Abstract
The role of sterol mutations in the resistance of Candida albicans to antifungal agents has not been thoroughly investigated. Previous work reported that clinical C. albicans strains resistant to both azole antifungals and amphotericin B were defective in ERG3, a gene encoding sterol Δ5,6-desaturase. It is also believed that a deletion of the lanosterol 14α-demethylase gene, ERG11, is possible only under aerobic conditions when ERG3 is not functional. We tested these hypotheses by creating mutants by targeted deletion of the ERG3 and ERG11 genes and subjecting those mutants to antifungal susceptibility testing and sterol analysis. The homozygous erg3/erg3 mutant created, DSY1751, was resistant to azole derivatives, as expected. This mutant was, however, slightly more susceptible to amphotericin B than the parent wild type. It was possible to generate erg11/erg11 mutants in the DSY1751 background but also, surprisingly, in the background of a wild-type isolate with functional ERG3 alleles under aerobic conditions. This mutant (DSY1769) was obtained by exposure of an ERG11/erg11 heterozygous strain in a medium containing 10 μg of amphotericin B per ml. Amphotericin B-resistant strains were obtained only from ERG11/erg11 heterozygotes at a frequency of approximately 5 × 10−5 to 7 × 10−5, which was consistent with mitotic recombination between the first disrupted erg11 allele and the other remaining functional ERG11 allele. DSY1769 was also resistant to azole derivatives. The main sterol fraction in DSY1769 contained lanosterol and eburicol. These studies showed that erg11/erg11 mutants of a C. albicans strain harboring a defective erg11 allele can be obtained in vitro in the presence of amphotericin B. Amphotericin B-resistant strains could therefore be selected by similar mechanisms during antifungal therapy.
Candida albicans can cause fungal diseases in immunocompromised patients, including cancer patients, transplant patients, and those with human immunodeficiency virus infections (6). The antifungal agents that are available for the treatment of C. albicans infections can be categorized into several chemical classes with different cellular targets. Enzymes of the ergosterol biosynthetic pathway are important targets of several classes of antifungals used to treat C. albicans infections, and among those, the polyenes and the azoles have a dominant position. Polyenes such as amphotericin B act at the level of ergosterol by binding tightly to this molecule. This effect damages the cell plasma membrane, thus resulting in leakage of intracellular ions. Azoles such as fluconazole, itraconazole, or voriconazole inhibit a cytochrome P450 (Erg11p) responsible for the 14α demethylation of lanosterol and thus block ergosterol biosynthesis. In the late steps of this ergosterol biosynthesis pathway, azoles inhibit also the Δ22 desaturation of the sterol moiety (Fig. 1). Other antifungal classes less relevant for the treatment of C. albicans infections, i.e., allylamines (terbinafine) and morpholines (amorolfine), inhibit ergosterol biosynthesis by blocking the activity of squalene epoxidase and sterol Δ14-reductase or Δ7-8-isomerase, respectively. Several mechanisms have been documented to be involved in the resistance to the azole and polyene antifungal classes in C. albicans. They include active efflux of the antifungals, target enzyme alterations, and the absence of the target enzyme (for amphotericin B resistance) (27, 28, 31, 34). The development of compensatory pathways that circumvent the inhibitory effects of