Abstract
Resistance to antimalarial drugs is a public health problem worldwide. Molecular markers for drug-resistant malaria, such as pfcrt and pfmdr1 polymorphisms, could serve as useful surveillance tools. To evaluate this possibility, sequence polymorphisms in pfcrt (position 76) and pfmdr1 (positions 86, 184, 1034, 1042, and 1246) and in vitro drug sensitivities were measured for 65 Plasmodium falciparum isolates from Thailand, Myanmar, Vietnam, and Bangladesh. The pfcrt Thr76 polymorphism was present in 97% of samples, consistent with observations that chloroquine resistance is well established in this region. Polymorphisms in pfmdr1 clustered into four specific patterns: the wild type (category I), a Tyr86 polymorphism only (category II), a Phe184 polymorphism only (category III), and Phe184 in combination with Cys1034 and/or Asp1042 (category IV). Isolates in categories I and III were more sensitive to chloroquine and more resistant to mefloquine, artesunate, and artemisinin than isolates in categories II and IV (P ≤ 0.01). Mefloquine resistance was significantly more common in category I and III isolates than in category II and IV isolates, with a prevalence ratio of 14.95 (95% confidence interval, 3.88 to 57.56). These categories identified mefloquine resistance with a sensitivity and a specificity of 94 and 91%, respectively. The pfmdr1 gene copy number was measured by real-time PCR as a ratio of the amount of pfmdr1 DNA to the amount of lactate dehydrogenase (ldh) DNA. Eight samples had pfmdr1 DNA/ldh DNA ratios ≥3. The isolates in all 8 samples fell into categories I and III and were significantly more resistant to mefloquine, quinine, artemisinin, and artesunate and more sensitive to chloroquine than the isolates in the 57 samples with <3 copies of the gene (P ≤ 0.001). Thus, measurement of pfmdr1 mutations and gene copy number may be useful for surveillance of mefloquine-resistant malaria in Southeast Asia.
Resistance to antimalarials is spreading throughout the world and is impeding efforts to control malaria, which causes 700,000 to 2.7 million deaths every year (3). Drug-resistant Plasmodium falciparum is a particularly serious problem in Southeast Asia, where strains are commonly resistant to chloroquine, antifolates, quinine, and mefloquine (20).
Surveillance for drug-resistant malaria is based at present on strict in vivo criteria for treatment failure and on measurement of the activities of antimalarial drugs against cultured parasites in vitro. Surveillance could be carried out more effectively by using molecular markers, once such markers have been validated. At present, there is good evidence that mutations in two genes (dhps and dhfr) correlate well with in vitro and in vivo resistance to sulfadoxine-pyrimethamine and that mutations in the gene pfcrt (especially at position 76) correlate well with in vitro and in vivo resistance to chloroquine (for a review, see reference 20). There is also evidence that mutations in pfmdr1 are associated with drug resistance, but the evidence is less conclusive. Many of the studies of this relationship were performed with laboratory strains of P. falciparum or with other eukaryotic models, so the results of these experiments might not be generalizable to naturally occurring P. falciparum isolates. Also, some field studies have suggested that mutations or amplification of this gene is associated with chloroquine resistance, while others have suggested that mutations or gene amplification is associated with increased chloroquine sensitivity (1, 6). On the other hand, there is more general agreement that mutations in pfmdr1 are associated with altered sensitivity to mefloquine and artemisinin derivatives in vitro, although the role of gene amplification is not clear (1, 4, 13, 14). However, most previous studies were small (n < 20 isolates) or limited to single geographical areas where specific polymorphisms may have been absent. Accordingly, we chose to determine the association of pfcrt and pfmdr1 mutations and pfmdr1 gene amplification with in vitro sensitivity to antimalarial drugs for a group of isolates from several areas in Southeast Asia where malaria is endemic.
MATERIALS AND METHODS
Patient isolates.
The Armed Forces Institute of Medical Sciences in Bangkok, Thailand, has been carrying out active surveillance for resistance to antimalarials in a variety of provinces of Thailand as well as in neighboring countries since 1992. Over 400 patient isolates have been grown in culture, tested for their drug sensitivities in vitro, and archived by storage in liquid nitrogen. This study received ethical clearance from the ethical review boards of the University of Michigan, the Walter Reed Army Institute of Research, and the Thai Ministry of Public Health.
