Abstract
We have characterized an early series of 5,6-bridged dioxinoquinolones which behaved strikingly different from typical quinolones. The 5,6-bridged dioxinoquinolones inhibited Escherichia coli DNA gyrase supercoiling activity but, unlike typical quinolones, failed to stimulate gyrase-dependent cleavable complex formation. Analogous unsubstituted compounds stimulated cleavable complex formation but were considerably less potent than the corresponding 5,6-bridged compounds. Consistent with a previous report (M. Antoine et al., Chim. Ther. 7:434-443, 1972) and contrary to established quinolone SAR trends, a compound with an N-1 methyl substitution (PGE-8367769) was more potent than its analog with an N-1 ethyl substitution (PGE-6596491). PGE-8367769 was shown to antagonize ciprofloxacin-mediated cleavable complex formation in a dose-dependent manner, suggesting an interaction with the gyrase-DNA complex that overlaps that of ciprofloxacin. Resistance to PGE-8367769 in E. coli was found to arise through missense mutations in gyrA, implicating DNA gyrase as the primary antibacterial target. Notably, only 1 of 15 distinct mutations selected on PGE-8367769 (D87G) has previously been implicated in quinolone resistance in E. coli. The remaining 14 mutations (E16V, G31V, R38L, G40A, Y50D, V70A, A84V, I89L, M135T, G173S, T180I, F217C, P218T, and F513C) have not been previously reported, and most were located outside of the traditional quinolone resistance-determining region. These novel GyrA mutations decreased sensitivity to 5,6-bridged dioxinoquinolones by four- to eightfold, whereas they did not confer resistance to other quinolones such as ciprofloxacin, clinafloxacin, or nalidixic acid. These results demonstrate that the 5,6-bridged quinolones act via a mechanism that is related to but qualitatively different from that of typical quinolones.
The recent increase in multiple-drug-resistant bacterial infections has created a critical need to develop novel antibacterial drugs that elude existing mechanisms of resistance. Although the quinolone class is the second largest group of medically important antibacterial drugs, their future utility in the clinic is threatened by the increased rate of emergence of resistant bacteria. Quinolones target two related but functionally distinct and essential type II topoisomerases, DNA gyrase and topoisomerase IV (11, 12, 22, 27). DNA gyrase introduces negative supercoils into DNA and is required to maintain the proper supercoiled state of the chromosome, whereas topoisomerase IV is required to decatenate interlinked replicated chromosomes. DNA gyrase is the primary target of most therapeutic quinolones in gram-negative bacteria, whereas topoisomerase IV is the primary target in gram-positive bacteria (10, 12). A defining feature of the quinolones is their ability to trap a covalent topoisomerase-DNA reaction intermediate termed the cleavable complex. These quinolone-topoisomerase-DNA ternary complexes block both DNA replication and RNA transcription and lead to the formation of lethal double-stranded DNA breaks (5, 14, 26, 28, 29).
An improved definition of the quinolone binding pocket within the topoisomerase-DNA complex may facilitate the rational design of more potent analogs. Because a crystal structure of the quinolone-topoisomerase-DNA ternary complex has not been elucidated, researchers have relied on data for quinolone-resistant mutants to help define this interaction. Spontaneous resistance to quinolones most often arises through point mutations in the topoisomerase-encoding genes. These mutations cluster within a small (∼40-amino-acid) region located in the amino-terminal portion of the GyrA (gyrase) and ParC (topoisomerase IV) subunits known as the quinolone resistance-determining region (QRDR) (15, 30). In Escherichia coli, the most common gyrase mutations occur at Ser-83 and Asp-87 of GyrA and generally lead to the largest increases in quinolone resistance. A crystal structure of the 59-kDa breakage-reunion fragment of GyrA (GyrA59) has revealed that these and other QRDR residues lie in the proposed DNA binding pocket and in close proximity to the active-site Tyr-122 residue (20). A small number of mutations in GyrB and ParE also contribute to quinolone resistance, and it has recently been proposed that the GyrB residues Lys-447 and Asp-426 form part of the quinolone binding pocket of DNA gyrase (13, 15, 31).
Although resistance to most quinolone drugs conforms to the pattern described above, recent reports have demonstrated that resistance to specific structural subfamilies can follow different patterns. For example, resistance to the nonfluorinated quinolones and other 8-methoxy quinolones can arise from mutations at a variety of previously unreported topoisomerase residues (16, 24). In this study, we have reinvestigated an early series of 5,6-bridged dioxinoquinolones originally reported to exhibit structure-activity relationships (SARs) that diverged from those of other quinolones (2). We demonstrate that these 5,6-bridged dioxinoquinolones target DNA gyrase in E. coli but that spontaneous resistance arises primarily through novel mutations in gyrA. In addition, we find that in vitro, these compounds inhibit the supercoiling activity of purified E. coli DNA gyrase but do not stimulate gyrase-dependent cleavable complex formation. We further demonstrate that the 5,6-bridged dioxinoquinolones antagonize ciprofloxacin-mediated cleavable complex formation, suggesting the presence of a binding site which overlaps that of conventional quinolones.
MATERIALS AND METHODS
Reagents and chemicals.
5,6-Bridged dioxinoquinolones, unsubstituted (5-H, 6-H) quinolones, ciprofloxacin, and clinafloxacin (see Table 3) were synthesized at Procter & Gamble Pharmaceuticals as described previously (2, 24). All other antibacterials were purchased from Sigma (St. Louis, Mo.). Wild-type and quinolone-resistant (GyrA, S83W) E. coli DNA gyrases were purchased from John Innes Enterprises Ltd., (Norwich, United Kingdom).
TABLE 3.
Antibacterial activities of 5,6-bridged dioxinoquinolones and comparator quinolones
E. coli, E. coli ATCC 25992; K. pne, K. pneumoniae KL618; M. cat, M. catarrhalis BC24; S. aur, S. aureus ATCC 29213; S. pne, S. pneumoniae ATCC 49619; S. pyo, S. pyogenes STA2.