antifungals may be another mechanism for resistance to antifungals (19). Compensatory pathways have been documented for the mechanisms of resistance to the azole and polyene classes and involve alterations of specific steps in ergosterol biosynthesis. For example, analysis of the sterol compositions of two separate azole-resistant C. albicans clinical isolates revealed the accumulation of ergosta-7,22-dienol, which is a feature consistent with the absence of sterol Δ5,6-desaturase activity, which is encoded by ERG3 (17, 18, 25). Azole resistance in these two cases was coupled with resistance to amphotericin B because of the absence of ergosterol in these cells. In Saccharomyces cerevisiae, mutants with mutations in ERG4 (35), ERG6 (7), and ERG3 (1) are also devoid of ergosterol and are resistant to polyene agents. The role of ERG3 in azole resistance originates from the observation that treatment of yeasts with azoles results in the accumulation of 14α-methylated sterols and 14α-methylergosta-8,24(28)-dien-3,6-diol. Formation of the latter sterol metabolite is thought to be catalyzed by sterol Δ5,6-desaturase; thus, inactivation of ERG3 can suppress toxicity and therefore causes azole resistance (19). In S. cerevisiae mutations of the ERG3 gene can result in azole resistance (33). However, inactivation of ERG3 does not always result in azole resistance: in Candida glabrata, a null mutation in ERG3 does not result in azole resistance (8). Another mutation potentially linked to azole resistance is the loss of function of ERG11, since it eliminates the target site of these agents. Unfortunately, yeasts with the ERG11 mutation are not viable under aerobic conditions. Viability with the ERG11 mutation is possible only when it is accompanied by the inactivation of ERG3, as reported for S. cerevisiae and C. glabrata (3, 15). Little is known about the effects of these mutations on azole or polyene resistance in C. albicans. A study has reported on the characterization of ERG3 from an azole-resistant strain known as the Darlington strain. This strain carries defective mutated ERG3 alleles, which thus contribute to azole resistance. Unfortunately, the effects of the mutated ERG3 alleles were masked in this strain by other azole resistance mechanisms (21). In this study, we therefore constructed erg3/erg3 and erg11/erg11 mutants and tested their susceptibilities to antifungal agents. Surprisingly, we were able to create erg11/erg11 mutants by positive selection on a polyene-containing medium under aerobic conditions without the need for ERG3 inactivation.
FIG. 1.
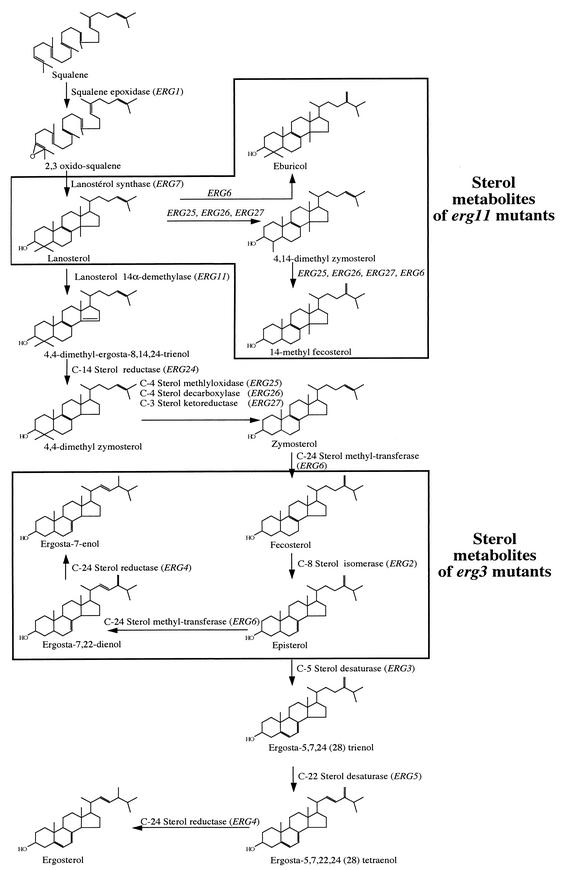
Schematic representation of the ergosterol biosynthetic pathway in C. albicans. Blocking of the functions of ERG11 and ERG3 results in the accumulation of the boxed metabolites. A detailed description of this pathway is also given by Parks et al. (26).
MATERIALS AND METHODS
Strains and media.
The C. albicans strains used in this study are listed in Table 1. The C. albicans strains were grown either in complete medium consisting of YEPD liquid medium (1% Bacto Peptone [Difco], 0.5% yeast extract [Difco], 2% glucose [Fluka]) or in minimal medium consisting of yeast nitrogen base (YNB medium; Difco) and 2% glucose (Fluka). When the strains were grown on solid media, 2% agar (Difco) was added to either medium. Escherichia coli DH5α (10) was used as a host for plasmid construction and propagation. DH5α cells were grown in Luria-Bertani (LB) broth or on LB plates, which were supplemented with ampicillin (0.1 mg/ml) when required.
TABLE 1.