Field isolates were cryopreserved in a dry liquid nitrogen shipping container and transported to the Armed Forces Institute of Medical Sciences, where they were thawed 1 to 3 months later. In vitro drug sensitivity assays were performed by a radioisotope microdilution technique slightly modified from that described previously (16). Briefly, the suspension of the malaria parasite culture (0.5% parasitemia and 1.5% hematocrit, 200 μl/well) was dispensed into the wells of a standard microtiter plate (a 96-well flat-bottom plate) predosed with duplicate serial dilutions of the antimalarial drugs. The microtiter plate was placed in a gas-tight box to maintain an atmosphere of 5% CO2, 5% O2, and 90% N2 and then in an incubator (37°C). At the end of the 24th hour, the plate was temporarily removed for pulsing with [3H]hypoxanthine. The contents of each plate were harvested after 42 to 44 h of incubation. The 50% inhibitory concentration (IC50), IC90, etc., were estimated by nonlinear regression analysis of the incorporated radioactivity data. The antimalarial drugs and the ranges of concentrations used were as follows: chloroquine diphosphate, 7.144 to 457.200 ng/ml; quinine citrate, 21.438 to 1,372.000 ng/ml; mefloquine hydrochloride, 1.948 to 124.700 ng/ml; artesunate, 0.265 to 16.93 ng/ml.
For the present study, 73 stabilates were selected to maximize the statistical power to detect differences in genotypes between sensitive and resistant isolates by achieving an even distribution of mefloquine-resistant and -sensitive isolates. Of these 73 stabilates, cultures were successfully obtained from 63. In addition, two laboratory strains originally obtained from Thai isolates, isolates W2 and PH6, were used. The isolates in these cultures were then retested for their sensitivities to chloroquine, mefloquine, quinine, artesunate, and artemisinin in vitro. There were good correlations between sensitivities before and after cryopreservation for mefloquine, quinine artesunate, and artemisinin, while the correlations were less good for chloroquine. Strains with the following assay results both in the initial test and in the repeat test were considered resistant to antimalarials: mefloquine, IC50 > 20 ng/ml or IC90 > 80 ng/ml; quinine, IC50 > 500 or IC90 > 1,000 ng/ml; and chloroquine, IC50 > 80.
Genetic polymorphisms.
DNA was obtained from each culture by using QIAamp DNA mini kits (Qiagen, Valencia, Calif.). Samples were genotyped for msp1, msp2, and glurp by the methods described by Snounou et al. (15). PCR amplification and direct sequencing were used to determine the sequences at a single pfcrt polymorphic site (position 76) and five pfmdr1 polymorphic sites (positions 86, 184, 1034, 1042, and 1246), as described previously (8). The sequences at all loci could be determined for 61 isolates plus laboratory strains W2 and PH6. For one isolate, the sequence at position 184 could not be obtained, and for a second isolate, the sequences at positions 1034 and 1042 could not be obtained.
Real-time PCR.
Real-time PCR was performed with an ABI PRISM 7000 sequence detection system (Applied Biosystems, Foster City, Calif.). This system uses a fluorescence-based PCR method that measures the rate of gene amplification; as more amplicons are made, the DNA-bound reporter dye and quencher are separated, generating fluorescence. The rate at which fluorescence increases is related to the number of gene copies initially present (2). Primers and fluorescence-labeled probes were designed by using Primer Express software (version 2.0; Applied Biosystems, Foster City, Calif.) to amplify the P. falciparum multidrug resistance (pfmdr1) and lactate dehydrogenase (ldh) genes (Table 1).
TABLE 1.
Primers and probes used for real-time PCR
| Primer or probe | Sequence |
|---|---|
| Primers | |
| LDH-F | ACG ATT TGG CTG GAG CAG AT |
| LDH-R | TCT CTA TTC CAT TCT TTG TCA CTC TTT C |
| PF-R | TCT CCT TCG GTT GGA TCA TAA AG |
| PF-F | TTA AGT TTT ACT CTA AAA GAA GGG AAA ACA TAT |
| Probes | |
| PF-FAM | FAM-CAT TTG TGG GAG AAT CAG GTT GTG GGA AAT-TAMRA |
| LDH-FAM | FAM-AGT AAT AGT AAC AGC TGG ATT TAC CAA GGC CCC A-TAMRA |
Probes were synthesized by Applied Biosystems and labeled in the standard manner with a reporter dye (6-carboxyfluorescein [FAM]) at the 5′ end and a quencher dye (6-carboxytetramethylrhodamine [TAMRA]) at the 3′ end. The primers were obtained from Qiagen. The primer and probe concentrations were optimized according to the protocol recommended for the TaqMan Universal PCR Master Mix (Applied Biosystems).