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. E. coli was routinely grown on LB broth at 37°C. E. coli D21, D22 (lpxC101), and CS1562 (tolC6::mini-Tn10) have been described previously (3, 23). A transducing lysate for tolC6::mini-Tn10 was prepared by growing bacteriophage P1 vir on CS1562 (19). This lysate was used to construct DM200 by transducing D21 to tetracycline resistance (10 μg/ml) and DM202 by transducing D22 to tetracycline resistance.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotype or description | Source or reference |
|---|---|---|
| E. coli | ||
| ATCC 25992 | Quality control strain (FDA strain Seattle 1946) | ATCCa |
| D21 | proA23 lac-28 tsx-81 trp-30 his-51 rpsL173 (Strr) tufA1 ampCp-1 | 23 |
| D22 | D21 lpxC101 | 23 |
| CS1562 | tolC6::miniTn10 | 25 |
| DM200 | D21 tolC6::mini-Tn10 [P1(CS1562) × D21] | This study |
| DM202 | D22 tolC6::mini-Tn10 [P1(CS1562) × D22] | This study |
| BW25113/pKD46 | Δ(araD-araB)567 ΔlacZ4787(::rmB-4) laclp-4000(lacIq) λ−rpoS396(Am) Δ(rhaD-rhaB)568 rmB-4 hsdR514 | 7 |
| CAG12183 gyrA+ | λ zfa-3145::Tn10kan rph-1 | 25 |
| DM232 | DM200 gyrA (D87N) | Macingab |
| DM236 | DM200 gyrB (K447N) | Macingab |
| DM237 | DM200 gyrA (D87H) | Macingab |
| DM316 | DM200 gyrB (G429V) | Macingab |
| DM318 | DM200 gyrB (E466D) | Macingab |
| DM327 | DM200 gyrB (D426N) | Macingab |
| DM340 | BW25113 gyrA (S83W) | This study |
| DM344 | DM200 gyrA (S83W) [P1(DM340 × DM200] | This study |
| K. pneumoniae KL618 | Respiratory pathogen | MRL Pharmaceutical Services |
| M. catarrhalis BC24 | UCLAc Medical Center throat isolate (strain UCLA 77) | J. Hindler |
| S. aureus ATCC 29213 | Quality control strain | ATCC |
| S. pneumoniae ATCC 49619 | Quality control strain | ATCC |
| S. pyogenes STA2 | Chenango Memorial Hospital isolate (strain 1016) | Norwich, N.Y. |
ATCC, American Type Culture Collection.
D. R. Macinga, unpublished data.
UCLA, University of California at Los Angeles.
Construction of an E. coli GyrA S83W chromosomal mutation.
To create a chromosomal GyrA S83W mutant, a hybrid of the bacteriophage λ Red recombinase procedures described by Datsenko and Wanner (7) and Ellis et al. (9) was used. A 71-bp oligonucleotide designated S83W-T, which corresponds to the gyrA template strand from bases 283 to 213 but which contains a G-to-C mutation at base 248, was synthesized (Table 2). E. coli strain BW25113/pKD46 made competent as described previously was electroporated with 0.2 μg of S38W-T, and transformants were selected on 4 μg of nalidixic acid per ml (7). Electroporations with a control oligonucleotide corresponding to the wild-type gyrA sequence from bases 283 to 213 did not yield transformants. The GyrA S83W mutation was confirmed by DNA sequence analysis with primers gyrA-QF and gyrA-QR (Table 2). The S83W allele was then moved into DM200 by P1 transduction, which was confirmed by DNA sequencing, to create DM344.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′)a |
|---|---|
| S83W-T | GCTGCGCCATGCGGACGATCGTGTCA TAGACCGCCCAGTCACCATGGGGAT GGTATTTACCGATTACGTCA |
| gyrA-QF | GACCTTGCGAGAGAAATTACAC |
| gyrA-QR | GATGTTGGTTGCCATACCTACG |
| gyrA-IF1 | TGCTGGTGAACGGTTCTTCC |
| gyrA-IR1 | GTACGACGGGTCACCACTTC |
| gyrA-IF2 | CATCGCGGCGTTTGTTCGTC |
| gyrA-IR2 | CTTAACGTAGCCCTGGTGAG |
| gyrA-IF3 | TCACCCAGGAAGATGTGGTC |
| gyrA-IR3 | GCCACGAGGCACGATCAGAG |
| gyrA-IF4 | GTATTCGCTTAGGTGAAGGC |
| gyrA-IR4 | CCAGACTTTGCAGCCTGGAC |
A point mutation is boldfaced.
MIC determinations.
Liquid MICs were determined in cation-adjusted Mueller-Hinton broth by twofold serial dilution in microtiter plates according to the guidelines of NCCLS (21). Agar MICs were determined by an agar dilution method on Luria-Bertani (LB) agar with twofold increasing concentrations of drug. The agar MIC was defined as the lowest concentration of drug that prevented the formation of single colonies.
Selection of spontaneous E. coli mutants resistant to PGE-8367769.
The agar MICs of PGE-8367769 and nalidixic acid were determined to be 0.5 and 1 μg/ml, respectively, for E. coli DM200. Four independent cultures of E. coli strain DM200 were grown in LB broth for approximately 8 h. Cultures were plated onto LB agar plates containing PGE-8367769 at 0.5 μg/ml (the MIC) and 1 μg/ml (two times the MIC). Three colonies were picked from each of the four cultures with each concentration of drug, for further analysis of a total of 24 mutants. As a control, 24 nalidixic acid-resistant mutants were isolated by a similar procedure with selection at 4 μg/ml (four times the MIC).
DNA sequence analysis.
Saturated cultures of E. coli strains were diluted 1:10 in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8]) and used as templates for PCR amplification by first heating the diluted cultures to 94°C for 5 min prior to the cycling reactions. The primers used for amplification of the gyrA QRDR were gyrA-QF and gyrA-QR (Table 2). The DNA sequences of both strands of the PCR products were determined with an Applied Biosystems automated DNA sequencer. For strains in which no mutation was found in the QRDR, overlapping fragments covering the entire gyrA gene were amplified, and both strands were sequenced. Primers covering the E. coli gyrA gene were as follows: gyrA-IF1, gyrA-IR1, gyrA-IF2, gyrA-IR2, gyrA-IF3, gyrA-IR3, gyrA-IF4, and gyrA-IR4 (Table 2).
Correlation of resistance phenotype and genotype by genetic mapping.
To confirm the genetic linkage of resistance to gyrA, selected quinolone (nalidixic acid or PGE-8367769)-resistant strains were transduced to neomycin resistance (30 μg/ml) with a P1 lysate prepared on CAG12383 gyrA+ (zfa-3145::Tn10kan) to cross in a linked wild-type gyrA gene (25). Neomycin-resistant transductants were then scored for the loss of quinolone resistance.
Bactericidal kinetics.
Log-phase cultures of E. coli strain DM200 (approximately 106 bacteria/ml) were incubated in cation-adjusted Mueller-Hinton broth at 37°C with shaking in the presence of test compounds at one-half, one, two, four, and eight times the MIC. At regular intervals, aliquots were removed and dilutions were prepared in 0.9% saline. The numbers of bacterial CFU were determined by the colony count method with a spiral plater (Spiral Biotech, Inc., Bethesda, Md.).
Topoisomerase catalytic and cleavable complex assays. (i) Gyrase supercoiling assays.
All assays comparing the wild-type and quinolone-resistant enzymes were conducted in parallel with the same reagent stock solutions. Reaction mixtures (20 μl) containing 35 mM Tris-HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 5 mM dithiothreitol, 1.8 mM spermidine, 0.36 μg of bovine serum albumin per ml, 6.5% glycerol, 0.14 mM EDTA, 1 mM ATP, 175 ng of relaxed pBR322 plasmid DNA, and 0.04 U of wild-type or quinolone-resistant E. coli DNA gyrase were incubated at 37°C for 30 min. The reactions were terminated with loading dye (G-2526; Sigma), and the products were separated on 1% agarose gels at 100 V in 1× TBE (89 mM Tris-borate, 2 mM EDTA [pH 8.3]). DNA was visualized after staining of the products with SYBR gold on a Kodak 440 CF image workstation. Supercoiled pBR322 bands were quantified with Kodak 1D analysis software, and 50% inhibitory concentrations (IC50s) were determined by use of a nonlinear regression curve fit on GraphPad Prism software.
