Abstract
A novel membrane-associated protein, MsrR, was identified in Staphylococcus aureus which affects resistance to methicillin and teicoplanin, as well as the synthesis of virulence factors. MsrR belongs to the LytR-CpsA-Psr family of cell envelope-related transcriptional attenuators and was shown to be inducible by cell wall-active agents, such as β-lactams, glycopeptides, and lysostaphin. The expression of msrR peaked in the early exponential growth phase and decreased sharply thereafter. msrR mutants showed increased sarA transcription and an earlier and higher expression of RNAIII, resulting in altered expression of virulence factors such as alpha-toxin and protein A. These observations suggest that MsrR is a new component involved in sarA attenuation and the regulatory network controlling virulence gene expression in S. aureus.
Staphylococcus aureus, an inhabitant of the skin and mucous membranes in 10 to 30% of the healthy population, easily survives in the environment and is, moreover, the most common cause of community-acquired and nosocomial infections. The pathogenicity of S. aureus is based on an impressive repertoire of virulence factors. The diseases caused range from superficial infections of the skin to life-threatening infections such as septicemia, endocarditis, osteomyelitis, and toxic shock syndrome. Virulence factors comprise surface-associated and extracellular proteins such as toxins and enzymes. Cell surface-associated proteins, the MSCRAMMs (microbial surface components recognizing adhesive matrix molecules), function in adhesion and colonization of the host and evasion of host defenses. Cell surface-associated factors are needed primarily during the initial stages of infection and are expressed mainly in the early exponential growth phase. The exoenzymes, which damage the host tissue and promote dissemination of the pathogen and most soluble exoproteins, have a role in a later stage of the infection and are produced in the postexponential phase. The coordinate and timely expression of those virulence factors during the growth cycle is governed by a complex network comprising two-component sensor transducers, global regulatory systems, and transcription factors, which are important for the pathogen to adapt to the changing host environment during different stages of infection (2, 21, 23, 26). One of the two major global regulatory systems is the agr (accessory gene regulator) regulon. This locus consists of two divergent transcripts originating from the promoters P2 and P3, which produce RNAII and RNAIII, respectively. The RNAII transcript encodes four proteins, AgrA to AgrD. AgrA and AgrC form a two-component regulatory system whereby AgrC acts as the signal receptor and AgrA acts as sensor regulator. The signal is a small peptide processed from AgrD by AgrB. This self-encoded autoinducing peptide determines the specificity group of S. aureus (29). The autoinducer leads from the phosphorylation of AgrC to the phosphorylation of AgrA, which upregulates both P2 and P3 promoters of the agr regulon. The RNAIII molecule is the effector molecule, which controls the expression of cell wall-associated and virulence proteins in a growth-dependent manner (36). The second major regulatory system is the sarA (staphylococcal accessory regulator A) locus (14, 18). SarA is a transcriptional regulator that binds to a consensus motif in the promoter of its target genes and has positive and negative regulatory effects on extracellular protein synthesis (48). SarA interacts with the agr system by binding to the agr promoter region, to stimulate the transcription of RNAII and RNAIII from the P2 and P3 promoters, respectively (4). SarA forms part of a family of SarA-related proteins, which participate in the SarA-RNAIII regulatory cascade and have regulatory functions as well (16).
Global regulators and effector molecules are important in the life cycle of S. aureus. They not only govern virulence factors but also are involved in the modulation of antibiotic resistance levels. Compromised agr function was found to help clinical isolates of S. aureus develop vancomycin heteroresistance (39). The sarA and agr operons were found to affect methicillin resistance (37), and high activity of transcription factor σB was shown to correlate with increased teicoplanin resistance (9) and high-level methicillin resistance (46). In this work we describe a novel potential sensor of cell wall damage, which influences sarA and agr transcription.
MATERIALS AND METHODS
Strains and culture conditions.