The reagents used for each unknown sample or standard were 1× TaqMan Universal Master Mix (2×; Applied Biosystems); 300 nM forward primer; 300 nM reverse primer; 250 nM TaqMan probe; and distilled, sterile water (Sigma, St. Louis, Mo.). The total reaction volume was 50 μl. The reaction mixtures were prepared at 4°C in a 96-well optical reaction plate (Applied Biosystems) covered with optical adhesive covers (Applied Biosystems). The thermal cycling conditions were 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The threshold was set to optimize the threshold cycle (CT) value for the first standardization reaction, and all subsequent CT values were obtained by using the same threshold value.
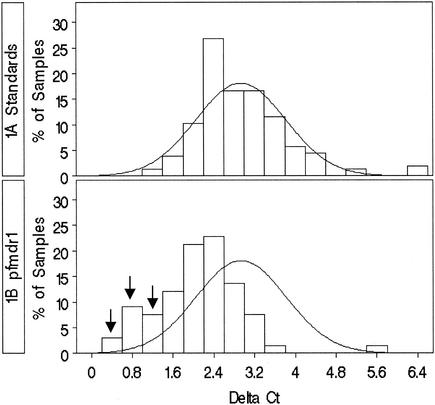
In order to determine the reproducibility of the real-time PCR, 160 amplifications of each gene were performed with P. falciparum strain 3D7 DNA, which was extracted and purified from culture in phosphate-buffered saline with Qiagen DNA Mini kits, as stated in the spin protocol for blood and body fluids. The P. falciparum gene copy number was calculated to be 8,000 copies/ng of DNA on the basis of a genome size of 1.06 × 106 bp (www.ncbi.nlm.nih.gov). Five 10-fold serial dilutions of P. falciparum 3D7 DNA were made, so that there were 8, 80, 800, 8,000, or 80,000 copies of genome per reaction mixture. The ldh DNA and pfmdr1 DNA in paired standard wells were amplified 40 times (quadruplicate reactions per plate times 10 reaction plates) for each of the four DNA concentrations (80 to 80,000 genome copies). Amplifications were successful for 157 of 160 reactions. The CT values for the reactions with ldh and pfmdr1 were calculated with ABI Prism 7000 SDS software, with a threshold value of 0.2000 and a reference baseline reading of cycles 6 to 15. The change in the CT value (ΔCT) was calculated by subtracting the CT for a given ldh DNA concentration from the paired CT for the same pfmdr1 DNA concentration. (Since the CT is proportional to the log of the concentration, ΔCT is equivalent to the log of the ratio.) A histogram (Fig. 1A) of the ΔCTs was made and used to calculate standard deviations and means by using SAS software (version 8; SAS, Inc., Cary, N.C.). The distribution shown is due to assay variability.
FIG. 1.
Frequency distribution of ΔCTs calculated from real-time PCR amplifications of pfmdr1 and ldh genes. (A) Histogram of 157 ΔCTs calculated from standard DNA, showing a normal distribution; (B) histogram of isolates. The arrows point to samples which are more than 2 standard deviations below the mean.
Since the ratio of the pfmdr1 gene copy number to the ldh gene copy number (pfmdr1/ldh ratios) is the ultimate end point, calculations were made to determine the minimal detectable ratio. ΔCTs were calculated for different pfrmdr1/ldh ratios (10,000:1, 1,000:1, 100:1, 10:1, 1:1) by pairing and subtracting the CT for a low concentration of ldh from the CT for a high concentration of pfmdr1 by using the same data obtained as described above plus the CT data for eight copies of ldh. For each ratio, mean ΔCTs were plotted against the copy number ratio and were found to fit to the equation ΔCT = −1.6145 ln(copy number ratio) + 2.8565 by using Excel Office 2000 software.
The pfmdr1 and ldh amplification reactions were run in duplicate by using 1-μl aliquots of DNA from each clinical sample. ΔCTs were calculated as described above, and the duplicate values were averaged. ΔCTs which were more than 2 standard deviations beyond the mean (95% confidence interval [CI]) calculated from the data in Fig. 1A were considered significantly different from 1.
Statistical analysis.
Statistical analyses were conducted with SAS software (version 8; SAS, Inc.). Nonnormally distributed IC50 data were assessed by Wilcoxon rank sum tests. Group differences in the percentage of resistant isolates and IC50s were assessed by the chi-square test. Prevalence ratios were estimated for the association between genetic markers and drug susceptibility phenotypes.
RESULTS
Polymorphisms.
