (suggesting no gross defect in HM silencing). Furthermore, when an esc4Δ mutation was introduced into a strain with a telomere reporter gene, no telomeric silencing defect was seen (data not shown). Thus, although Esc4 binds to Sir3, Esc4 does not appear to be a protein required for Sir protein-mediated silencing.
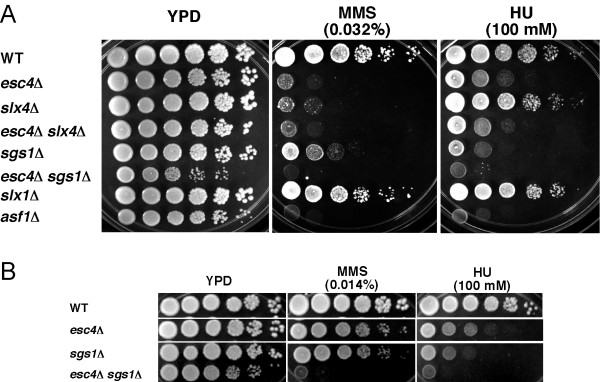
The BRCT motif was originally identified in the human BRCA1 tumor suppressor protein [23]. BRCA1 functions in DNA repair and DNA damage-sensing in cell cycle checkpoints (reviewed in [56]). As shown in Figure 5, strains deleted for ESC4 grew significantly less well than wild type on medium containing either MMS or HU. This result confirms reports that have since been published [18,21,22,46], as does our observation that esc4Δ mutants are not sensitive to ultraviolet radiation [18,21] (data not shown).
Figure 5.
Genetic evidence that ESC4 and SLX4 function together in a parallel pathway to SGS1 for repair of damaged DNA. Strains were grown in YPD medium to saturation and then ten-fold serial dilutions were spotted onto YPD, YPD + HU (100 mM), or YPD + MMS (0.032% in part A and 0.014% in part B) media. Cells were incubated at 30°C for 2 days (SC and SC + MMS) or 3 days (SC + HU) before being photographed. WT, wild type. An asf1Δ mutant was used as a positive control for MMS and HU sensitivity.
As shown in Figure 5, like esc4Δ mutants, slx4Δ mutants were sensitive to 0.032% MMS. Furthermore, the esc4Δ slx4Δ double mutant did not exhibit a greater MMS sensitivity than either single mutant (Figure 5A and data not shown), suggesting that they cooperate in providing resistance to MMS. Another group reported that an esc4Δ slx4Δ double mutant was more sensitive than either single mutant but the difference was very slight [22]. In contrast, an esc4Δ, but not a slx4Δ mutant, was significantly HU-sensitive, and the double mutant was no more sensitive than the esc4Δ strain. Thus, Esc4 appears to act independently of Slx4 in providing resistance to HU.
Because Esc4 bound to Slx4 and because the mutant was sensitive to MMS, this suggested that Esc4 might function in the same pathway as Slx4. SLX4 was first identified in a screen for genes required for viability of yeast cells deleted for SGS1 [39]. Therefore, we tested if esc4 was also synthetically lethal with sgs1. To do this, an esc4Δ mutant was crossed with an sgs1Δ mutant, the diploid was sporulated and dissected and meiotic progeny were analyzed. Haploid esc4Δ sgs1Δ cells were viable, but were noticeably slower-growing that either single mutant (e.g., see Figure 5, YPD plate). While this work was in progress, this genetic interaction was also observed in genome-wide studies [41,46].
An sgs1Δ mutant showed sensitivity to both MMS and HU (Figure 5), as expected based on previously published results. An asf1Δ mutant was used as a control and displayed sensitivity to both DNA damaging chemicals, as expected [57,58]. Interestingly, an esc4Δ sgs1Δ mutant displayed MMS and HU sensitivity that was much more pronounced than that of either single mutant (Figure 5A and 5B). The enhanced sensitivity of this double mutant (10,000-fold on 0.014% MMS plates) seemed to be due to a synergistic repair defect and not entirely due to the growth defect (10-fold difference in colony number on YPD control plates) that was also observed in the esc4Δ sgs1Δ strain.