Genotypes of C. albicans mutants used in this study
| Strain | Parent strain | Genotype | Reference |
|---|---|---|---|
| CAF4-2 | CAF2-1 | ura3Δ::imm434/ura3Δ::imm434 | 5 |
| DSY1216 | CAF4-2 | erg11Δ::hisG-URA3-hisG/ERG11 | This study |
| DSY1217 | DSY1216 | erg11Δ::hisG/ERG11 | This study |
| DSY1336 | DSY1217 | erg3AΔ::hisG-URA3-hisG/ERG3 erg11Δ::hisG/ERG11 | This study |
| DSY1338 | DSY1336 | erg3AΔ::hisG/ERG3 erg11Δ::hisG/ERG11 | This study |
| DSY1751 | DSY1338 | erg3AΔ::hisG/erg3BΔ::hisG-URA3-hisG erg11Δ::hisG/ERG11 | This study |
| DSY1769 | DSY1216 | erg11Δ::hisG-URA3-hisG erg11Δ::hisG-URA3-hisG | This study |
| DSY1758 | DSY1751 | erg3AΔ::hisG/erg3BΔ::hisG erg11Δ::hisG/ERG11 | This study |
| DSY1764 | DSY1758 | erg3AΔ::hisG/erg3BΔ::hisG erg11Δ::hisG/erg11Δ::hisG-URA3-hisG | This study |
| DSY1431 | DSY1338 | erg3AΔ::hisG/ERG3 erg11Δ::hisG/erg11Δ::hisG | This study |
Construction of mutants with ERG3 and ERG11 mutations.
The deletion of ERG3 from C. albicans was performed by using two different constructs based on comparisons with the S. cerevisiae ERG3 gene and available C. albicans genome data. For the first construct, a fragment amplifying the first ERG3 allele (ERG3A) was generated by PCR with C. albicans genomic DNA, Pwo polymerase (Roche), and primers ERG3-A (5′-AAATTCATTCTTTTCACCGAT-3′) and ERG3-B (5′-ATCTGGTCTTCTGTAAGATT-3′), whose sequences matched the sequence between positions +637 and +1037 with respect to the first ATG codon of ERG3. The PCR fragment generated was subcloned into the compatible HincII and SmaI sites of pBluescript KS to obtain pDS545. A deletion between nucleotides +833 and +850 was created by PCR with primers ERG3-SALI (5′-GCGCAAAGTCGACGCAATGGGAATAATAATGGGTAAAGATGGT-3′) and ERG3-BGLII (5′-GCGCAAAGATCTTTTTGTTCACTTTTGTTAACTTTTGGACTG-3′) and pDS545 as the template. The product of this PCR was digested with SalI and BglII and ligated to a 3.7-kb SaII-BglII fragment from pMB7 containing the hisG-URA3-hisG “Ura blaster” cassette (5) to yield pDS546. The first ERG3 disruption cassette was liberated by ApaI and SacI digestion, and this linear fragment was used for disruption of the first ERG3 allele. For the second ERG3 disruption cassette, a fragment corresponding to the second ERG3 allele (ERG3B) was amplified from C. albicans genomic DNA with primers ERG3-XBA (5′-TACAATCTAGATATCTTTGGACATTCT-3′) and ERG3-XHO (5′-GCGCAAACTCGAGAAGATTATTTTCAA TTATCAACACCAAATTG-3′) and cloned into a compatible restriction site of pBluescript KS(+) to yield pDS749. A deletion between nucleotides +390 and +1028 with respect to the first ATG codon was created by PCR with primers ERG3-PST (5′-GCGCAAAGACGTCCCAACCAAATATAGTAGTGATAATG-3′) ERG3-BGL (5′-GCGCAAGATCTCAGATGATTCATTGTTTGT-3′) and with pDS749 as a template. The product of this PCR was digested with PstI and BglII and ligated to a 3.7-kb PstI-BglII fragment from pMB7 to yield pDS765. The second ERG3 disruption cassette was liberated by ApaI and SacI digestion, and this linear fragment was used for disruption of the remaining ERG3B allele after marker regeneration, as described by Fonzi and Irwin (5). Schematic restriction maps are also given in Fig. 2.
FIG. 2.