Of the 65 isolates successfully cultured, amplified, and sequenced, 39 were from Thailand, 14 were from Myanmar (18), 7 were from Bangladesh (11), and 5 were from Vietnam (19). There were no associations between the msp1, msp2, or glurp types of the strains or the degree of clonality and drug resistance (data not shown).
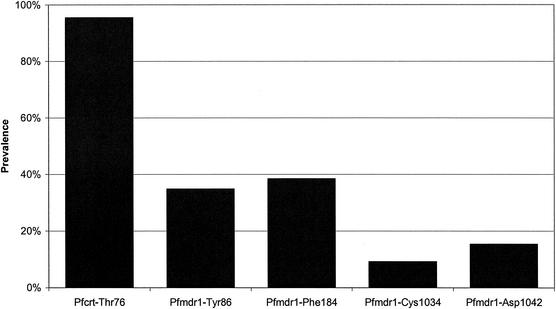
Mutations in both pfcrt and pfmdr1 were quite common (Fig. 2). Ninety-seven percent (63 of 65) of the samples contained the Thr76 polymorphism in pfcrt. pfmdr1 polymorphisms Tyr86 and Phe184 were each present in about one-third of the samples. Cys1034 and Asp1042 were present in 9 and 15% of the samples, respectively, while no polymorphisms were found at position 1246. None of the amplicons gave mixed sequences at any of these loci.
FIG. 2.
Prevalence of specific point mutations in pfcrt and pfmdr1. No mutations were observed at pfmdr1 position 1246 (data not shown).
Polymorphisms in pfmdr1 clustered into four specific patterns without variation. We arbitrarily named these specific patterns (Table 2). The wild-type pattern (Asn86, Tyr184, Ser1034, Asn1042) was classified as category I. Isolates which had the Tyr86 polymorphism (and which had no polymorphisms at any of the other sites) were classified as category II. Isolates with the Phe184 polymorphism alone were classified as category III. The only multiple polymorphism observed among these isolates was Phe184 in combination with Cys1034 and/or Asp1042; isolates with this pattern were classified as category IV (Table 2). The two laboratory strains, strains W2 and PH6, fit into categories II and IV, respectively. Both isolates with incomplete sequences had the Tyr86 polymorphism, and no other polymorphism was detected. Thus, they were both presumed to be in category II.
TABLE 2.
Patterns of mutations observed in pfmdr1
| Category | Codon at positiona
|
No. of isolates | |||
|---|---|---|---|---|---|
| 86 | 184 | 1034 | 1042 | ||
| I | Asn | Tyr | Ser | Asn | 17 |
| II | Tyr | Tyr | Ser | Asn | 23 |
| III | Asn | Phe | Ser | Asn | 15 |
| IV | Asn | Phe | Cys | Asp | 10 |
| Asn | Phe | Cys | Asn | ||
| Asn | Phe | Ser | Asp | ||
The mutated amino acids (in boldface) and the number of isolates in each category are shown.
There were significant differences in the IC50s between the different categories. The IC50s of mefloquine, artemisinin, and artesunate were significantly higher for the isolates in categories I and III than for the isolates in categories II and IV (one-sided P values, <0.0001, 0.0006, and <0.0001, respectively) (Table 3). The opposite was true for chloroquine (P = 0.007), while the quinine IC50 data did not follow any specific pattern. Thus, isolates in categories I and III are relatively more resistant to mefloquine, artemisinin, and artesunate and relatively more sensitive to chloroquine.
TABLE 3.
Median IC50s and percent resistance for isolates in four catagories
| Category | Mefloquine
|
Quinine
|
Chloroquine
|
Median IC50 (interquartile range)b
|
||||
|---|---|---|---|---|---|---|---|---|
| % Ra | Median IC50 (interquartile range) | % R | Median IC50 (interquartile range) | % R | Median IC50 (interquartile range) | Artemisinin | Artesunate | |
| I (n = 17) | 100 | 60.68 (49.64-69.90) | 35 | 188.62 (104.44-271.88) | 76 | 56.40 (47.08-73.92) | 1.74 (1.10-2.04) | 4.51 (2.47-4.84) |
| II (n = 23) | 0 | 15.79 (11.43-19.56) | 4 | 94.68 (77.79-107.83) | 91 | 88.19 (73.09-107.83) | 0.73 (0.54-1.40) | 1.43 (1.06-2.69) |
| III (n = 15) | 80 | 59.51 (30.87-73.96) | 31 | 148.87 (70.07-239.88) | 81 | 70.92 (19.38-92.08) | 2.57 (1.80-3.85) | 2.57 (1.80-3.85) |
| IV (n = 10) | 20 | 19.44 (10.01-24.70) | 11 | 184.36 (141.28-233.41) | 89 | 70.10 (63.69-98.97) | 0.97 (0.80-1.30) | 0.85 (0.64-0.93) |
| P value for differencec | <0.001 | <0.001 | 0.266 | 0.002 | <0.001 | 0.007 | <0.001 | 0.011 |
% R, percentage of resistant isolates.