(ii) Gyrase cleavable complex assays.
Reaction mixtures (20 μl) containing 35 mM Tris-HCl (pH 7.5), 24 mM KCl, 4 mM MgCl2, 2 mM dithiothreitol, 1.8 mM spermidine, 100 μg of bovine serum albumin per ml, 6.5% glycerol, 9 μg of E. coli tRNA per ml, 2 mM ATP, 240 ng of relaxed pBR322 plasmid DNA, and 2.5 U of wild-type E. coli DNA gyrase were incubated at 37°C for 60 min. The reactions were stopped by the addition of 2 μl of 10% sodium dodecyl sulfate and 1 μl of 1.8 mg of proteinase K per ml was added. The reaction mixtures were incubated for 30 min at 37°C, the reactions were stopped with loading dye, and separation was done on 1% agarose gels containing 0.5 mg of ethidium bromide per ml. The cleaved complex (linear pBR322) was quantified, and the concentration of inhibitor that produced 50% of the maximum cleavage (CC50) was determined by use of a nonlinear regression curve fit.
RESULTS
Antibacterial activities of 5,6-bridged dioxinoquinolones.
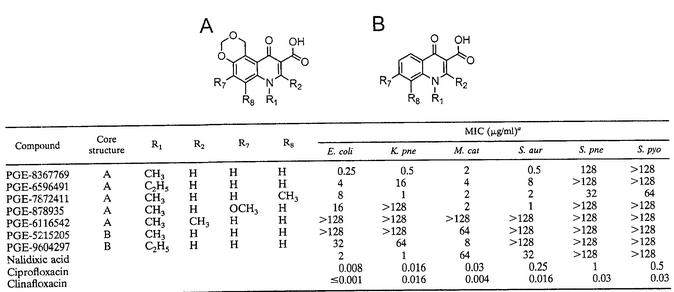
An early study of quinolones described the synthesis of a series of 5,6-bridged dioxinoquinolones (2). The investigators reported an atypical SAR at the N-1 position in which the methyl substitution was more potent than the ethyl substitution. This result was contrary to the general rule for quinolones containing N-1 alkyl substitutions, in which the ethyl substitution generally yields compounds with the greatest activities (1). Because this work was performed before the antibacterial target for quinolones was known, we have reinvestigated these compounds to identify the molecular mechanisms responsible for their apparent atypical behavior. We synthesized a small series of 5,6-bridged dioxinoquinolones and first examined their activities against both gram-negative and gram-positive bacteria (Table 3). The N-1 methyl compound, PGE-8367769, exhibited moderate activity against gram-negative organisms, with MICs for E. coli, Klebsiella pneumoniae, and Moraxella catarrhalis being 0.25, 0.5, and 2 μg/ml, respectively. These MICs were 2- to 32-fold lower than those of the early quinolone nalidixic acid but were significantly higher than those of the later quinolones ciprofloxacin and clinafloxacin. PGE-8367769 also demonstrated activity against Staphylococcus aureus (MIC = 0.5 μg/ml) and displayed only marginal activity against other gram-positive organisms such as Streptococcus pneumoniae and Streptococcus pyogenes. Although the factors responsible for this marked loss of potency against the streptococci are not clear, our results confirm the previously reported antibacterial activity profiles for the 5,6-bridged quinolones (2). The analog with the N-1 ethyl substitution, PGE-6596491, was significantly less potent than PGE-8367769 against all of the organisms tested, confirming the observations of Antoine et al. (2). When corresponding unsubstituted (5-H, 6-H) compounds were examined, the N-1 ethyl compound (PGE-9604297) was more potent than the partner with the methyl substitution (PGE-5215205), in agreement with established SAR trends at the N-1 position. The unsubstituted compounds were significantly less potent than their 5,6-bridged analogs (compare PGE-5215205 to PGE-8367769 and PGE-9604297 to PGE-6596491 in Table 3), demonstrating the importance of the 5,6-dioxino bridge for activity.
We also investigated the effects of various other substitutions to the 5,6-bridged dioxinoquinolone backbone. Consistent with established quinolone SAR trends (1), substitution at the C-2 position completely abolished the activities of the 5,6-bridged series (Table 3, PGE-6116542). A methyl substitution at the C-8 position (PGE-7872411) resulted in increased activities against S. pneumoniae and S. pyogenes relative to those of PGE-8367769 but decreased activities against S. aureus as well as the gram-negative organisms E. coli and K. pneumoniae. A C-7 methoxy substitution (PGE-878935) had little effect on activities against gram-positive bacteria but resulted in significantly diminished activity against gram-negative bacteria relative to the activities of PGE-8367769.
Contribution of outer membrane permeability and efflux to intrinsic resistance of E. coli to 5,6-bridged dioxinoquinolones.
Although spontaneous resistance to quinolones in E. coli most often arises through mutation of the topoisomerase-encoding genes, low-level quinolone resistance may also occur via non-target-site mutations. Mutations leading to the increased expression of the AcrAB-TolC efflux pump or mutations at the mar (multiple antibiotic resistance) locus have been identified in quinolone-resistant isolates (17, 18). It has also been observed that mutations in lpxC, a gene involved in lipid A synthesis, cause increased sensitivity to the quinolones (32, 33). To examine the contribution of efflux and permeability to the intrinsic resistance of E. coli to the 5,6-bridged dioxinoquinolones, we constructed a set of isogenic strains containing mutations at the lpxC or tolC locus and determined the MICs of PGE-8367769, nalidixic acid, and ciprofloxacin. As demonstrated in Table 4, the lpxC101 allele conferred fourfold increased sensitivities to both PGE-8367769 and nalidixic acid, and the tolC6::mini-Tn10 allele conferred eightfold increased sensitivities to these quinolones. When the lpxC and tolC mutations were combined, the sensitivities increased twofold compared to that of the tolC single mutant. Sensitivities to ciprofloxacin were only marginally affected by single mutations (twofold), whereas the sensitivities increased significantly (greater than eightfold) in the lpxC tolC double mutant. Similar results were also observed for other 5,6-bridged dioxinoquinolones and benchmark quinolones (data not shown). These results demonstrate that the outer membrane permeability barrier and active efflux both contribute to the intrinsic resistance of E. coli to classical quinolones as well as the 5,6-bridged dioxinoquinolones.
TABLE 4.
Activities of quinolones against hyperpermeable E. coli strains
| Strain | Relevant genotype | MIC (μg/ml)
|
||
|---|---|---|---|---|
| PGE-8367769 | Nalidixic acid | Ciprofloxacin | ||
| D21 | Wild type | 2 | 4 | 0.008 |
| D22 | lpxC101 | 0.5 | 1 | 0.004 |
| DM200 | tolC6::mini-Tn10 | 0.25 | 0.5 | 0.004 |
| DM202 | lpxC101 tolC6::mini-Tn10 | 0.125 | 0.25 | ≤0.001 |
Spontaneous resistance to PGE-8367769 in E. coli arises through novel gyrA mutations.