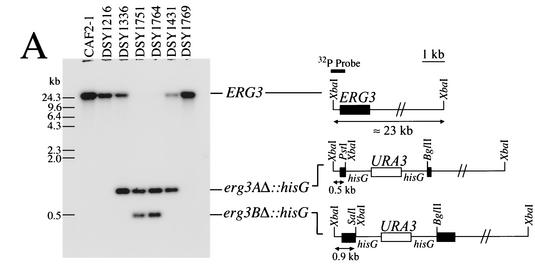
Disruption and expression of ERG3 in C. albicans. (A) Southern blot analysis of the ERG3 deletion in C. albicans. Five micrograms of genomic DNA was extracted from each strain, digested with XbaI, separated by electrophoresis, blotted onto a GeneScreen membrane (Dupont NEN), and probed with an XbaI-PstI fragment from pDS546, as indicated in the schematic restriction map. The following restriction fragment sizes are expected for each hybridization signal: wild-type allele, approximately 23 kb; erg3AΔ::hisG-URA3-hisG and erg3AΔ::hisG alleles, 0.9 kb; erg3BΔ::hisG-URA3-hisG and erg3BΔ::hisG alleles, 0.5 kb. These sizes correspond to those obtained by Southern blotting for each mutant constructed. (B) ERG3 expression in diverse erg mutants. Total RNA was extracted from C. albicans, and 5 μg of total RNA was separated by electrophoresis and blotted onto GeneScreen membranes. After hybridization with the same probe used for the experiment whose results are shown in panel A and washing steps, the membranes were exposed to Fuji X-ray film and were revealed after exposure to −80°C. The TEF3-specific probe was hybridized to the same membrane after removal of the ERG3-specific probe, as described previously (31).
For the disruption of ERG11, this gene was first cloned from pDS271, a plasmid that has been reported by Sanglard et al. (29) and that contains ERG11. A 3.5-kb EcoRV-ClaI fragment containing ERG11 was cloned into compatible sites of pMTL21, yielding pDS501. After the removal of an internal 1-kb XbaI-AccI fragment in pDS501 and blunt ending of the XbaI site, the Ura blaster cassette was inserted as a BglII-AccI fragment to create pDS506. A linear ApaI-SacI fragment was obtained from pDS506 for transformation into CAF4-2. Schematic restriction maps are also given in Fig. 3.
FIG. 3.
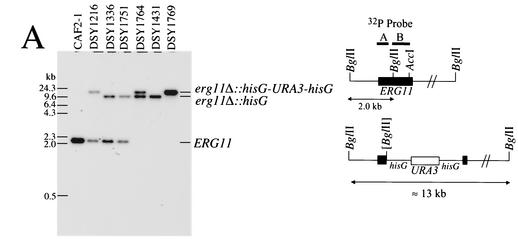
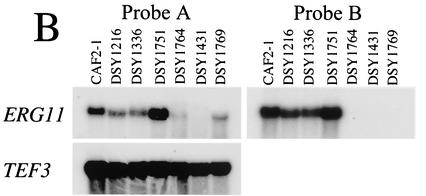
Disruption and expression of ERG11 in C. albicans. (A) Southern blotting analysis of ERG11 deletion in C. albicans. Five micrograms of genomic DNA was extracted from each indicated strain, digested with BglII, separated by electrophoresis, blotted onto a GeneScreen membrane (Dupont NEN), and probed with a fragment (probe A) comprising the first 418 bp of the ERG11 open reading frame, as indicated in the schematic restriction map. The following restriction fragment sizes are expected for each hybridizing signal: wild-type allele, 2 kb; erg11Δ::hisG-URA3-hisG and erg11Δ::hisG alleles, 13 and 10.3 kb, respectively. These sizes correspond to those obtained by Southern blotting for each mutant constructed. (B) ERG11 expression in diverse erg mutants. Total RNA was extracted from C. albicans, and 5 μg of total RNA was separated by electrophoresis and blotted onto GeneScreen membranes. After hybridization with labeled probe A and probe B as an 800-bp AccI-BglII internal fragment corresponding to the ERG11 deletion, the membranes were exposed to Fuji X-ray film and revealed after exposure to −80°C. The TEF3-specific probe was hybridized to the same membrane after removal of the ERG11-specific probe, as described previously (31).
Northern and Southern blotting.
Northern blotting and Southern blotting were carried out as described previously (31).