The strains and plasmids used are listed in Table 1. Strains were grown in Luria-Bertani broth (Becton Dickinson, Sparks, Md.) at 37°C unless specified otherwise. Transductions were performed with phage 85 (5). Erythromycin at 10 μg ml−1, tetracycline at 10 μg ml−1, or kanamycin at 20 μg ml−1 was added to the medium when needed. MICs of antibiotics were determined by Etest (AB-Biodisk, Solna, Sweden) on Mueller-Hinton agar plates with an inoculum of an 0.5 McFarland standard after 24 h of incubation at 35°C or by broth microdilution as recommended by the NCCLS (35). MICs of teicoplanin and vancomycin were determined on brain heart infusion agar after 48 h of incubation at 35°C. Antibiotic resistance levels were compared by swabbing an 0.5 McFarland standard suspension of overnight cultures along an antibiotic gradient in rectangular agar plates.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype and phenotypea | Reference or source |
|---|---|---|
| Escherichia coli | ||
| DH5α | F− φ80d/acZΔM15 recA1 | Invitrogen |
| Staphylococcus aureus | ||
| RN4220 | NCTC8325-4 r− m+; recipient for electroporations | 25 |
| J126 | RN4220 msrR::ermB | This study |
| BB270 | Essentially the same as NCTC8325; rsbU mec | 6 |
| BB1259 | BB270 Ω2020chr::Tn551 | 44 |
| J141 | BB270 msrR::ermB | This study |
| J190 | J141[pAW17(msrAR)] | This study |
| J191 | J141(pAW17) | This study |
| BB938 | Essentially the same as 8325; Tcr | 10 |
| J161 | BB938 msrR::ermB | This study |
| MSSA1112 | Clinical isolate; bla | 20 |
| J156 | MSSA1112 msrR::ermB | This study |
| J175 | MSSA1112 msrR::pEC1(msrRp-luc+) msrR+ | This study |
| J198 | MSSA112[pBUS(mecAp-luc+)] | This study |
| J192 | J156[pBUS(mecAp-luc+)] | This study |
| J209 | J156(pAW17) | This study |
| J210 | J156[pAW17(msrAR)] | This study |
| Plasmids | ||
| pAW17 | S. aureus-E. coli shuttle vector ori pAMα1-ori ColE1 aac-aph; Gmr Kmr | This study |
| pAW17 (msrAR) | 1.5-kb fragment covering msrAR cloned in pAW17 | This study |
| pBUS1 | S. aureus-E. coli shuttle vector pAW8 (42) with multicloning site from pBluescript II SK (Stratgene) and the rrnT14 terminator sequence from pLL2443 (24) | S. Burger, unpublished data |
| pJR3 | pBT with a 3.45-kb insert containing the ermB cassette from pEC1 substituting for msrR, flanked on both sides by 1-kb chromosomal sequences from BB270; Tetr | This study |
| pEC1(msrRp-luc+) | 2.3-kb KpnI-EcoRI msrRp-luc+ fragment cloned in pEC1; Apr Ermr | This study |
| pBUS1(mecAp-luc+) | 2.5-kb KpnI-EcoRI mecAp-luc+ fragment cloned in pBUS1; Tetr | This study |
Abbreviations: Apr, ampicillin resistant; Ermr, erythromycin resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant; Mcr, methicillin resistant; Tcr, teicoplanin resistant; Tetr, tetracycline resistant.
Molecular biological methods.
Standard molecular biology procedures were performed essentially according to the protocols of Sambrook et al. (40) and Ausubel et al. (3). Total RNA was isolated as described by Cheung et al. (12), with a FastRNA kit and a Fastprep reciprocating shaker (Bio 101, Vista, Calif.). Digoxigenin-labeled DNA was produced using the PCR DIG Probe synthesis kit (Roche, Rotkreuz, Switzerland). Primers used are listed in Table 2. Probes for sarA and RNAIII were those described earlier (9, 22). For Northern blots, 6 μg of total RNA was loaded per lane. Chromosomal sequencing was performed according to the protocol of A. Wada (45) on an ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, Calif.). For sequencing the msr region a 1.8-kb fragment was amplified using primers JR1 and JR2. Homology searches were performed with the program Blast (http://www.ncbi.nm.nih.gov/BLAST) (1). Sequence data were analyzed with the GCG sequence analysis software package, version 9 (Genetics Computer Group, University of Wisconsin, Madison). Primer extension was performed using 40 μg of total RNA from early exponential growth and 3 pmol of the γ-32P-labeled primer JR7 or JR8 for msrR or JR11 and JR12 for msrA, respectively, with Omniscript reverse transcriptase (Qiagen, Hilden, Germany), according to the instructions of the manufacturer. The sequencing reaction was performed with the Thermo Sequenase cycle sequencing kit (U.S. Biochemicals, Cleveland, Ohio).
TABLE 2.