For artemisinin and artesunate, there are no established in vitro cut offs for resistance and sensitivity.
P-values were determined by chi-square analysis for percentage of resistant isolates and analysis of variance for IC50s.
We reasoned that under mefloquine pressure, isolates in the wild-type category (category I) could have developed the category II mutation; in the same way, the isolates with the category III mutation present under chloroquine pressure could have developed additional polymorphisms under mefloquine pressure, resulting in the category IV pattern. Because the isolates in categories I and III showed similar resistance patterns, while the isolates in categories II and IV also had similar resistance patterns, we collapsed these two sets of categories to test the hypothesis that there is an association between pfmdr1 polymorphisms and drug resistance.
The estimated risk of resistance was higher for isolates in categories I and III than for isolates in categories II and IV (Table 3), with prevalence ratios of 14.95 (95% CI, 3.88 to 57.56) for mefloquine and 1.44 for quinine (95% CI, 0.51 to 4.08). In contrast, the estimated risk of resistance to chloroquine was lower for isolates in categories I and III than for isolates in categories II and IV (prevalence ratio, 0.94; 95% CI, 0.86 to 1.03). Thus, there was a strong association between mutation category and the prevalence of mefloquine resistance and a weaker association between mutation category and the prevalence of quinine and chloroquine resistance.
Only two isolates (3%) had the wild-type pfcrt genotype (Lys76). Both of these isolates were chloroquine sensitive, quinine sensitive, and mefloquine resistant. One isolate fell into pfmdr1 category I, and the other isolate fell into category III.
Real-time PCR to determine pfmdr1/ldh ratio.
A total of 157 ΔCTs were determined from the concentration-response experiments run with standard DNA (Fig. 1A), yielding a mean ΔCT of 2.82 with a 95% CI of 1.05 to 4.57. ΔCT values that fall below the lower CI are indicative of high pfmdr1/ldh ratios. pfmdr1/ldh ratios yielding ΔCT values that fall within the CI are not considered statistically significantly different from 1. Only pfmdr1/ldh ratios of 3.06 or more would yield clinical ΔCT values less than 1.05. Thus, the sensitivity limit of this assay is a pfmdr1/ldh ratio of 3.
The pfmdr1 and ldh genes were successfully amplified from all DNA samples. The frequency distributions for ΔCT values are shown in Fig. 1B. The isolates in 8 of 65 samples (12.3%) had ΔCT values which were below the 95% confidence limit (i.e., less than 1.05), which is indicative of pfmdr1/ldh ratios between 3 and 4.2. Five of these isolates were in category I, and three were in category III. The IC50s were compared for three groups: categories I and III with amplification, categories I and III without amplification, and categories II and IV without amplification. The IC50s of all drugs examined were significantly different across these three categories (Table 4). The mefloquine, quinine, artemisinin, and artesunate IC50s were the highest for isolates in categories I and III with amplification, moderate for isolates in categories I and III without amplification, and lowest for isolates in categories II and IV without amplification (P values, <0.0001, 0.001, 0.0008, and <0.0001, respectively) (Table 4). Chloroquine exhibited the opposite pattern, with the lowest chloroquine IC50s being for isolates in categories I and III with amplification and the highest chloroquine IC50s being for isolates in categories II and IV without amplification.
TABLE 4.
Median IC50s and percent resistance for isolates categorized by pfmdr1 gene amplification
| Category | Mefloquine
|
Quinine
|
Chloroquine
|
Median IC50 (interquartile range)
|
||||
|---|---|---|---|---|---|---|---|---|
| % Ra | Median IC50 (interquartile range) | % R | Median IC50 (interquartile range) | % R | Median IC50 (interquartile range) | Artemisinin | Artesunate | |
| II and IV, no amplification (n = 33) | 6.1 | 16.86 (11.13-20.45) | 15.2 | 112.01 (80.98-143.74) | 100 | 84.37 (68.22-107.12) | 0.82 (0.64-1.09) | 1.32 (0.83-2.13) |
| I and III, no amplification (n = 24) | 87.5 | 53.01 (34.15-62.79) | 16.7 | 144.35 (71.96-199.15) | 91.7 | 69.06 (50.87-86.92) | 1.18 (0.74-1.92) | 2.48 (1.71-4.60) |
| I and III, amplification (n = 8) | 100 | 72.13 (64.77-99.21) | 37.5 | 289.31 (253.44-360.07) | 100 | 57.55 (43.51-64.92) | 1.89 (1.28-2.76) | 5.63 (3.33-8.33) |
| P value for differenceb | <0.0001 | <0.0001 | 0.3280 | 0.0010 | 0.3654 | 0.0029 | 0.0008 | <0.0001 |
% R, percentage of resistant isolates.