C. albicans transformations.
The different C. albicans strains were transformed by the lithium acetate procedure adapted by Sanglard et al. (30). After transformation with a linear DNA fragment, the cells were plated on YNB selective medium and incubated for 2 to 3 days at 30°C.
PCR.
Pwo polymerase (Roche) was used in the PCR to generate DNA fragments for use in cloning steps or as probes. The PCR buffers and conditions were those suggested by the manufacturer. PCR was carried out in a Thermal Cycler 480 instrument (Perkin-Elmer), with a first cycle at 94°C for 2 min, followed by 30 cycles of annealing at 54°C for 2 min. Elongation was performed at 72°C for 2 min. Yeast DNA templates for PCR were prepared from overnight cultures by mechanical breakage with glass beads, as described previously (30).
Susceptibility assays.
The C. albicans strains were cultivated overnight in YEPD liquid medium at 30°C with constant shaking. Saturated cultures were diluted to an inoculum size of 104 cells per ml. Susceptibility assays were performed in YEPD liquid medium in microtiter plates and were inspired by the standard protocol approved by the National Committee of Clinical Laboratory Standards (24). Antifungal agents were diluted by stepwise twofold dilutions in a total volume of 50 μl. One hundred fifty microliters of the diluted culture was next added to each antifungal agent-containing well. A drug-free culture and a sterile control were included in each microtiter plate. The microtiter plate was then sealed with Parafilm, and incubation was carried out at 35°C for 24 h. After this incubation period, the optical density of each well of the microtiter plate was read with a microplate reader (Bio-Rad) at 540 nm. The drug MIC was defined as the drug concentration needed to decrease the optical density by at least 50% compared with the optical density of the drug-free culture.
Sterol analysis.
C. albicans strains were cultured to saturation in YEPD medium, and the cells were harvested by centrifugation. The frozen cell pellets were thawed on ice and resuspended in ice-cold H2O at 200 mg/ml. One milliliter of each suspension was transferred to glass tubes containing 1 ml of 15% KOH in 90% (vol/vol) ethanol. The samples were heated at 80°C for 45 min and then cooled on ice. The sterols were extracted two times with 5 ml of heptane each time, and the extracts were evaporated to dryness under N2 at 55°C. The extracts were dissolved in 0.25 ml of toluene, and 0.25 ml of the BSTFA reagent (T-5634; Sigma) was added. The samples were heated at 60°C for 1 h and then evaporated to dryness. The residues were dissolved in 0.25 ml of heptane and stored at −20°C prior to analysis by gas chromatography-mass spectrometry (GC-MS). GC-MS analyses were performed with a Fison CE 8000 instrument with an AS800 autosampler and a Trio-1000-4559. The gas chromatograph was equipped with a Chrompak fused-silica column (25 m by 0.25 mm; CP-SIL 5CB). The following settings were used: source, positive electron ionization; scan, 50 to 650 Da in 0.9 s; interscan delay, 0.1 s; multiplier voltage, 450 V; solvent delay, 2.0 min (this may be extended up to 4.0 min); temperatures of zones 1 and 2, 220 and 80°C, respectively; gas chromatograph temperature ramp, 0.0 min at 180°C, 3.0 min at 180°C, 7.8 min at 300°C (25°C/min), 12.8 min at 325°C (5°C/min), and 14.8 min at 325°C; analyzer pressure, −3.95 mbar; filament current, 3.0 A; source current, 1124 μA; trap current, 341 μA; and He pressure, 10 lb/in2. Sterols were identified from their retention times and specific mass spectrometric patterns.
RESULTS
Phenotypic analysis of mutants with ERG3 and ERG11 mutations.