Primers
| Primer | Sequencea (5′-3′) |
|---|---|
| JR1 | GGGATCCTGAGCTAAAGTTAAGTCGCC |
| JR2 | TGAATTCAAACCATTATGGTGTGGCTGG |
| JR3 | CGGATCCTGGCATATTCTACACCGC |
| JR4 | GGAATTCAGTTATGCCTGATGCGCTTGG |
| JR5 | CATTGACTGCAGCAATCAAGAACTCATACGAAG |
| JR6 | GACTAAGCTTCGCTTCAACATCCATAGC |
| JR7 | CGTTGTCATTAGTTTCTTTATC |
| JR8 | TCAGAAGAATGATAGGTAATTTC |
| JR9 | ATTTCAGATCTGTACCGTATTTTAATATTG |
| JR10 | ATTTCAGATCTGAGTATGATAGAAATCG |
| JR11 | GCAAAATAAGCTGTATTAATATTC |
| JR12 | CAAATGGTTTCGTCATACAC |
| JR13 | GGGTACCTGAGCTAAAGTTAAGTCGCC |
| JR14 | TATCCATGGTTACCTACCTTATATCTTC |
| msrR | AATGGACCAGTAAAAAATGATG |
| msrR for rev | CCTCGGATACCAAAACTCAAAC |
| msrA | GGGATCCTGAGCTAAAGTTAAGTCGCC |
| msrA for rev | GAGGTACCTTTTGGTGTATGACGAAACC |
| sarR | CTTCTAATTCTGAAATCAG |
| sarR for rev | GACATTAATGATTTAGTCAAC |
| hla | GTGATGGAAAATAGTTGATGAG |
| hla for rev | GTGAATTTGTTCACTGTGTCG |
| spa | TGAATTCGTAAACTAGGTGTAGG |
| spa for rev | CGGTACCAGGCTTGTTATTGTCTTCC |
Restriction enzyme sites used in this work are underlined.
Construction of mutants and plasmids.
For allelic replacement of msrR, a 1-kb fragment covering the 5′ region of msrR was amplified from strain BB270 with primers JR3 and JR4 and cloned 5′ to the ermB resistance cassette of vector pEC1 (11). A 1-kb fragment covering the 3′ region of msrR amplified with primers JR5 and JR6 was subsequently cloned downstream of the ermB cassette. The insert was then subcloned into the suicide vector pBT (22), resulting in plasmid pJR3, which was electroporated into RN4220. Mutants with the msrR::ermB allelic replacement were selected for Ermr and screened for loss of Tetr. The msrR::ermB construct was then transduced into different genetic backgrounds. The plasmid pAW17(msrAR) used for complementation was constructed by cloning a 1.7-kb DNA fragment generated with primers JR9 and JR10, covering msrAR, into pAW17. Luciferase reporter gene fusions were constructed by amplifying the desired promoter regions and cloning them 5′ to the firefly luciferase gene luc+ of the pSP-luc+ vector (Promega, Madison, Wis.). For msrRp-luc+, a DNA fragment covering 709 bp of the msrR promoter region of strain MSSA1112 was generated by PCR with upstream primer JR13, including a KpnI linker, and downstream primer JR14, including an NcoI site. For mecAp-luc+, a DNA fragment covering 895 bp of the mecA promoter region of the SCCmec element of S. aureus BB270 was generated by PCR with upstream primer JR15, including a KpnI site, and downstream primer JR16, including an NcoI site. The PCR products obtained were digested with KpnI and NcoI and cloned in frame with the 5′ end of the luciferase gene of plasmid pSP-luc+. Sequence analysis confirmed the identity of the constructs. The 2.3-kb KpnI-EcoRI fragment, including the msrR promoter region fused to the luciferase coding region, was subsequently cloned into suicide plasmid pEC1 to obtain plasmid pEC1(msrRp-luc+), which was electroporated into RN4220 and transduced into MSSA1112, resulting in strain J175. Plasmid pBUS1(mecAp-luc+) was generated by cloning the 2.5-kb KpnI-EcoRI fragment, including the mecA promoter region fused to the luciferase coding region, into the Escherichia coli-S. aureus shuttle vector pBUS1. All constructs were verified by restriction analysis, Southern blotting, and pulsed-field gel electrophoresis.
Enzymatic assays.
Luciferase activity was determined as described earlier (9) using the luciferase assay substrate and a Turner Designs TD-20/20 luminometer (Promega). Spontaneous autolysis was measured in cells that were grown at 37°C, harvested at mid-exponential phase, washed twice with cold phosphate-buffered saline, and resuspended in 0.05 M Tris-HCl (pH 7.5), by determining turbidity loss as optical density at 600 nm (OD600) when incubated with shaking (200 rpm) at 37°C. Penicillin-binding proteins (PBPs) from membrane preparations of exponentially growing cells were labeled with [3H]penicillin and separated on sodium dodecyl sulfate-polyacrylamide gels as described earlier (8).
Nucleotide sequence accession number.
The nucleotide sequence was deposited in the EMBL and GenBank nucleotide sequence databases under accession number AF311784.
RESULTS
Insertion site characterization.