P values were determined by Fisher's exact chi-square test for percentage of resistant isolates and the Wilcoxon rank sum test for IC50s.
Evaluation for the presence of category I and III genotypes may be useful tool in surveillance for mefloquine resistance. The presence of these genotypes has a positive predictive value of 91% for this group of isolates, with a sensitivity of 94% and a specificity of 91% for the detection of in vitro mefloquine resistance. For quinine resistance and chloroquine sensitivity, on the other hand, the values are much lower. Because all isolates whose DNA was amplified were mefloquine resistant, the pfmdr1 gene copy number has a positive predictive value and a specificity of 100% for the detection of mefloquine resistance; however, it is less useful than the genotype for surveillance because of a sensitivity of only 26%.
DISCUSSION
Among 65 cultured isolates from Southeast Asia, almost all contained the pfcrt polymorphism at position 76. Polymorphisms were found at four loci in the pfmdr1 gene and occurred in four specific patterns (Table 2). The isolates with two patterns (categories I and III) tended to be more resistant to mefloquine, artesunate, and artemisinin, while the isolates with two others (categories II and IV) tended to be more resistant to chloroquine. Our data also demonstrate that isolates with increased pfmdr1 copy numbers tend to be more resistant to mefloquine, quinine, artesunate, and artemisinin and more sensitive to chloroquine than isolates without increased pfmdr1 copy numbers.
An association between the polymorphism that comprised category II (Tyr86) and mefloquine susceptibility had previously been suggested, but the strength of this association was never precisely quantified (for a review, see reference 20). Attempts to answer the question related to pfmdr1 gene amplification also have a history of conflicting results. Some studies have reported increased copy numbers with selection for mefloquine resistance (5, 12), while others have not (9, 10).
In this study, isolates that were in categories I or III were 15-fold more likely to be mefloquine resistant in vitro. This is the strongest association ever reported between pfmdr1 and resistance to antimalarials. The presence of these genotypes correctly identifies 94% of the mefloquine-resistant isolates in vitro, while the absence of these genotypes correctly identifies 91% of the mefloquine-sensitive isolates in vitro.
We also found a robust association between pfmdr1 copy number and drug resistance. Increased pfmdr1 copy numbers were found only among isolates in the two categories (categories I and III) in which isolates already displayed increased mefloquine resistance. Within categories I and III, isolates with pfmdr1 amplification were significantly more resistant to mefloquine, quinine, artemisinin, and artesunate than isolates without pfmdr1 amplification. This suggests that pfmdr1 amplification may occur secondarily as a way to achieve greater levels of resistance. However, pfmdr1 amplification appears to be less useful as a surveillance tool, with a sensitivity of only 26%. One possible reason for this is the fact that our real-time PCR assay could only reliably detect pfmdr1/ldh ratios ≥3. Improvements to the assay might improve its sensitivity.
The distribution of pfmdr1 polymorphisms found here can be compared constructively with those found in a previous study, in which the pfmdr1 genotypes of 54 isolates from the northwestern border of Thailand and Myanmar were reported (13). Of the 54 sequences published, 52 fit into the categories described in this paper: 28 (54%) were category I, 5 (10%) were category II, and 19 (37%) were category III or IV. That study did find increased pfmdr1 copy numbers in mefloquine-resistant parasites; similar to the present study, isolates with increased copy numbers were all wild type at position 86 (corresponding to possible category I or III isolates) (13).
The lack of a polymorphism at position 1246 in this study supported previous reports from Asia, suggesting that this polymorphism may not be important for isolates from this region (4, 13, 17). Also, the coexistence of elevations in mefloquine, artemisinin, and artesunate IC50s for the same categories of isolates is also consistent with previous observations (1, 7, 16, 21).