To identify the primary target of the 5,6-bridged dioxinoquinolones in E. coli, we isolated and characterized a number of spontaneous PGE-8367769-resistant mutants. tolC-negative strain DM200 was chosen as the parental strain to maximize sensitivity to PGE-8367769 and to potentially eliminate non-target-site mutations (Table 4). Because initial selections with PGE-8367769 at four times the MIC did not yield resistant colonies, we isolated resistant mutants at the MIC and two times the MIC of PGE-8367769. The frequencies at which resistant colonies arose at the MIC and two times the MIC of PGE-8367769 were 2 × 10−9 and 4 × 10−9, respectively. A total of 24 PGE-8367769-resistant isolates were selected for characterization. As a control, 24 spontaneous nalidixic acid-resistant mutants selected in a similar manner were also characterized.
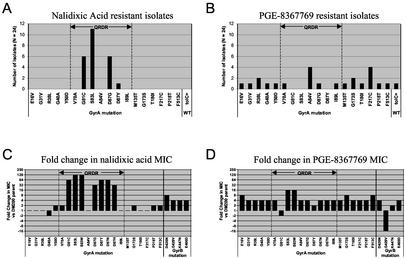
To determine the identities of the resistance-conferring mutations, we sequenced the QRDR of gyrA from each of the PGE-8367769- and nalidixic acid-resistant mutants. For strains that did not contain mutations in this region, the entire gyrA gene was sequenced. The results of the DNA sequence analysis are summarized in Fig. 1A and B. Selection for nalidixic acid resistance produced typical GyrA QRDR hotspot mutations, including S83L, D87G, D87Y, and G81C (Fig. 1A). The mutation pattern for PGE-8367769 was strikingly different (Fig. 1B). Twenty-three of the 24 strains harbored mutations in GyrA, but the mutations were located inside the traditional QRDR (between residues 67 and 106) in only 7 strains. These included V70A, A84V, D87G, and I89L mutations. To our knowledge, the V70A and I89L mutations have not been previously implicated in quinolone resistance in E. coli, and A84V represents a novel amino acid substitution at position 84 (30). Notably, no mutations were observed at Ser-83, a residue known to play a critical role in quinolone resistance development. Several novel mutations were identified outside of the traditional QRDR. These included E16V, G31V, R38L, G40A, Y50D, V70A, M135T, G173S, T180I, F217C, P218T, and F513C. A single PGE-8367769-resistant strain did not contain a mutation in either gyrA or gyrB. Analysis of the tolC locus from this strain revealed that a perfect excision of the tolC6::mini-Tn10 insertion had occurred, producing a functional wild-type tolC gene and confirming the role of efflux in the intrinsic susceptibility of E. coli to PGE-8367769 (Fig. 1B, tolC+).
FIG. 1.
Characterization of spontaneous quinolone-resistant E. coli mutants. A total of 24 nalidixic acid-resistant (A) or 24 PGE-8367769-resistant (B) E. coli isolates were selected, and the gyrA gene was sequenced as described in Materials and Methods. The QRDR, spanning from residue 66 to residue 106, is delineated by dashed line; tolC+, perfect excision of the mini::Tn10 element regenerating the wild-type tolC gene (see text). The fold change in the nalidixic acid MICs (C) and the PGE-8367769 MICs (D) for quinolone-resistant GyrA and GyrB mutants relative to those for the DM200 parental strain are also shown.
To demonstrate genetically that each gyrA mutation was responsible for the observed resistance phenotype, representative strains containing each unique mutation were subjected to transduction linkage analysis. Strains were transduced to neomycin resistance with a P1 lysate containing the zfa-3145::Tn10kan marker, which is transductionally linked to gyrA. Transductants were then scored for the loss of quinolone resistance. For each representative strain, quinolone resistance was found to be linked to the zfa-3145::Tn10kan marker with linkage frequencies ranging from 57 to 93%, demonstrating that quinolone resistance is linked to gyrA (data not shown).
Unique gyrA mutations selected with PGE-8367769 confer resistance to 5,6-bridged dioxinoquinolones but not typical quinolones.
To measure the effect of each GyrA mutation on the antibacterial activities of PGE-8367769, we determined the MICs for a representative strain containing each unique mutation, as well as for other gyrA and gyrB mutant strains (D. R. Macinga, unpublished data, 2002). Figure 1D and Table 5 illustrate that the novel gyrA mutations conferred four- to eightfold increased resistance to PGE-8367769 relative to the resistance of isogenic parent DM200. Although Ser-83 mutations were not isolated with PGE-8367769 (Fig. 1B), isogenic S83L and S83W mutations each conferred 16-fold increased resistance to PGE-8367769. With the exception of G81C and D87Y, other GyrA QRDR mutations led to fourfold increases in PGE-8367769 resistance; G81C and D87Y produced nonsignificant changes. GyrB D426N, G429V, and E466D mutants, which were isolated with other topoisomerase inhibitors, were fourfold more resistant to PGE-8367769 than the parent strain, DM200 (Macinga, unpublished data). Unexpectedly, a strain harboring a novel GyrB G429V mutation was hypersusceptible (16-fold) to PGE-8367769. As a control, we determined the nalidixic acid MICs for the same strains (Fig. 1C). Common QRDR mutations at Ser-83, Asp-87, and Gly-81 conferred 32- to 128-fold-increased nalidixic acid resistance, and GyrB mutations conferred four- to eight-fold-increased nalidixic acid resistance. In contrast, the novel GyrA mutations did not confer significant changes in sensitivity to nalidixic acid. We also tested the potencies of a number of other 5,6-bridged and control quinolones against the same panel of gyrA and gyrB mutants (Table 5). All of the 5,6-bridged quinolones were found to be four- to eightfold less potent against strains containing novel PGE-8367769-selected GyrA mutations. In contrast, the potencies of the unsubstituted compounds PGE-5215205 and PGE-9604297, as well as those of the benchmark quinolones nalidixic acid, ciprofloxacin, and clinafloxacin, were not significantly affected by these novel GyrA mutations.
TABLE 5.