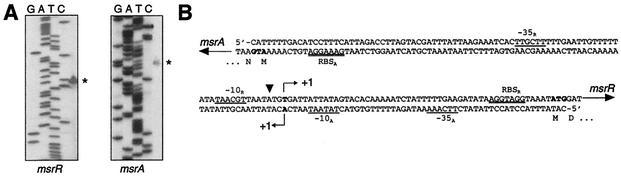
The insertion Ω2020::Tn551 was reported to increase susceptibility to β-lactams in S. aureus, but its nature remained unsolved (44). Sequencing of the insertion site revealed that the transposon had integrated into the intercistronic region of two divergently transcribed genes, tentatively termed msrA and msrR, which are separated by 136 nucleotides (Fig. 1). The msrA gene was shown to code for a peptide methionine sulfoxide reductase, MsrA2, specific for the S isomer of methionine sulfoxide (34). The msrR gene has not yet been characterized; the deduced amino acid sequence includes a hypothetical protein of 327 amino acids with 44% identity and 66% similarity to Enterococcus faecalis Psr (U42211) and 42% identity and 66% similarity to Bacillus subtilis LytR, which belong to the LytR-CpsA-Psr family of cell envelope-related transcriptional attenuators. A predicted transmembrane domain in the N-terminal part of MsrR divides MsrR into a shorter, presumably intracellular domain and a larger, extracellular domain. Analysis of the MsrR amino acid sequences obtained by the public genome sequencing projects revealed that the protein is strictly conserved among S. aureus strains.
FIG. 1.
Genetic organization of the msrAR region. (A) Transcriptional start points (*) for msrR and msrA were determined by primer extension as outlined in Materials and Methods. (B) msrA-msrR intercistronic region. Transposon Tn551 integration site (inverted triangle), transcriptional starts of msrA and msrR (+1), ribosomal binding sites (RBS), and −10 and −35 boxes are indicated.
The open reading frames of msrA and msrR are each terminated by a structure resembling a putative rho-independent terminator, suggesting that they lie on monocistronic transcripts. Interestingly, primer extension showed that msrA and msrR share the same nucleotide for the transcriptional start (Fig. 1). The Tn551 insertion mapped 2 nucleotides upstream of the msrA transcriptional start. Northern blots showed that Ω2020::Tn551 abolished the transcription of both msrA and msrR (data not shown).
Phenotype of msrR inactivation.
To determine if only one of the two inactivated genes was responsible for the increased β-lactam susceptibility, we replaced msrR with an ermB resistance cassette in strain RN4220 and transduced the inactivated msrR::ermB gene into the methicillin-resistant strain BB270. The resulting strain, J141, showed a fourfold-lower resistance to oxacillin than did the parent (Fig. 2), suggesting that msrR inactivation was responsible for the reduced resistance. Oxacillin resistance could be restored by complementing strain J141 with plasmid pAW17(msrAR), supporting the MsrR effect on oxacillin resistance levels (Fig. 2). Transduction of msrR::ermB into the teicoplanin-intermediate-resistant strain BB938 reduced the teicoplanin MIC for the strain twofold, from 16 to 8 μg ml−1, showing that MsrR had an effect on teicoplanin resistance as well.
FIG. 2.
Effects of msrR inactivation and complementation of oxacillin resistance. The methicillin-resistant strain BB270, its msrR mutant J141, strain J190 [J141 complemented by pAW17(msrAR)], and control strain J191 [J141(pAW17)] were swabbed along a plate containing an oxacillin gradient. The corresponding MICs of oxacillin (in micrograms per milliliter) for the strains are indicated.
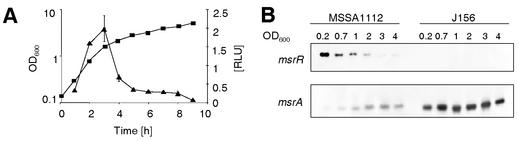
Expression of msrR during growth.
The transcription profile of msrR during growth in liquid culture was determined by two different methods. Both the Northern blots with the wild-type strain MSSA1112 and the msrRp-luciferase reporter activity in the isogenic strain J175 showed that msrR transcription peaked in the early exponential growth phase and decreased towards the late exponential and stationary phases (Fig. 3). The transcription profile of msrA was the reverse, lower in the exponential phase and increasing towards the stationary phase. Interestingly, in the msrR mutant J156, msrA transcription was strongly enhanced over the entire growth cycle compared to that in the wild type (Fig. 3B). The sizes of the msrA and msrR mRNAs, 0.5 and 1 kb, respectively, indicated that both genes are located on monocistronic transcripts, as predicted from the nucleotide sequence.
FIG. 3.
Expression profiles of msrA and msrR. (A) Expression of msrRp-luc+ during growth of S. aureus strain J175 was determined by quantifying the luciferase activity (triangles). Bacterial growth was monitored by measuring the OD600 (squares). RLU, relative light units. (B) Northern blot of msrA and msrR. Total RNA of strain MSSA1112 and its msrR mutant J156 was harvested at different growth stages indicated as OD600 values above the lanes.