Since the isolates in category IV contain the one mutation found among the isolates in category III plus other mutations, category IV isolates could have derived from category III isolates. Different geographic distributions were found for the different classes and may support this idea. The most striking geographic differences were between Yala (in southern Thailand) and Borai (on the Thai-Cambodian border): eight of eight isolates in samples selected from among those that originated in Yala were category II, while five of five isolates in samples from Borai were category III or IV. Circumstantial support for the evolution of genotype is offered by the only two chloroquine-sensitive isolates, which were mefloquine resistant and which were in categories I and III.
The interpretation of our results must bear certain considerations. First, isolates were chosen to provide the broadest range of in vitro sensitivities to mefloquine. Thus, the power for discriminating between mefloquine-sensitive and -resistant isolates was high. This study was designed to test the hypothesis that there is an association between mefloquine resistance and the pfmdr1 genotype. Because of this, the prevalence of sensitive and resistant isolates in this study does not represent the prevalence in the Southeast Asian populations from which they were drawn. Our observation that polymorphisms were more significantly associated with resistance to mefloquine than with resistance to other drugs may simply be due to our selection criterion. Second, the observation that polymorphisms fell into only four patterns may not be generalizable; studies with larger numbers of samples from Southeast Asia or samples from other regions might reveal other patterns. Third, like other studies in Southeast Asia, this study was not able to assess the relative importance of pfcrt mutations, given the high percentage of isolates with the Thr76 polymorphism. Fourth, it is possible that pfmdr1 mutations may not be responsible for mefloquine resistance but may only be in linkage disequilibrium with other determinants that do cause resistance. Finally, the in vitro resistance studied here may not correlate with in vivo resistance. Future studies are needed to determine the association between pfmdr1 polymorphisms and in vivo resistance.
In summary, our data suggest that categories of polymorphisms in pfmdr1 exist in Southeast Asia and that isolates resistant to mefloquine and chloroquine have opposite genetic patterns. Chloroquine use may have selected for pfmdr1 polymorphism categories II and IV, but when mefloquine became widely used, the selection pressure was put on isolates in categories I and III. The strong association observed suggests that categories I and III, along with pfmdr1 gene amplification, could be key determinants of resistance to mefloquine in Southeast Asia. Future studies should more rigorously study this association. Furthermore, the patterns of the polymorphisms reported here have high degrees of sensitivity and specificity for the detection of mefloquine resistance and might serve as useful epidemiological tools.
Acknowledgments
We thank Charles Poole and Donglin Zheng of the University of North Carolina School of Public Health for advice.
This work was supported by NIH grant AI 45426, the U.S. Department of Defense Global Emerging Infections Surveillance and Response System (DoD-GEIS), and grant B-13576006 from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Basco, L. K., J. Le Bras, Z. Rhoades, and C. M. Wilson. 1995. Analysis of pfmdr1 and drug susceptibility in fresh isolates of Plasmodium falciparum from sub-Saharan Africa. Mol. Biochem. Parasitol. 74:157-166. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A. S., and L. C. Ranford-Cartwright. 2002. Real-time quantitative PCR in parasitology. Trends Parasitol. 18:337-342. [PubMed] [Google Scholar]
- 3.Breman, J. G. 2001. The ears of the hippopotamus: manifestations, determinants, and estimates of the malaria burden. Am. J. Trop. Med. Hyg. 64:1-11. [DOI] [PubMed] [Google Scholar]
- 4.Chaiyaroj, S. C., A. Buranakiti, P. Angkasekwinai, S. Looressuwan, and A. F. Cowman. 1999. Analysis of mefloquine resistance and amplification of pfmdr1 in multidrug-resistant Plasmodium falciparum isolates from Thailand. Am. J. Trop. Med. Hyg. 61:780-783. [DOI] [PubMed] [Google Scholar]