Fold changes in MICs of quinolones for GyrA and GyrB mutants relative to MICs for DM200 parental strain
| Strain | GyrA mutation | GyrB mutation | Fold change in MIC relative to MIC for DM200a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PGE- 8367769 (0.25)b | PGE-6596491 (0.5) | PGE-7872411 (0.25) | PGE-878935 (0.125) | PGE-5215205 (64) | PGE-9604297 (8) | Nalidixic acid (0.5) | Cipro- floxacin (0.004) | Clina- floxacin (0.002) | |||
| DM289 | E16V | 8 | 8 | 8 | 4 | 2 | 1 | 1 | 1 | 1 | |
| DM291 | G31V | 4 | 4 | 8 | 4 | 2 | 1 | 1 | 1 | 1 | |
| DM276 | R38L | 4 | 4 | 8 | 8 | 2 | 1 | 1 | 1 | 1 | |
| DM275 | G40A | 4 | 4 | 4 | 4 | 1 | 1 | −2 | −2 | 1 | |
| DM279 | Y50D | 4 | 4 | 4 | 4 | 2 | −2 | 1 | −2 | −2 | |
| DM283 | V70Ac | 4 | 8 | 8 | 8 | 2 | 2 | 2 | 1 | 1 | |
| DM225 | G81Cc | −2 | 1 | ≤−4 | ≤−2 | 1 | 16 | 64 | 8 | 4 | |
| DM218 | S83Lc | 16 | 32 | 16 | 16 | 2 | 4 | 128 | 16 | 16 | |
| DM344 | S83Wc | 16 | 16 | 8 | 16 | 2 | 8 | 128 | 32 | 16 | |
| DM270 | A84Vc | 4 | 8 | 8 | 8 | 1 | −4 | 1 | 1 | 4 | |
| DM219 | D87Gc | 4 | 4 | 8 | 4 | 2 | 2 | 32 | 8 | 4 | |
| DM221 | D87Yc | 2 | 2 | 4 | 2 | 2 | 2 | 64 | 8 | 4 | |
| DM232 | D87Nc | 4 | 4 | 4 | 4 | 2 | 2 | 64 | 8 | 8 | |
| DM237 | D87Hc | 4 | 4 | 8 | 8 | 2 | 2 | 32 | 8 | 4 | |
| DM280 | I89Lc | 4 | 4 | 4 | 4 | 2 | −4 | 1 | 1 | 1 | |
| DM281 | M135T | 4 | 4 | 8 | 8 | 2 | −4 | 1 | 1 | 1 | |
| DM282 | G173S | 8 | 4 | 4 | 8 | 1 | 1 | 2 | 1 | 1 | |
| DM290 | T180I | 4 | 4 | 8 | 8 | 2 | −4 | 1 | 2 | 2 | |
| DM271 | F217C | 4 | 4 | 8 | 4 | 2 | −8 | 1 | 1 | 2 | |
| DM288 | P218T | 4 | 4 | 8 | 8 | 2 | −4 | 2 | 1 | 1 | |
| DM293 | F513C | 8 | 8 | 8 | 4 | 1 | 1 | 2 | 1 | 1 | |
| DM327 | D426N | 4 | 1 | 2 | 2 | 1 | 1 | 8 | 2 | 2 | |
| DM316 | G429V | −16 | −4 | ≤−4 | ≤−2 | −4 | 1 | 4 | 2 | 4 | |
| DM236 | K447N | 2 | 1 | −2 | 1 | 1 | −16 | 4 | ≤−4 | −2 | |
| DM318 | E466D | 4 | 4 | 8 | 4 | ≥4 | −2 | 4 | 4 | 4 | |
Fold change was calculated by dividing the MIC for the mutant strain by the MIC for strain DM200. For ratios less than 1, the negative reciprocal is displayed. Changes of fourfold or greater are in boldface, and fold changes of negative fourfold or less are in italics.
The values in parentheses are the MICs for DM200 (in micrograms per milliliter).
Residues inside the GyrA QRDR.
Bactericidal kinetics.
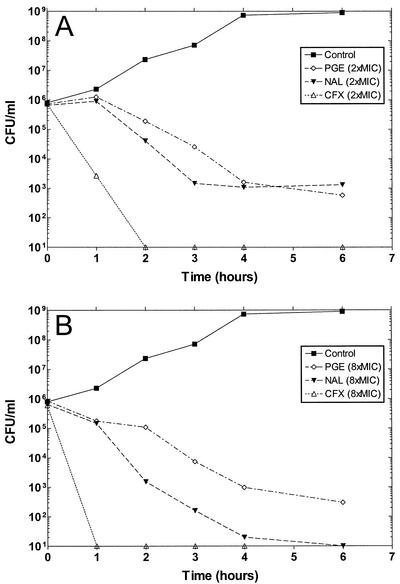
To determine whether the 5,6-bridged dioxinoquinolones were bactericidal, the bactericidal kinetics of PGE-8367769 against E. coli strain DM200 were compared to those of nalidixic acid and ciprofloxacin. The results from experiments with two and eight times the MICs are shown in Fig. 2. At two times the MIC, PGE-8367769 and nalidixic acid each produced an approximately 3-log reduction in the number of viable CFU per milliliter by 6 h (Fig. 2A). However, the time of killing by PGE-8367769 was delayed by approximately 1 h compared to the time of killing by nalidixic acid. When PGE-8367769 and nalidixic acid were tested at eight times the MIC, the effects of PGE-8367769 and nalidixic acid were easily distinguishable (Fig. 2B). In this experiment, nalidixic acid produced a 3-log reduction in the number of CFU per milliliter by 3 h and a reduction in viable counts below detectable limits by 6 h. In contrast, PGE-8367769 required 6 h to reduce the number of CFU per milliliter by 3 log units and did not reduce the viable counts below detectable levels. Ciprofloxacin was significantly better than either PGE-8367769 or nalidixic acid at killing DM200, reducing the number of CFU per milliliter to below the detection limit by 2 h at two times the MIC (Fig. 2A) and by 1 h at eight times the MIC (Fig. 2B).
FIG. 2.
Bactericidal kinetics of PGE-8367769, nalidixic acid, and ciprofloxacin against E. coli DM200. The results obtained with two times the MIC (A) and four times the MIC (B) are shown. The MICs of the drugs for DM200 are shown in Table 5. PGE, PGE-8367768; NAL, nalidixic acid; CFX, ciprofloxacin. Datum points below the detection limit of the assay (20 CFU/ml) are plotted as 101 CFU/ml.
5,6-Bridged dioxinoquinolones inhibit E. coli DNA gyrase supercoiling activity.
To investigate the molecular mechanisms behind the atypical SAR trends of the dioxinoquinolones, we tested the abilities of the compounds to inhibit the supercoiling activity of purified E. coli DNA gyrase. As shown in Table 6, PGE-8367769 (N-1 methyl) inhibited gyrase supercoiling activity with an IC50 of 7.5 μg/ml. This compound was about twofold more potent than the benchmark nalidixic acid but was significantly less potent than ciprofloxacin and clinafloxacin. Consistent with the potencies observed against E. coli (Table 3), PGE-8367769 was approximately 8-fold more potent at gyrase supercoiling inhibition than the N-1 ethyl analog, PGE-6596491 (IC50 = 62.3 μg/ml), and 63-fold more potent than its unsubstituted (5-H, 6-H) partner, PGE-5215205 (IC50 = 219.7 μg/ml). PGE-6596491 was approximately threefold more potent than its unsubstituted partner, PGE-9604297 (IC50 = 179.5 μg/ml). The C-8 methyl (PGE-7872411) and C-7 methoxy (PGE-878935) compounds exhibited potencies against DNA supercoiling activity that were similar to those of PGE-837769. In agreement with the lack of antibacterial activity, the compound with the C-2 methyl substitution, PGE-6116542, did not inhibit supercoiling at concentrations as high as 500 μg/ml.
TABLE 6.
Inhibition of E. coli DNA gyrase supercoiling by 5,6-bridged dioxinoquinolones and comparator quinolones
| Quinolone | IC50 (μg/ml)a
|
QR/WT ratiob | |
|---|---|---|---|
| WT gyrase | QR gyrase | ||
| PGE-8367769 | 7.5 | 81.3 | 10.8 |
| PGE-6596491 | 62.3 | >500 | — |
| PGE-7872411 | 6.5 | 102.1 | 15.7 |
| PGE-878935 | 6.6 | 77.1 | 11.6 |
| PGE-6116542 | >500 | >500 | — |
| PGE-5215205 | 219.7 | >500 | — |
| PGE-9604297 | 179.5 | >500 | — |
| Nalidixic Acid | 15.9 | >500 | — |
| Ciprofloxacin | 0.12 | 16.1 | 134.3 |
| Clinafloxacin | 0.032 | 0.45 | 13.8 |
WT, wild type; QR, quinolone resistant (GyrA S83L).