Induction of msrR by antibiotic stress.
Knowing that msrR inactivation resulted in lower oxacillin resistance, we analyzed the response of msrR to different cell wall-active antibiotics by measuring the msrRp-luciferase reporter activity in strain J175. The antibiotics were added at concentrations corresponding to their MICs to exponentially growing cultures. Since staphylococci are known to show a strong inoculum effect with glycopeptides, vancomycin and teicoplanin were added at 5× their MICs. Transcription of msrR was found to be induced approximately two- to fivefold by (i) β-lactam derivatives such as oxacillin, cefoxitin, and imipenem; (ii) vancomycin, and to a lesser extent teicoplanin; (iii) antibiotics interfering with peptidoglycan precursor formation, such as fosfomycin and bacitracin; and (iv) the glycyl-glycine endopeptidase lysostaphin, which specifically cleaves the characteristic staphylococcal peptidoglycan pentaglycine interpeptide (Table 3). Northern blots showed that induction started immediately after addition of the antibiotic. Induction of msrR reached its maximum after 30 min and decreased thereafter, presumably due to the antibiotic's effect on cell growth. The extent of the induction was found to be concentration dependent and was higher when higher oxacillin concentrations were added (data not shown).
TABLE 3.
Induction of msrR in strain J175 by antibiotics
| Antibiotic | MIC (μg ml−1) | Fold inductiona |
|---|---|---|
| Oxacillin | 0.125 | 3.2 ± 0.50 |
| Cefoxitin | 3 | 3 ± 0.59 |
| Imipenem | 0.032 | 3 ± 0.97 |
| Vancomycinb | 1.5 | 4.5 ± 1.30 |
| Teicoplaninb | 1.5 | 1.8 ± 0.28 |
| Bacitracin | 48 | 5 ± 1.48 |
| Fosfomycin | 1.5 | 2.5 ± 1.27 |
| Lysostaphin | 0.008 | 4.7 ± 1.84 |
| Trimethoprim | 0.75 | 0.96 ± 0.09 |
| Ciprofloxacin | 0.25 | 0.98 ± 0.09 |
Fold increase in msrR transcription in strain J175 after 30 min of exposure to antibiotics, determined by measuring the luciferase activity of msrRp-luc+. The values shown represent the means of three independent assays.
Glycopeptides were added at 5× MICs due to scavenging of glycopeptides by the cell wall, resulting in an inoculum effect.
Non-cell wall-active antibiotics such as trimethoprim, which acts on folate synthesis, or ciprofloxacin, which inhibits DNA gyrase, had no effect on msrR expression (Table 3). Similar results were obtained by Northern blot analyses when protein synthesis inhibitors such as tetracycline or erythromycin were tested for their effects on msrR expression (data not shown).
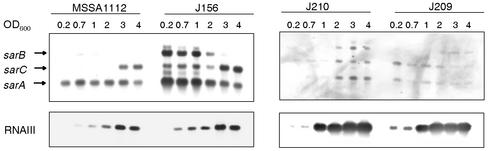
Effects of msrR on sarA and agr.
SarA is a global regulator which controls the synthesis of virulence factors and stimulates the transcription of the agr operon from the P2 and P3 promoters (17; see review by Arvidson and Tegmark [2]). The sar locus is composed of three overlapping transcripts, designated sarA (0.56 kb), sarC (0.8 kb), and sarB (1.2 kb), originating from the P1, P3, and P2 promoters, respectively, all encoding the SarA protein (30). An impressive upregulation of the three sarA transcripts was observed in msrR mutants, particularly during the early exponential growth phase, up to an OD600 of 1 to 2, as shown in Fig. 4. The increase of sarA transcripts in the mutant coincided precisely with the time of the otherwise highest msrR transcription in the wild type (Fig. 3A).
FIG. 4.
Effect of msrR on the expression of the global regulators sarA and RNAIII. Shown are Northern blots of sarA and RNAIII transcripts of the wild-type strain MSSA1112 and its derivatives J156 (msrR), J210 [J156 pAW17(msrAR)], and J209 (J156 pAW17) harvested during growth at the OD600 values indicated.
The SarA protein acts as an activator of the agr operon. We therefore measured transcription of RNAIII, the effector molecule of the agr operon. RNAIII expression, which usually begins in the stationary phase, started to increase in the msrR mutant already during the exponential growth phase and appeared also slightly stronger than in the wild type, as shown in Fig. 4.
Effects of msrR on virulence factors.