Given that most erg mutations in C. albicans are of clinical origin or have not been generated by targeted mutagenesis, we undertook the sequential disruption of ERG3 and ERG11 in C. albicans strain CAF4-2. The disruption of ERG3 was performed with strain DSY1217, a Ura− derivative in which one ERG11 allele had first been inactivated. The resulting erg3/erg3 homozygous mutant, DSY1751, did not contain a copy of the wild-type ERG3 gene, as deduced by Southern blotting analysis (Fig. 2A). Consistent with the deletion of both ERG3 alleles, no ERG3 mRNA could be observed in DSY1751 (Fig. 2B). Attempts to disrupt the second allele of ERG11 in DSY1217 were not successful in the first phase. Since erg11/erg11 homozygous mutants should lack ergosterol and therefore, in theory, should be resistant to amphotericin B, we plated the ERG11 Ura+ heterozygote DSY1216 onto amphotericin B-containing medium. Amphotericin B-resistant colonies appeared at a high frequency (5 × 10−5 to 7 × 10−5) after 3 days of incubation at 30°C compared to the frequency of appearance of wild-type parent CAF4-2 (1 × >10−8). Southern blot analysis of an amphotericin B-resistant isolate, DSY1769, indicated that no wild-type ERG11 alleles were present in this strain and that two alleles corresponding to erg11Δ::hisG-URA3-hisG alleles could be observed (Fig. 3A). This result suggests that the second allele disruption was obtained by mitotic recombination between the first disrupted allele and the remaining wild-type ERG11 allele. Such events have been reported to occur in C. albicans at frequencies ranging from 10−5 to 10−6 (9), which are very similar to the values obtained by selection for amphotericin B resistance. Deletion of both ERG11 alleles in DSY1769 was accompanied by the loss of signals corresponding to ERG11 mRNA. The residual ERG11 mRNA signal that was still detected with a labeled ERG11 probe (probe A in Fig. 3B, including the first 400 bp of the ERG11 open reading frame) does not likely correspond to intact mRNA, since no ERG11 mRNA could be observed by use of a labeled probe whose sequence corresponded to that of the fragment deleted from the C. albicans ERG11 genomic locus (Fig. 3B). Deletion of homozygous ERG11 alleles was obtained in two other strains created in this study. Strain DSY1431 was obtained by selection on amphotericin B-containing medium and originated from DSY1336, which is heterozygous for ERG3. Strain DSY1764 was obtained from a homozygous erg3/erg3 mutant (DSY1751) by targeted deletion of the remaining ERG11 allele by the Ura blaster method. No wild-type ERG11 allele could be detected in either DSY1431 or DSY1764 (Fig. 3A), and no signals corresponding to ERG11 mRNA were observed with ERG11-specific probe B (Fig. 3B). The ERG3 mRNA signals were markedly increased in erg11/erg11 deletion mutants still carrying a functional ERG3 allele (Fig. 2B). On the other hand, the ERG11 mRNA signals were significantly increased only in the erg3/erg3 homozygote, DSY1751 (Fig. 3B). Analysis of ERG11 and ERG3 expression by the mutants constructed in this study gave results that agree with the idea that, in the absence of ergosterol or when the ergosterol pathway is inhibited, ERG genes can be upregulated. Upregulation of both ERG3 and ERG11 in mutants lacking either gene had an effect comparable to that from the inhibition of sterol biosynthesis by the addition of azoles or other ergosterol biosynthesis inhibitors. This phenomenon has been described by the use of microarrays in S. cerevisiae (12) and in C. albicans exposed to itraconazole (4, 11).
The deletion of the ERG3 and ERG11 alleles affected the growth of C. albicans in YEPD medium to different degrees. We calculated the doubling times (tds) to be between 102 and 106 min for strains CAF2-1, DSY1216, and DSY1336, all of which carry functional ERG11 and ERG3 alleles, and for the mutant with the homozygous ERG3 mutation, DSY1751. The tds were 120 min for DSY1764 and 160 min for DSY1431 and DSY1769, thus showing that deletion of ERG11 had a negative effect on the C. albicans growth rate. In addition to the fact that these differences in growth rates were similar in synthetic YNB medium, the ability of all strains to grow in this type of medium shows that these strains are not auxotrophic for ergosterol. In practical terms, the differences in the growth rates between these different strains means that if they are not under amphotericin B selection pressure, the mutants with higher tds will be not competitive in vitro or in vivo, unlike other normally growing C. albicans isolates.