- 5.Cowman, A. F., D. Galatis, and J. K. Thompson. 1994. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc. Natl. Acad. Sci. USA 91:1143-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duraisingh, M. T., L. V. von Seidlein, A. Jepson, P. Jones, I. Sambou, M. Pinder, and D. C. Warhurst. 2000. Linkage disequilibrium between two chromosomally distinct loci associated with increased resistance to chloroquine in Plasmodium falciparum. Parasitology 121(Pt 1):1-7. [DOI] [PubMed] [Google Scholar]
- 7.Huong, N. M., S. Hewitt, T. M. Davis, L. D. Dao, T. Q. Toan, T. B. Kim, N. T. Hanh, V. N. Phuong, D. H. Nhan, and L. D. Cong. 2001. Resistance of Plasmodium falciparum to antimalarial drugs in a highly endemic area of southern Viet Nam: a study in vivo and in vitro. Trans. R. Soc. Trop. Med. Hyg. 95:325-329. [DOI] [PubMed] [Google Scholar]
- 8.Ittarat, W., A. L. Pickard, P. Rattanasinganchan, P. Wilairatana, S. Looareesuwan, K. Emery, J. Low, R. Udomsangpetch, and S. R. Meshnick. 2003. Recrudescence in artesunate-treated patients with falciparum malaria is dependent on parasite burden not on parasite factors. Am. J. Trop. Med. Hyg. 68:147-152. [PubMed]
- 9.Lim, A. S., D. Galatis, and A. F. Cowman. 1996. Plasmodium falciparum: amplification and overexpression of pfmdr1 is not necessary for increased mefloquine resistance. Exp. Parasitol. 83:295-303. [DOI] [PubMed] [Google Scholar]
- 10.Mungthin, M., P. G. Bray, and S. A. Ward. 1999. Phenotypic and genotypic characteristics of recently adapted isolates of Plasmodium falciparum from Thailand. Am. J. Trop. Med. Hyg. 60:469-474. [DOI] [PubMed] [Google Scholar]
- 11.Noedl, H., M. A. Faiz, E. B. Yunus, M. R. Rahman, M. A. Hossain, R. Samad, R. S. Miller, L. W. Pang, and C. Wongsrichanalai. 2003. Drug-resistant malaria in Bangladesh: an in vitro assessment. Am. J. Trop. Med. Hyg. 68:140-142. [PubMed] [Google Scholar]
- 12.Peel, S. A., P. Bright, B. Yount, J. Handy, and R. S. Baric. 1994. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 51:648-658. [DOI] [PubMed] [Google Scholar]
- 13.Price, R. N., C. Cassar, A. Brockman, M. Duraisingh, M. van Vugt, N. J. White, F. Nosten, and S. Krishna. 1999. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob. Agents Chemother. 43:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed, M. B., K. J. Saliba, S. R. Caruana, K. Kirk, and A. F. Cowman. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906-909. [DOI] [PubMed] [Google Scholar]
- 15.Snounou, G., X. Zhu, N. Siripoon, W. Jarra, S. Thaithong, K. N. Brown, and S. Viriyakosol. 1999. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans. R. Soc. Trop. Med. Hyg. 93:369-374. [DOI] [PubMed] [Google Scholar]
- 16.Webster, H. K., S. Thaithong, K. Pavanand, K. Yongvanitchit, C. Pinswasdi, and E. F. Boudreau. 1985. Cloning and characterization of mefloquine-resistant Plasmodium falciparum from Thailand. Am. J. Trop. Med. Hyg. 34:1022-1027. [DOI] [PubMed] [Google Scholar]
- 17.Wilson, C. M., S. K. Volkman, S. Thaithong, R. K. Martin, D. E. Kyle, W. K. Milhous, and D. F. Wirth. 1993. Amplification of pfmdr1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol. Biochem. Parasitol. 57:151-160. [DOI] [PubMed] [Google Scholar]
- 18.Wongsrichanalai, C., K. Lin, L. W. Pang, M. A. Faiz, H. Noedl, T. Wimonwattrawatee, A. Laoboonchai, and F. Kawamoto. 2001. In vitro susceptibility of Plasmodium falciparum isolates from Myanmar to antimalarial drugs. Am. J. Trop. Med. Hyg. 65:450-455. [DOI] [PubMed] [Google Scholar]
- 19.Wongsrichanalai, C., T. D. Nguyen, N. T. Trieu, T. Wimonwattrawatee, P. Sookto, D. G. Heppner, and F. Kawamoto. 1997. In vitro susceptibility of Plasmodium falciparum isolates in Vietnam to artemisinin derivatives and other antimalarials. Acta Trop. 63:151-158. [DOI] [PubMed] [Google Scholar]
- 20.Wongsrichanalai, C., A. L. Pickard, W. H. Wernsdorfer, and S. R. Meshnick. 2002. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. 2:209-218. [DOI] [PubMed] [Google Scholar]
- 21.Wongsrichanalai, C., T. Wimonwattrawatee, P. Sookto, A. Laoboonchai, D. G. Heppner, D. E. Kyle, and W. H. Wernsdorfer. 1999. In vitro sensitivity of Plasmodium falciparum to artesunate in Thailand. Bull. W. H. O. 77:392-398. [PMC free article] [PubMed] [Google Scholar]