The global regulators sarA and agr control a large number of cell wall-associated and excreted proteins. We therefore analyzed the influence of msrR inactivation on spa (coding for protein A) and hla (coding for alpha-toxin) transcription, respectively. The spa transcription appeared to be reduced in the mutant compared to its parent, especially in the stationary phase. In the very early exponential phase, at an OD600 of 0.2, though, there appeared to be more spa transcripts in the mutant than in the wild type. The transcription of hla, in contrast, was enhanced and started earlier during growth in the mutant than in the wild type (Fig. 5).
FIG. 5.
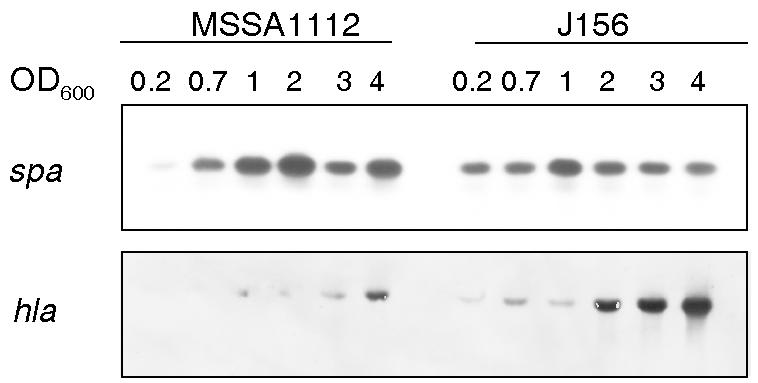
Northern blots of spa and hla transcripts in the wild-type strain MSSA1112 and its msrR mutant J156, harvested during growth at the OD600 values indicated and probed with hla or spa, respectively.
Effects of MsrR on mecA transcription and autolytic activities.
Since msrR inactivation reduced oxacillin resistance, the transcription profile of mecA, the gene encoding the low-affinity PBP2′, which is responsible for methicillin resistance, was analyzed in a wild-type strain and in the corresponding msrR mutant. For this purpose, a plasmid containing a mecA promoter-luciferase reporter gene fusion was introduced in parent strain MSSA1112, yielding strain J198, and into the corresponding msrR mutant J156, yielding strain J192. The mecA transcription, though, was not affected in either strain by msrR inactivation (data not shown). The effect of msrR inactivation was further analyzed for the expression pattern of the endogenous PBPs in two genetically distinct methicillin-resistant S. aureus strains. However, the PBP patterns were identical for the methicillin-resistant strains and their corresponding msrR mutants. There was also no change in the rate of spontaneous autolysis in parent and mutant strains (data not shown).
DISCUSSION
MsrR shares high sequence identity and similarity with Psr, the PBP5 synthesis repressor of E. faecalis and Enterococcus hirae, and with LytR of B. subtilis. All belong to the LytR-CpsA-Psr family of cell envelope-related transcriptional attenuators, whose exact function has yet to be defined. Psr of E. hirae, postulated to be a global regulator of cell wall synthesis genes, was shown to alter the autolytic response by reducing the cell wall's content of rhamnose, a sugar occurring in nonpeptidoglycan polysaccharides and teichoic acids, and to repress PBP5 synthesis (31). Interestingly, the enterococcal PBP5, which is associated with increased levels of ampicillin resistance, has sequence similarity with the staphylococcal methicillin resistance-determining PBP2′, encoded by mecA. The B. subtilis LytR was shown to act as an attenuator of the expression of its own gene and of the divergently transcribed lytABC operon, which codes for a lipoprotein, a modifier of an N-acetylmuramoyl-l-alanine amidase, and the amidase itself (28). It is worth noting that MsrR seemed to influence its own expression (M. Bischoff and J. Rossi, unpublished results) and that of the divergently transcribed msrA as well, with the latter being strongly upregulated in msrR mutants.
The most important finding was that MsrR appeared to act as an attenuator of sarA transcription. This is strongly supported by the remarkable sarA upregulation in msrR mutants occurring during exponential phase, precisely the period of otherwise peak msrR activity in the wild type. It is further supported by the downregulation of sarA transcription in the mutants to wild-type levels through msrAR trans-complementation (data not shown). The effect, though, is presumably not mediated directly by MsrR, since no DNA-binding motifs were found in the short predicted internal N-terminal fragment of MsrR. Additional factors have therefore to be postulated as intermediates leading from MsrR to sarA attenuation. The same may be true for Psr, which was postulated to be a global regulator of cell wall synthesis in E. hirae but seemed to act as neither a transcriptional repressor nor an activator in Enterococcus faecium (38), suggesting that Psr, like MsrR, may be a mediator for the regulatory effects.
Since SarA was known to be an activator of the agr operon (18), the drastic changes in sarA transcription levels in the msrR mutants were expected to influence agr expression. The premature initiation and increased RNAIII transcription observed in the msrR mutant may thus be explained by an increased and earlier SarA production.