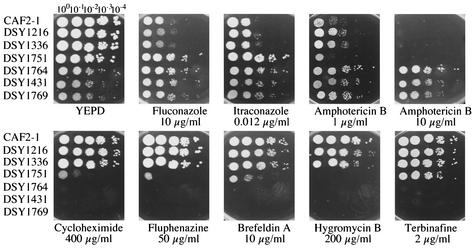
The susceptibilities of these mutants to different classes of antifungal and metabolic inhibitors were first determined by a visual spotting assay on YEPD medium (Fig. 4) with different drug concentrations and were then determined by a regular NCCLS methodology, with the exception that YEPD medium was used in place of RPMI medium (Table 2). The results presented in Fig. 4 indicate that, in general, the mutants with single and double ERG3 and ERG11 mutations were more susceptible than the wild type to metabolic inhibitors (i.e., cycloheximide, fluphenazine, brefeldin A, or hygromycin B). The erg3/erg3 mutant with the single mutation, DSY1751, was resistant to azoles but remained as susceptible to amphotericin B and terbinafine as the wild type (Fig. 4). DSY1751 was even slightly more susceptible to amphotericin B than the wild type, as judged by growth differences in spotting assays and the MICs measured by the standard NCCLS method (Table 2). The erg11/erg11 mutants were resistant to azoles as well as to amphotericin B in either the wild-type or the erg3/erg3 mutant background. The data shown in Fig. 4 were again consistent with the MICs measured by the NCCLS method: fluconazole MICs increased from 0.25 μg/ml for the wild type to more than 128 μg/ml for the erg3/erg3 mutants, the erg11/erg11 single mutants, and the erg3/erg3 erg11/erg11 double mutants. The amphotericin B MICs increased from 0.25 μg/ml for the wild type to more than 16 μg/ml for the erg11/erg11 single mutants and the erg3/erg3 erg11/erg11 double mutants.
FIG. 4.
Susceptibility assays with C. albicans erg3 and erg11 mutants. Each strain was diluted as indicated from a starting solution containing 107 cells per ml, and 5 μl of each dilution was spotted onto the different agar plates containing the different drugs at the indicated concentrations. The plates were incubated for 48 h at 35°C.
TABLE 2.
Susceptibilities of C. albicans erg3 and erg11 mutants to different antifungal agents
| Strain | Genotype | MIC (μg/ml)
|
|
|---|---|---|---|
| Fluconazole | Amphotericin B | ||
| CAF2-1 | Wild type | 0.25 | 0.5 |
| DSY1216 | erg11/ERG11 | 0.06 | 0.5 |
| DSY1336 | erg11/ERG11 erg3/ERG3 | 0.06 | 0.5 |
| DSY1751 | erg11/ERG11 erg3/erg3 | >128 | 0.25 |
| DSY1764 | erg11/erg11 erg3/erg3 | >128 | >16 |
| DSY1431 | erg11/erg11 erg3/ERG3 | >128 | >16 |
| DSY1769 | erg11/erg11 | >128 | >16 |
Sterol analysis profiles.
The wild type and the mutants created during this work were subjected to GC-MS analysis for the identification of sterols. As summarized in Table 3, ergosterol was the major sterol in strains still carrying functional ERG3 or ERG11 alleles, i.e., strains DSY1216 and DSY1336. The major difference between these two strains and the parent, CAF2-1, was the increase in the lanosterol fraction (from 6.7 to 22.3 and 18.2%, respectively) at the expense of the ergosterol fraction, which decreased from 67 to 60 and 51%, respectively. As expected, the erg3/erg3 mutant accumulated large amounts of 14α-demethylated sterols that lacked desaturation between the C-5 and C-6 sterol moieties. In this type of strain, ergosta-7,22-dienol (61% of all sterols), ergosta-7-enol (7% of all sterols), and also fecosterol or episterol (26.8% of all sterols) were identified. The detection of ergosta-7,22-dienol and ergosta-7-enol can be explained by the action of the C-24 sterol methyltransferase and C-24 sterol reductase (Fig. 1). The detection of these sterols is consistent with a blockage of Δ5,6 desaturation of the sterol moiety. In the other mutants lacking ERG11 alleles, 14α-methylated sterols in the form of eburicol and lanosterol, 4,14-dimethylzymosterol, and 14α-methyl-fecosterol were the dominant sterols. The last two sterol metabolites could be formed by removal of the C-4 methyl group by the actions of ERG25, ERG26, and ERG27. An additional methylation step (by the action of ERG6) is required for the formation of 14α-methyl-fecosterol. The larger proportion of lanosterol or eburicol is consistent with the absence of 14α demethylation (Fig. 1). Even if ERG3 alleles were intact in strain DSY1769, the presence of a 14α-methyl group was apparently inhibiting the further processing of sterol downstream of the action of ERG6 in the ergosterol biosynthesis pathway.