The QR/WT ratio were calculated by dividing the IC50 for the wild-type gyrase by the IC50 for the quinolone-resistant gyrase. —, could not be calculated.
To investigate the influence of a GyrA Ser-83 mutation on the potencies of the 5,6-bridged dioxinoquinolones, supercoiling inhibition assays were performed with a quinolone-resistant mutant E. coli DNA gyrase (GyrA S83W). PGE-8367769 was approximately 11-fold less potent against the quinolone-resistant enzyme than against the wild-type enzyme (Table 3). This result correlates well with the MIC data shown in Fig. 1D and Table 5, in which the S83W mutant strain DM344 was 16-fold less sensitive to PGE-8367769 than the isogenic parent DM200. Similar results were observed for PGE-7872411 and PGE-878935. For comparison, clinafloxacin and ciprofloxacin were 14- and 134-fold less potent against the quinolone-resistant enzyme than against the wild-type enzyme, and nalidixic acid was inactive against the quinolone-resistant enzyme (Table 6).
5,6-Bridged dioxinoquinolones do not stimulate DNA gyrase-dependent cleavable complex formation.
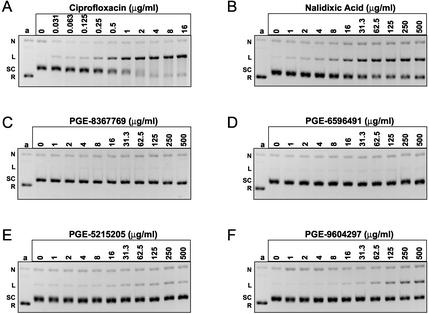
We have used an in vitro assay to measure the cleavable complex-stimulating activities of both benchmark quinolones and 5,6-bridged dioxinoquinolones using relaxed pBR322 as a DNA template (see Materials and Methods). As demonstrated in Fig. 3A and B, respectively, ciprofloxacin and nalidixic acid stimulated gyrase-dependent cleavable complex formation in a dose-dependent manner. Ciprofloxacin stimulated a maximum conversion of 79% of the template into the linear product (cleaved complex), and the CC50 was 0.69 μg/ml. Nalidixic acid stimulated 47% conversion of the template to the linear product, with a CC50 of 13.9 μg/ml. It is also evident from the gels that ciprofloxacin and nalidixic acid inhibited gyrase supercoiling activity under the cleavage reaction conditions. In contrast, PGE-8367769 and PGE-6596491 did not stimulate the cleavable complex above the background level at concentrations as high as 500 μg/ml (Fig. 3C and D, respectively). Strikingly, the corresponding unsubstituted analogs, PGE-5215205 and PGE-9604297, respectively, were able to stimulate cleavable complex formation (Fig. 3E and F, respectively). The absolute levels of cleavage were considerably lower than those produced by benchmark compounds, with PGE-5215205 and PGE-9604297 producing only 13 and 27% maximum cleavage, respectively. Moreover, none of the 5,6-bridged dioxinoquinolones stimulated cleavable complex formation when they were tested with purified human DNA topoisomerase IIα (unpublished data), and PGE-8367769 induced no micronucleus formation when it was tested for genetic toxicity with Chinese hamster ovary cells (6), suggesting that the activities of the present compounds are relatively selective toward bacterial topoisomerases.
FIG. 3.
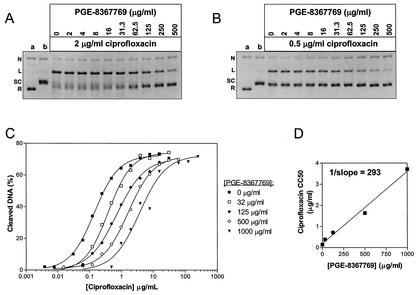
Comparison of quinolone-stimulated cleavable complex formation by E. coli DNA gyrase. (A to F) Cleavable complex assays were performed as described in the Materials and Methods and contained the quinolones at the indicated concentrations. Gel images have been inverted for easier visualization of the DNA species. Lane a, relaxed pBR322 DNA; N, nicked DNA; L, linear DNA; SC, supercoiled DNA; R, relaxed DNA.
PGE-8367769 antagonizes quinolone-mediated cleavable complex formation.
Because PGE-8367769 inhibited DNA gyrase supercoiling activity but did not simulate cleavable complex formation, we hypothesized that PGE-8367769 may interact with DNA gyrase in a manner qualitatively different from that for conventional quinolones. We reasoned that if PGE-8367769 binds to the gyrase-DNA complex at the same site as typical quinolones, then PGE-8367769 would be expected to compete with quinolones for binding to the complex. We first tested the ability of PGE-8367769 to antagonize the cleavable complex stimulated by a constant concentration of ciprofloxacin. As demonstrated in Fig. 4A, titration of PGE-8367769 into reaction mixtures containing 2 μg of ciprofloxacin per ml resulted in a dose-dependent inhibition of cleavable complex formation. Cleavage was reduced from 60% in the presence of ciprofloxacin alone to 18% in the presence of ciprofloxacin and 500 μg of PGE-8367769 per ml. The results of a similar experiment in which ciprofloxacin was included at 0.5 μg/ml are depicted in Fig. 4B. In this experiment, inhibition of ciprofloxacin-mediated cleavable complex formation was even more evident, with cleavage being reduced from 38% in the presence of ciprofloxacin alone to 6% in the presence of ciprofloxacin and 500 μg of PGE-8367769 per ml. In contrast, titration of the GyrB inhibitor novobiocin into cleavage reactions containing ciprofloxacin did not result in a reduction in the amount of cleaved pBR322 (data not shown). These experiments suggest that PGE-8367769 competes with ciprofloxacin for a similar binding site on the gyrase-DNA binary complex. To further test this hypothesis, we performed a series of cleavage assays in which ciprofloxacin was titrated in the presence of constant concentrations of PGE-8367769. Figure 4C demonstrates that the ciprofloxacin dose response was shifted in the presence of PGE-8367769, resulting in increased apparent CC50s. The CC50 of ciprofloxacin was 0.14 μg/ml in the absence of PGE-8367769 and was shifted to 0.39, 0.72, 1.64, and 3.71 μg/ml in the presence of 32, 125, 500, and 1,000 μg of PGE-8367769 per ml, respectively. The maximum amount of cleavage stimulated by ciprofloxacin (approximately 70%) remained unchanged regardless of the PGE-8367769 concentration. Taken together, these results strongly suggest that ciprofloxacin and PGE-8367769 share an overlapping binding site. In Fig. 4D, the apparent CC50 of ciprofloxacin has been plotted against the concentration of PGE-8367769 and fit by linear regression. The inverse slope of this line, 293, gives an estimate of the relative amount of PGE-8367769 required to outcompete a given amount of ciprofloxacin for binding to the gyrase-DNA complex.