The altered activity profile of the global regulators sarA and agr upon msrR inactivation was predicted to have consequences for the synthesis of a variety of virulence factors (19). We therefore investigated whether the msrR mutation influenced the transcription of spa, known to be repressed by SarA and RNAIII (13, 19), and of hla, which, on the other hand, is upregulated by these two regulators (14). Consistently with the previously demonstrated effects of msrR inactivation on sarA and RNAIII expression, we could observe a clear increase in hla expression in the msrR mutant, which was again not only stronger but appeared also earlier in growth. Expression of spa seemed to be downregulated in the mutant, especially during later growth stages, again consistent with the already-demonstrated effect of the two global regulators. However, the reduction of spa transcripts in the mutant was less pronounced than expected, pointing out the complex regulation of this cell wall-associated protein. It has been shown that further regulatory elements, such as SarS (SarH1) (15, 43), Rot (32), and ArlSR (21), are involved in regulation of spa transcription.
Since msrR was expressed mainly during early exponential growth in wild-type strains, effects of inactivation are expected to manifest mainly during this growth phase and to a lesser extent over the remaining growth cycle. The early growth phase is thought to mimic some of the conditions needed in the first steps in the infection cycle of S. aureus, namely, the adhesion to and colonization of the host. MsrR may therefore play a more important role in vivo than in vitro, since except for reduced resistance to certain antibiotics, the doubling time and growth yield in vitro seemed not to be affected by msrR inactivation.
How the MsrR function correlates with glycopeptide and methicillin resistance remains unclear. Many factors are known to influence methicillin resistance levels. Resistance can be lowered by preventing the production of PBP2′ and/or by reducing the expression of the endogenous PBPs. Peptidoglycan precursor formation or changes in the peptidoglycan composition, as well as control of autolytic activities, can affect resistance levels (reviewed in reference 7). The global regulators sarA and agr were also shown to be involved in some way in methicillin resistance (37). Glycopeptide resistance depends, in most instances, on an abundant production of cell wall precursors (27, 33) and genes involved in cell wall synthesis (41). Since neither peptidoglycan composition and cross-linking (K. Ehlert, personal communication) nor changes in the PBP patterns or autolytic activities were apparent upon msrR inactivation, other reasons for the reduced resistance have to be sought. The effect of MsrR on resistance levels may be attributed to its effect on the regulation of the global regulators, which, besides virulence factors, may affect cell wall metabolism. Alternatively, changes in cell wall constituents other than peptidoglycan, such as teichoic acids, polysaccharides, or capsule, are conceivable, since Streptococcus mutans LytR, similarly to the B. subtilis LytR, was shown to be involved in biofilm formation (47). An analogous function for MsrR may be postulated. Predicated upon the upregulation of msrR by inhibition of cell wall synthesis or degradation of the peptidoglycan, the primary function of MsrR may be in the response to signals following cell wall damage or restructuring. It is conceivable that MsrR acts as a sensor that relays an external signal, possibly generated by cell wall metabolism during growth or upon damage, to the cytoplasm. MsrR may be an intrinsic low-level defense mechanism against cell wall damage that may occur during infection and may affect the ability of S. aureus to survive in different environments. Further experiments will have to show if MsrR is of importance in adhesion to and invasion and infection of the host, since its inactivation disturbs the timing of virulence factor production.
Acknowledgments
We thank K. Ehlert for peptidoglycan analysis, S. Burger for sequencing, E. Huf for construction and restriction analysis of strains, and S. Projan and P. Dunman at Wyeth USA for providing the microarray facilities.