Discussion
By screening a library of factors that could function in place of the HMR-E silencer when targeted to DNA, we identified Esc4 for its ability to establish silent chromatin [16]. Protein sequence analysis showed that Esc4 protein contains six BRCT motifs; four are found in tandem at the amino-terminus and two more are at the carboxy-terminus. The entire Esc4 protein was present in the hybrid identified in the targeted silencing screen. Since targeted silencing by Esc4 at HMR was found to be SIR-dependent, it seemed likely that some region within Esc4 was attracting a silencing protein complex to DNA. We tested subsets of the BRCT motifs, as well as the linker between them, for targeted silencing at HMR. These experiments demonstrated that the C-terminal two BRCTs caused targeted silencing that was nearly as strong as with full-length Esc4. Because silencing by this pair of BRCT motifs of Esc4 was also SIR-dependent, it seemed very likely that this region was recruiting a Sir protein when tethered to DNA. Therefore, we tested the C-terminal BRCT motifs for interactions with known silencing proteins by two-hybrid analysis. We identified a specific interaction with Sir3 (aa 252–978). We conclude that binding of Sir3 by Esc4 is likely to be responsible for the SIR-dependent targeted silencing activity.
In some cases BRCT motifs have been shown to bind to phosphorylated serine residues. Specifically, they have been shown to bind to phosphopeptides with the following consensus: pSxxF [32]. Interestingly, Sir3 has an SxxF sequence (aa 583–586) within the Esc4-interacting region that we describe here (aa 252–978) and, furthermore, Sir3 protein has been shown to be phosphorylated [59,60], suggesting that an Esc4 BRCT motif or perhaps the combination of the two in the C-terminus may bind to phospho-Sir3. However, not all proteins bound by BRCT motifs have the SxxF motif [61], so the precise BRCT-interacting region of Sir3 could be elsewhere.
In addition to binding Sir3 via C-terminal tandem BRCT motifs, Esc4 also binds to Slx4 via four tandem N-terminal BRCTs, as we have shown here by two-hybrid screening. This two-hybrid result demonstrates that these four BRCTs are sufficient for binding Slx4 and agrees with a recent report showing that the N-terminal BRCT motifs are required for this interaction [22]. It seems quite possible that Esc4 could bind Sir3 and Slx4 concurrently, given that these nuclear proteins' binding sites within Esc4 map to BRCT clusters separated by a long linker. Slx4 has been shown to heterodimerize with the endonuclease Slx1 to cleave DNA containing 5' -flap structures, such as in stalled replication forks, to facilitate their repair [62]. Thus, Esc4 binds the silencing protein Sir3 and also to Slx4, an important DNA repair complex component. Esc4 may play a role in facilitating repair of aberrant DNA structures, perhaps specifically within silent chromatin. Esc4 is a Mec1 kinase target [21] and this phosphorylation is required for its repair function. It is possible that phosphorylation of Esc4 by Mec1, which occurs just N-terminal to the Sir3-binding BRCTs, regulates association with Sir3 or other factors required for its ability to repair particular chromosomal loci in S phase.
We analyzed Esc4 protein alignments (such as that shown in Figure 3) for evidence of conserved regions in the protein other than BRCT motifs. One region of interest was the SQ/TQ motifs between amino acids 743 and 807, which were shown to be important for function in DNA repair [21]. We did not find that these motifs were well-conserved, suggesting that the specific site of phosphorylation is not particularly critical in proteins with otherwise similar overall BRCT domain architecture (i.e., [BRCT]4-linker-[BRCT]2 arrangement). This may be because of some differences in Esc4 functions in diverse yeasts or may suggest that flexibility is tolerated in positioning of the phosphorylation sites, and therefore the exact relative location of kinase target sites has not been constrained during evolution.
Future structural and genome sequencing studies are likely to unveil similarities and differences between multi-BRCT domain containing proteins. Whether these proteins play largely protein-scaffolding roles or also contain intrinsic enzymatic properties will be interesting to discover.
Conclusion
We have shown that Esc4 caused targeted silencing when tethered at a weakened HMR locus. The targeted silencing activity was primarily due to the C-terminal two tandem BRCT motifs in Esc4, which bound to Sir3, probably through a direct interaction. This interaction led to the recruitment of the Sir complex and hence caused targeted silencing. The N-terminal BRCT domains were sufficient for binding to Slx4, which functions with Esc4 in DNA repair. Thus, the nuclear Esc4 protein uses its six BRCT motifs to connect diverse proteins involved in DNA repair and silent chromatin.