FIG. 4.
Antagonism of ciprofloxacin-stimulated cleavable complex formation by PGE-8367769. Cleavable complex assays containing ciprofloxacin at 2 μg/ml (A) or 0.5 μg/ml (B) and PGE-8367769 at the indicated concentrations were performed as described in the Materials and Methods. Gel images have been inverted for easier visualization of the DNA species. Lane a, relaxed pBR322 DNA; lane b, control reaction with no ciprofloxacin; N, nicked DNA; L, linear DNA; SC, supercoiled DNA; R, relaxed DNA. (C) Ciprofloxacin dose-response curves in the presence of constant concentrations of PGE-8367769. (D) Ciprofloxacin CC50 versus the concentration of PGE-8367769 included in the reaction.
DISCUSSION
In the present study, we have reinvestigated a series of early quinolones containing a 5,6-dioxino bridge (5,6-bridged dioxinoquinolones) (2). The original study by Antoine et al. (2) was performed before type II DNA topoisomerases were demonstrated to be the antibacterial targets of quinolones, and thus, only the antibacterial potencies of the compounds were reported. We have now further demonstrated that the 5,6-bridged dioxinoquinolones are inhibitors of purified E. coli DNA gyrase supercoiling activity and that spontaneous resistance to these compounds in E. coli arises through point mutations in gyrA. Taken together, these data implicate DNA gyrase as the primary antibacterial target in E. coli. Our results confirm the atypical finding that the derivative with the N-1 methyl substitution has greater antibacterial activity than its analog with an N-1 ethyl substitution. In E. coli, this difference was 16-fold and was in good agreement with the observed potencies against purified E. coli DNA gyrase supercoiling activity (8.3-fold difference). Thus, the atypical SAR at position N-1 is most likely caused by the differences in the relative potencies against DNA gyrase. To address the contribution of the 5,6-dioxino bridge to the biological properties, we synthesized unsubstituted (5-H, 6-H) analogs of the N-1 methyl and N-1 ethyl dioxinoquinolones. The unsubstituted compounds exhibited weak to no antibacterial activity and were considerably less potent than the corresponding bridged compounds at gyrase supercoiling inhibition, demonstrating that the 5,6-dioxino bridge contributes significantly to the activities of these compounds. Consistent with established SAR trends, the unsubstituted N-1 ethyl compound was more potent than its partner with the N-1 methyl substitution. These results suggest that the 5,6-dioxino bridge is directly responsible for the atypical SAR trend at the N- 1 position.
The 5,6-bridged dioxinoquinolones PGE-8367769 and PGE-6596491 failed to stimulate gyrase-dependent cleavable complex formation at concentrations as high as 500 μg/ml. Because PGE-8367769 was approximately twofold more potent than nalidixic acid at gyrase supercoiling inhibition and because nalidixic acid stimulated the cleavable complex with a CC50 of 13.9 μg/ml, it is highly unlikely that higher concentrations of PGE-8367769 would have led to cleavable complex formation. The 5,6-bridged quinolones may function as gyrase supercoiling inhibitors without stimulating cleavable complex formation by inhibiting the enzyme prior to DNA cleavage. This mechanism would be in contrast to that of typical quinolones such as ciprofloxacin, which are thought to stimulate cleavage and inhibit religation (8). Stabilization of the cleavable complex by quinolones is thought to be a prerequisite for double-stranded DNA break formation and bacterial death. We have found PGE-8367769 to be significantly less effective at killing E. coli than nalidixic acid and ciprofloxacin. This decreased killing may be directly related to the inability of PGE-8367769 to stabilize the cleavable complex in the bacterium. Further experiments will need to be conducted to determine whether PGE-8367769 fails to block DNA synthesis or to produce double-stranded DNA breaks in vivo. Such experiments may provide insights into the mechanisms of bacterial killing by quinolones.
PGE-8367769 was shown to antagonize ciprofloxacin-mediated cleavable complex formation, suggesting competition between PGE-8367769 and ciprofloxacin for binding to the gyrase-DNA complex. The coumarin antibacterial novobiocin, a potent inhibitor of GyrB ATPase activity, also inhibits gyrase supercoiling activity without simulating cleavable complex formation (4). Novobiocin did not antagonize ciprofloxacin-mediated cleavable complex formation, demonstrating that inhibition of gyrase supercoiling activity is not sufficient to block this effect. In dose-response experiments with ciprofloxacin (Fig. 4C), the inclusion of increasing PGE-8367769 concentrations caused a corresponding increase in the apparent ciprofloxacin CC50s, whereas the maximum level of cleavage stimulated by ciprofloxacin remained unchanged. From Fig. 4D, the affinity of ciprofloxacin for the gyrase-DNA complex was estimated to be approximately 300-fold greater than that of PGE-8367769. Overall, these results demonstrate that binding of ciprofloxacin and PGE-8367769 to DNA gyrase are mutually exclusive and strongly suggest that both compounds compete for a common, overlapping binding site within the gyrase-DNA complex.
The analysis of spontaneous mutants resistant to PGE-8367769 identified DNA gyrase as the primary antibacterial target in E. coli. However, the GyrA residues implicated in resistance development were dramatically different from those conferring resistance to typical quinolones. Of the 15 different mutations identified, only the D87G mutation has been described previously. Several isolates were found to harbor a conservative alanine-to-valine change at position 84 (A84V), which differs from a previously described A84P mutation (30). According to the GyrA59 fragment crystal structure, Ala-84 is located in α helix 4 and is situated between the key residues in quinolone resistance development, Ser-83 and Asp-87 (20). The A84P mutation is thought to confer resistance to quinolones by disrupting α helix 4 and perturbing the quinolone binding site (8), whereas the conservative A84V mutation would be expected to have a more subtle effect on the structure of α helix 4. The A84V mutation as well as a second conservative α helix 4 mutation, I89L, conferred resistance only to the 5,6-bridged quinolones. A previously described α helix 4 mutation, G81C (30), conferred resistance to nalidixic acid, ciprofloxacin, and clinafloxacin. In contrast, G81C did not confer resistance to the 5,6-bridged quinolones and led to significant hypersusceptibility to PGE-7872411 and PGE-878935 (Table 5). These data provide strong genetic evidence that the 5,6-dioxinoquinolones interact with DNA gyrase at a site similar to that at which typical quinolones interact, but do so in a qualitatively unique manner.
The majority of the mutations conferring PGE-8367769 resistance arose at residues outside of the QRDR and have not been previously reported. Like the A84V and I89L mutations, these conferred resistance only to the 5,6-bridged quinolones. The GyrA59 crystal structure demonstrates that the key residues involved in quinolone resistance development (Ser-83 and Asp-87) are located in close proximity to the active-site tyrosines (Tyr-122) at the dimer interface (20). A model with double-stranded DNA bound to this domain places Ser-83 and Asp-87 at the point where distortion of the DNA is predicted to occur. We speculate that the non-QRDR mutations may cause long-range conformational changes that alter the binding site of the 5,6-bridged quinolones. These conformational changes may also alter the distortion of the double-stranded DNA bound to the enzyme. The observation that these mutations confer resistance only to the 5,6-bridged dioxinoquinolones suggests that the dioxino moiety contributes directly to the binding of the drugs to the gyrase-DNA complex. This view is further supported by the data showing that unsubstituted analogs of the 5,6-bridged dioxinoquinolones are much less potent, as measured by both antibacterial activity and gyrase supercoiling inhibition. Alternatively, the 5,6-dioxino moiety may cause indirect inhibition of gyrase by instead promoting drug binding to the DNA substrate (rather than binding to the gyrase-DNA complex, as occurs for classical quinolones). However, this interpretation seems unlikely because these compounds did not show a significant affinity for DNA when they were tested in parallel experiments for direct binding to calf thymus DNA (data not shown).