This study was supported by the Swiss National Science Foundation grant NF31-63552.00 and the Forschungskredit der Universität Zürich 560030.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 4.Bayer, M. G., J. H. Heinrichs, and A. L. Cheung. 1996. The molecular architecture of the sar locus in Staphylococcus aureus. J. Bacteriol. 178:4563-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger-Bächi, B. 1983. Increase in transduction efficiency of Tn551 mediated by the methicillin resistance marker. J. Bacteriol. 154:533-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger-Bächi, B. 1983. Insertional inactivation of staphylococcal methicillin resistance by Tn551. J. Bacteriol. 154:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger-Bächi, B., and S. Rohrer. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178:165-171. [DOI] [PubMed] [Google Scholar]
- 8.Berger-Bächi, B., A. Strässle, and F. H. Kayser. 1989. Natural methicillin resistance in comparison with that selected by in vitro drug exposure in Staphylococcus aureus. J. Antimicrob. Chemother. 23:179-188. [DOI] [PubMed] [Google Scholar]
- 9.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bischoff, M., M. Roos, J. Putnik, A. Wada, P. Glanzmann, P. Giachino, P. Vaudaux, and B. Berger-Bächi. 2000. Involvement of multiple genetic loci in Staphylococcus aureus teicoplanin resistance. FEMS Microbiol. Lett. 194:77-82. [DOI] [PubMed] [Google Scholar]
- 11.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 12.Cheung, A. L., K. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 13.Cheung, A. L., K. Eberhardt, and J. H. Heinrichs. 1997. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect. Immun. 65:2243-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung, A. L., and G. Zhang. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7:1825-d1842. [DOI] [PubMed]
- 17.Chien, Y., A. C. Manna, and A. L. Cheung. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30:991-1001. [DOI] [PubMed] [Google Scholar]
- 18.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 19.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Entenza, J. M., J. Vouillamoz, M. P. Glauser, and P. Moreillon. 1977. Levofloxacin versus ciprofloxacin, flucloxacillin, or vancomycin for treatment of experimental endocarditis due to methicillin-susceptible or -resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1662-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 22.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 24.Jana, M., T. T. Luong, H. Komatsuzawa, M. Shigeta, and C. Y. Lee. 2000. A method for demonstrating gene essentiality in Staphylococcus aureus. Plasmid 44:100-104. [DOI] [PubMed] [Google Scholar]
- 25.Kreiswirth, B., J. Kornblum, R. D. Arbeit, W. Eisner, J. N. Maslow, A. McGeer, D. E. Low, and R. P. Novick. 1993. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science 259:227-230. [DOI] [PubMed] [Google Scholar]
- 26.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 28.Lazarevic, V., P. Margot, B. Soldo, and D. Karamata. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon—a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J. Gen. Microbiol. 138:1949-1961. [DOI] [PubMed] [Google Scholar]
- 29.Lyon, G. J., J. S. Wright, T. W. Muir, and R. P. Novick. 2002. Key determinants of receptor activation in the agr autoinducing peptides of Staphylococcus aureus. Biochemistry 41:10095-10104. [DOI] [PubMed] [Google Scholar]
- 30.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massidda, O., R. Kariyama, L. Daneo-Moore, and G. D. Shockman. 1996. Evidence that the PBP 5 synthesis repressor (psr) of Enterococcus hirae is also involved in the regulation of cell wall composition and other cell wall-related properties. J. Bacteriol. 178:5272-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreira, B., S. Boyle-Vavra, B. L. M. De Jonge, and R. S. Daum. 1997. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moskovitz, J., V. K. Singh, J. Requena, B. J. Wilkinson, R. K. Jayaswal, and E. R. Stadtman. 2002. Purification and characterization of methionine sulfoxide reductases from mouse and Staphylococcus aureus and their substrate stereospecificity. Biochem. Biophys. Res. Commun. 290:62-65. [DOI] [PubMed] [Google Scholar]
- 35.NCCLS. 2000. Performance standards for antimicrobial susceptibility testing; 10th informational supplement M100-SID(M7). NCCLS, Wayne, Pa.
- 36.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piriz Duran, S., F. H. Kayser, and B. Berger-Bächi. 1996. Impact of sar and agr on methicillin resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 141:255-260. [DOI] [PubMed] [Google Scholar]
- 38.Rice, L. B., L. L. Carias, R. Hutton-Thomas, F. Sifaoui, L. Gutman, and S. D. Rudin. 2001. Penicillin-binding protein 5 and expression of ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 45:1480-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Sieradzki, K., and A. Tomasz. 1998. Suppression of glycopeptide resistance in a highly teicoplanin-resistant mutant of Staphylococcus aureus by transposon inactivation of genes involved in cell wall synthesis. Microb. Drug Resist. 4:159-168. [DOI] [PubMed] [Google Scholar]
- 42.Strandén, A. M., M. Roos, and B. Berger-Bächi. 1996. Glutamine synthetase and heteroresistance in methicillin-resistant Staphylococcus aureus. Microb. Drug Resist. 2:201-207. [DOI] [PubMed] [Google Scholar]
- 43.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 44.Vasilieva, E., H. Labischinski, and B. Berger-Bächi. 1997. Identification and mapping of new chromosomal sites affecting response to β-lactams in Staphylococcus aureus. Int. J. Antimicrob. Agents 8:13-21. [DOI] [PubMed] [Google Scholar]
- 45.Wada, A. 2001. An improved method for purifying bacterial genomic DNAs for direct sequencing by capillary automated sequencer. Tech. Tips Online. http://research.bmn.com/tto.
- 46.Wu, S. W., H. de Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida, A., and H. K. Kuramitsu. 2002. Multiple. Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 68:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziebandt, A. K., H. Weber, J. Rudolph, R. Schmid, D. Hoper, S. Engelmann, and M. Hecker. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics 1:480-493. [DOI] [PubMed] [Google Scholar]