Methods
Targeted silencing
Esc4 was identified in a targeted silencing screen that has been described previously [16]. Briefly, a Gal4 DNA binding domain (GBD) library was screened for hybrid proteins capable of establishing silencing of a URA3 reporter gene integrated in place of mating-type genes at an HMR locus that had the HMR-E silencer replaced by a Gal4 DNA binding site (G) (see Figure 1A). A full length GBD-Esc4 (1–1070) clone, aeb15, was identified as being capable of establishing targeted silencing of hmr::URA3, causing resistance to 5-fluororotic acid (5-FOA). This GBD-Esc4 clone was subsequently transformed into strain YEA76 (HMR-E Aeb::G hmr::URA3 gal4::LEU2) and YEA77 (HMR-E aeB::G hmr::URA3 gal4::LEU2) (see Table 1 and Yeast Strain section for details) and tested for targeted silencing. To test SIR-dependence of targeted silencing by Esc4 at HMR, shown in Figure 4, targeted silencing in strain YEA76 (SIR+) was compared with that in sir mutant derivatives YAM7 (sir2Δ), YEA 118 (sir3Δ), and YKAB17 (sir4Δ).
For the targeted silencing experiments shown in Figures 2 and 4, assays were carried out as follows: strains were transformed with plasmids expressing the appropriate GBD hybrid protein, grown at 30°C for two days in SC -Trp medium (to select for the GBD plasmid), serially diluted ten-fold five times and spotted on SC -Trp + 5-FOA plates (to assay for silencing at HMR) or on SC -Trp control plates.
Plasmids
Plasmid aeb15, expressing GBD-Esc4(1–1070) was isolated in the targeted silencing screen. This plasmid was recovered from a library based on pGBT9.C (ADH1UAS-GBD, TRP1, CEN4/ARS1; see Acknowledgements). To generate plasmids for use in the two-hybrid system, ESC4 sequences were amplified from genomic DNA and subcloned into plasmid pSTT91, a derivative of pBTM116 that contains the ADE2 gene ([63]; see also [64]). Plasmids pAM2 (LexA-Esc4N (a.a. 1–480) and pAM7 (LexA-Esc4C (a.a. 818–1070) were used for two-hybrid experiments. To test for LexA-Esc4C binding to GAD-Sir3 and GAD-Sir4, plasmid pEDA195, GAD-Sir3(252–978), and pCTC36, GAD-Sir4(839–1358), were used. pCTC36 expresses the same region of SIR4 as plasmid pCTC18 [55] except that the Sir4 hybrid is expressed from pGAD-R. pEDA195 was constructed by cloning a PstI fragment of the SIR3 gene into vector pGAD424. Plasmid pAM2, expressing LexA-Esc4(1–480), and a clone expressing GAD-Slx4(383–748) isolated in the two-hybrid screen were used as a positive control in two-hybrid experiments summarized in Table 3.
Yeast strain construction
All strains are listed in Table 1. To make slx1Δ, slx4Δ, sir2Δ, sir4Δ, and sgs1Δ mutants, PCR primers with 5' homology to sequences flanking these ORFs and 3' homology to sequences in a plasmids harboring selectable marker genes were used for PCR, generating targeting cassettes that were transformed into yeast as has been previously described [65]. esc4Δ ::his5+ mutants were generated by the same method, but using a different plasmid as a template for PCR (gift of N. Dean, Stony Brook University). Strain YEA76 and its derivatives are derived from strain YSB35 [13].
Two-hybrid screening and direct tests
Screening was performed essentially as described [54,63]. Plasmid pAM2, which expresses LexA-Esc4(1–480), (ADE2, TRP1, 2μ) was co-transformed with approximately 1 μg of GAD library (LEU2, 2μ; [66]) into strain L40 [54], which contains LexA binding sites upstream of both the HIS3 gene and the LacZ gene. The following specificity tests were performed: (1) His+ candidates were assayed for lacZ expression both after curing candidates of either the LexA or the GAD plasmid, (2) GAD hybrids were tested for interactions with nonspecific LexA hybrids (e.g. LexA-lamin) by mating candidates cured of the bait plasmid to LexA-containing AMR70 [54], and (3) the GAD hybrid plasmids which passed the aforementioned tests were subsequently retransformed into L40 along the original bait and tested for LacZ expression.