In summary, the 5,6-bridged dioxinoquinolones were found to target DNA gyrase in E. coli but differed significantly in their interaction with this target relative to those of conventional quinolones, as determined by (i) SARs, (ii) resistance development, (iii) bactericidal kinetics, and (iv) cleavable complex stabilization. The results of this study demonstrate that modifications of the quinolone backbone can lead to qualitative differences in drug-target interactions and suggest that compounds can be designed to circumvent existing topoisomerase-mediated resistance. Although the compounds investigated in this study were considerably less potent than present therapeutic fluoroquinolones, it should be noted that their substitution patterns were rather simple. Because the initial compound, PGE-8367769, did not stimulate enzyme-mediated cleavable complex formation with E. coli gyrase or human topoisomerase IIα and showed no genetic toxicity against Chinese hamster ovary cells, this series of 5,6-bridged quinolones may represent an opportunity to design new analogs with improved selectivities and safety profiles in humans. Although the existing knowledge of the SARs of this series is limited, further investigation of well-known substituents at the N-1, C-7, and C-8 positions may yield new derivatives with significantly improved potencies, spectra, and/or selectivities. At a minimum, even the present limited SARs for these unique compounds strongly suggests that there is still significant unexploited chemical and biological space for the invention of new antibacterials based on modifications of the traditional quinolone scaffold.
Acknowledgments
We thank Anthony Maxwell for critical comments on the manuscript.
REFERENCES
- 1.Albrecht, R. 1977. Development of antibacterial agents of the nalidixic acid type. Prog. Drug Res. 21:9-104. [DOI] [PubMed] [Google Scholar]
- 2.Antoine, M., S. Chabassier, S. Geiger, J. Le Blevec, J. Le Coent, M. Pesson, D. Richer, E. Horvath, M. P. de Lajudie, S. Patte, and B. Pradeau. 1972. Acides m-dioxino quinoleine carboxyliques a action antibacterienne. I. Chim. Ther. 7:434-443. [Google Scholar]
- 3.Austin, E. A., J. F. Graves, L. A. Hite, C. T. Parker, and C. A. Schnaitman. 1990. Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli K-12: insertion mutagenesis of the rfa locus. J. Bacteriol. 172:5312-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett, J. F., J. I. Bernstein, H. M. Krause, J. J. Hilliard, and K. A. Ohemeng. 1993. Testing potential gyrase inhibitors of bacterial DNA gyrase: a comparison of the supercoiling inhibition assay and “cleavable complex” assay. Anal. Biochem. 214:313-317. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. R., M. Malik, M. Snyder, and K. Drlica. 1996. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J. Mol. Biol. 258:627-637. [DOI] [PubMed] [Google Scholar]
- 6.Ciaravino, V., M. J. Suto, and J. C. Theiss. 1993. High capacity in vitro micronucleus assay for assessment of chromosome damage: results with quinolone/naphthyridone antibacterials. Mutat. Res. 298:227-236. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drlica, K. 1999. Mechanism of fluoroquinolone action. Curr. Opin. Microbiol. 2:504-508. [DOI] [PubMed] [Google Scholar]
- 9.Ellis, H. M., D. Yu, T. DiTizio, and D. L. Court. 2001. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 98:6742-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 11.Fournier, B., and D. C. Hooper. 1998. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob. Agents Chemother. 42:121-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gellert, M., K. Mizuuchi, M. H. O'Dea, T. Itoh, and J. I. Tomizawa. 1977. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. USA 74:4772-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heddle, J., and A. Maxwell. 2002. Quinolone-binding pocket of DNA gyrase: role of GyrB. Antimicrob. Agents Chemother. 46:1805-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiasa, H., D. O. Yousef, and K. J. Marians. 1996. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J. Biol. Chem. 271:26424-26429. [DOI] [PubMed] [Google Scholar]
- 15.Hooper, D. C. 1999. Mechanisms of fluoroquinolone resistance. Drug. Resist. Update 2:38-55. [DOI] [PubMed] [Google Scholar]
- 16.Ince, D., and D. C. Hooper. 2000. Mechanisms and frequency of resistance to premafloxacin in Staphylococcus aureus: novel mutations suggest novel drug-target interactions. Antimicrob. Agents Chemother. 44:3344-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jellen-Ritter, A. S., and W. V. Kern. 2001. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob. Agents Chemother. 45:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maneewannakul, K., and S. B. Levy. 1996. Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Morais Cabral, J. H., A. P. Jackson, C. V. Smith, N. Shikotra, A. Maxwell, and R. C. Liddington. 1997. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388:903-906. [DOI] [PubMed] [Google Scholar]
- 21.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 22.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1996. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Normark, S., H. G. Boman, and E. Matsson. 1969. Mutant of Escherichia coli with anomalous cell division and ability to decrease episomally and chromosomally mediated resistance to ampicillin and several other antibiotics. J. Bacteriol. 97:1334-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roychoudhury, S., T. L. Twinem, K. M. Makin, M. A. Nienaber, C. Li, T. W. Morris, B. Ledoussal, and C. E. Catrenich. 2001. Staphylococcus aureus mutants isolated via exposure to nonfluorinated quinolones: detection of known and unique mutations. Antimicrob. Agents Chemother. 45:3422-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder, M., and K. Drlica. 1979. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J. Mol. Biol. 131:287-302. [DOI] [PubMed] [Google Scholar]
- 27.Sugino, A., C. L. Peebles, K. N. Kreuzer, and N. R. Cozzarelli. 1977. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. USA 74:4767-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wentzell, L. M., and A. Maxwell. 2000. The complex of DNA gyrase and quinolone drugs on DNA forms a barrier to the T7 DNA polymerase replication complex. J. Mol. Biol. 304:779-791. [DOI] [PubMed] [Google Scholar]
- 29.Willmott, C. J., S. E. Critchlow, I. C. Eperon, and A. Maxwell. 1994. The complex of DNA gyrase and quinolone drugs with DNA forms a barrier to transcription by RNA polymerase. J. Mol. Biol. 242:351-363. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida, H., M. Bogaki, M. Nakamura, L. M. Yamanaka, and S. Nakamura. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young, K., and L. L. Silver. 1991. Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. J. Bacteriol. 173:3609-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young, K., L. L. Silver, D. Bramhill, P. Cameron, S. S. Eveland, C. R. Raetz, S. A. Hyland, and M. S. Anderson. 1995. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J. Biol. Chem. 270:30384-30391. [DOI] [PubMed] [Google Scholar]