Two-hybrid tests done using the C-terminal BRCT motifs in Esc4 were performed by co-transforming L40 with plasmid pAM7, expressing LexA-Esc4(818–1070), and various GAD hybrids to silencing proteins, such as Sir1, Sir2, Sir3, Sir4, and Rap1. To test whether Sir3 bridged binding of LexA-Esc4(818–1070) to Sir4, a sir3Δ::kanMX6 derivative of L40 was generated (see yeast strains section for details), strain YRJ3, and the interaction was then retested in this strain. The control sir4Δ-derivative of L40 used was strain YJL03.
DNA damage sensitivity tests
Cultures were grown in YPD medium ~18 h at 30°C and then serially diluted ten-fold, five times before being spotted onto plates containing YPD medium with MMS, HU, or no chemical. Cells were then incubated at 30°C and, in the case of MMS media, the plates were wrapped in aluminum foil.
Authors' contributions
RS and DCZ initiated and oversaw the course of the study, designed the experiments and wrote the paper. DCZ did the targeted silencing tests, protein sequence alignments and made the figures. ASRM and DCZ performed the two-hybrid screening and direct two-hybrid tests. JJC and RAJ did some of the genetic analyses. All authors have read and approved this manuscript.
Acknowledgments
Acknowledgements
We thank Carlos Evangelista and Stan Fields for the GBD library and Phil James for the two-hybrid GAD library. Thanks to Joe Landry for strain YJL103, Erik Andrulis for plasmid pEDA195 and to Cheng-ting Chien for pCTC36. Aaron Neiman and Doug Theobald provided helpful suggestions. This work was supported by NIH grant GM28220 and a Carol Baldwin Breast Cancer Research Support Grant.
Contributor Information
David C Zappulla, Email: david.zappulla@gmail.com.
Arindel SR Maharaj, Email: arindel_maharaj@student.hms.harvard.edu.
Jessica J Connelly, Email: jconnell@chg.duhs.duke.edu.
Rebecca A Jockusch, Email: rjockusc@chem.utoronto.ca.
Rolf Sternglanz, Email: rolf@life.bio.sunysb.edu.
References
- Gasser SM, Cockell MM. The molecular biology of the SIR proteins. Gene. 2001;279:1–16. doi: 10.1016/S0378-1119(01)00741-7. [DOI] [PubMed] [Google Scholar]
- Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Moretti P, Yuan P, Sternglanz R, Shore D. Abf1 recruits Sir3 to initiate transcriptional silencing in yeast
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-X. [DOI] [PubMed] [Google Scholar]
- Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/S0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr Biol. 1998;8:653–656. doi: 10.1016/S0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Abraham J, Nasmyth KA, Strathern JN, Klar AJ, Hicks JB. Regulation of mating-type information in yeast. Negative control requiring sequences both 5' and 3' to the regulated region. J Mol Biol. 1984;176:307–331. doi: 10.1016/0022-2836(84)90492-3. [DOI] [PubMed] [Google Scholar]
- Feldman JB, Hicks JB, Broach JR. Identification of sites required for repression of a silent mating type locus in yeast. J Mol Biol. 1984;178:815–834. doi: 10.1016/0022-2836(84)90313-9. [DOI] [PubMed] [Google Scholar]
- Chien CT, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- Buck SW, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Zappulla DC, Alexieva-Botcheva K, Evangelista C, Sternglanz R. One-hybrid screens at the Saccharomyces cerevisiae HMR locus identify novel transcriptional silencing factors. Genetics. 2004;166:631–635. doi: 10.1534/genetics.166.1.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Zappulla DC, Ansari A, Perrod S, Laiosa CV, Gartenberg MR, Sternglanz R. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol Cell Biol. 2002;22:8292–8301. doi: 10.1128/MCB.22.23.8292-8301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanway D, Chin JK, Xia G, Oshiro G, Winzeler EA, Romesberg FE. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc Natl Acad Sci USA. 2002;99:10605–10610. doi: 10.1073/pnas.152264899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci USA. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J. Esc4p, a new target of Mec1p (ATR), promotes resumption of DNA synthesis after DNA damage. Embo J. 2004;23:1188–1197. doi: 10.1038/sj.emboj.7600129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TM, Kobor MS, Bastin-Shanower SA, Ii M, Horte SA, Gin JW, Emili A, Rine J, Brill SJ, Brown GW. Slx4 Regulates DNA Damage Checkpoint-dependent Phosphorylation of the BRCT Domain Protein Rtt107/Esc4. Mol Biol Cell. 2006;17:539–548. doi: 10.1091/mbc.E05-08-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Altschul SF, Bork P. BRCA1 protein products ... Functional motifs. Nat Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- Zhang X, Morera S, Bates PA, Whitehead PC, Coffer AI, Hainbucher K, Nash RA, Sternberg MJ, Lindahl T, Freemont PS. Structure of an XRCC1 BRCT domain: a new protein-protein interaction module. Embo J. 1998;17:6404–6411. doi: 10.1093/emboj/17.21.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Green R, Glover JN. Crystal structure of the BRCT repeat region from the breast cancer-associated protein BRCA1. Nat Struct Biol. 2001;8:838–842. doi: 10.1038/nsb1001-838. [DOI] [PubMed] [Google Scholar]
- Joo WS, Jeffrey PD, Cantor SB, Finnin MS, Livingston DM, Pavletich NP. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev. 2002;16:583–593. doi: 10.1101/gad.959202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire DJ, Basu BP, Serpell LC, Joo WS, Date T, Iwabuchi K, Doherty AJ. Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor. Embo J. 2002;21:3863–3872. doi: 10.1093/emboj/cdf383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan VV, Thornton KH, Thelen MP, Cosman M. Solution structure and backbone dynamics of the human DNA ligase IIIalpha BRCT domain. Biochemistry. 2001;40:13158–13166. doi: 10.1021/bi010979g. [DOI] [PubMed] [Google Scholar]
- Lee JY, Chang C, Song HK, Moon J, Yang JK, Kim HK, Kwon ST, Suh SW. Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications. Embo J. 2000;19:1119–1129. doi: 10.1093/emboj/19.5.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol Cell. 2004;14:405–412. doi: 10.1016/S1097-2765(04)00238-2. [DOI] [PubMed] [Google Scholar]
- Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Lu J, Mullen JR, Brill SJ, Kleff S, Romeo AM, Sternglanz R. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Conte D, Jr, Curcio MJ. The Sgs1 helicase of Saccharomyces cerevisiae inhibits retrotransposition of Ty1 multimeric arrays. Mol Cell Biol. 2001;21:5374–5388. doi: 10.1128/MCB.21.16.5374-5388.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EVs. Prokaryotic homologs of the eukaryotic DNA end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Hishida T, Iwasaki H, Ohno T, Morishita T, Shinagawa H. A yeast gene, MGS1, encoding a DNA-dependent AAA(+) ATPase is required to maintain genome stability. Proc Natl Acad Sci USA. 2001;98:8283–8289. doi: 10.1073/pnas.121009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Johnson RE, Yu SL, Prakash L, Prakash S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/S0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. Faseb J. 1997;11:68–76. [PubMed] [Google Scholar]
- Chin JK, Bashkirov VI, Heyer WD, Romesberg FE. Esc4/Rtt107 and the control of recombination during replication. DNA Repair (Amst) 2006;5:618–628. doi: 10.1016/j.dnarep.2006.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2004:D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Website title http://pfam.janelia.org
- Jowsey PA, Doherty AJ, Rouse J. Human PTIP facilitates ATM-mediated activation of p53 and promotes cellular resistance to ionizing radiation. J Biol Chem. 2004;279:55562–55569. doi: 10.1074/jbc.M411021200. [DOI] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- Stone EM, Pillus L. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J Cell Biol. 1996;135:571–583. doi: 10.1083/jcb.135.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Hector RE, Roy N, Song JH, Berkner KL, Runge KW. Sir3p phosphorylation by the Slt2p pathway effects redistribution of silencing function and shortened lifespan. Nat Genet. 2003;33:522–526. doi: 10.1038/ng1132. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J Biol Chem. 2003;278:52914–52918. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ. Slx1–Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1–Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A, Heller RC, Landry J, Choy JS, Sirko A, Sternglanz R. A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol Cell Biol. 2001;21:3514–3522. doi: 10.1128/MCB.21.10.3514-3522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P, Chien CT, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]