TABLE 3.
Sterol compositions of C. albicans sterol mutants
| Sterol identified | Total sterol fraction (%) in strain:
|
||||||
|---|---|---|---|---|---|---|---|
| CAF2-1 (wild type) | DSY1216 (erg11/ERG11) | DSY1336 (erg11/ERG11 erg3/ERG3) | DSY1751 (erg11/ERG11 erg3/erg3) | DSY1764 (erg11/erg11 erg3/erg3) | DSY1431 (erg11/erg11 erg3/ERG3) | DSY1769 (erg11/erg11) | |
| Lanosterol | 6.7 | 22.3 | 18.2 | 2.4 | 26.9 | 21.4 | 57.0 |
| Eburicol (24-methylene-lanosterol) | ND | ND | ND | ND | 37.9 | 29.6 | 18.0 |
| Zymosterol | 7.2 | 5.7 | 6.2 | ND | ND | ND | ND |
| 4,14-Dimethyl-zymosterol | ND | ND | ND | ND | 27.6 | 31.5 | 15.1 |
| Episterol or fecosterol | 3.8 | ND | 3.3 | 26.8 | ND | ND | ND |
| 14-Methyl-fecosterol | ND | ND | ND | ND | 3.9 | 2.5 | 4.1 |
| Ergosta-5,8,22-trienol | 5.3 | 4.3 | 2.5 | ND | ND | ND | ND |
| Ergosta-7,22-dienol | 7.0 | 4.8 | 10.8 | 61.0 | ND | ND | ND |
| Ergosta-7-enol | ND | ND | ND | 7.1 | ND | ND | ND |
| Ergosterol | 67.9 | 60.8 | 51.6 | ND | ND | ND | ND |
| Unidentified | 2.1 | 2.1 | 7.4 | 2.7 | 3.7 | 15.0 | 5.8 |
ND, not detected.
DISCUSSION
In this study we created several mutants with mutations in the ERG pathway either by targeted mutagenesis or by selection in amphotericin B-containing medium. The drug susceptibility profile for erg3/erg3 deletion mutant DSY1751 was resistance to fluconazole, as expected. This mutant was also more susceptible than the wild type to other categories of metabolic inhibitors, including, for example, cycloheximide, fluphenazine, brefeldin A, and hygromycin B. We did not systematically test all types of C. albicans growth inhibitors in this study; however, the phenotypes of susceptibility to the growth inhibitors tested mirrored well the hypersusceptibility profiles observed for other C. albicans erg mutants in which mutations were constructed by targeted mutagenesis. For example, the C. albicans erg6 and erg24 mutants constructed by Jensen-Pergakes et al. (13) and Jia et al. (14), respectively, showed hypersusceptibility to substances identical to those used in this study, such as cycloheximide, fluphenazine, and brefeldin A. Only the erg24 mutant showed a slight increase in azole resistance, while the erg6 mutant was not affected by this class of antifungal agents (13, 14). The hypersusceptibilities to several unrelated drugs and metabolic inhibitors are believed to be the result of alterations in membrane fluidity, which thus contributes to enhanced drug permeability.
In our study, we observed that erg3 mutant DSY1751 was not more resistant to amphotericin B, but it was even slightly more susceptible to this drug than the wild type (Fig. 4, agar plate with a concentration of 1 μg/ml). This suggests that amphotericin B could have additional cellular targets in C. albicans. This was unexpected, given that no ergosterol was found in this mutant and that this defect is generally linked to amphotericin B resistance (20). Moreover, comparisons with erg3 mutants of other yeast species in which erg3 mutations were constructed showed some discrepancies with the results obtained here. In C. glabrata, the deletion of ERG3 does not lead to azole or amphotericin B resistance (8). In S. cerevisiae, targeted deletion of ERG3 is coupled with resistance to both azoles and nystatin, the latter of which is structurally related to amphotericin B. The reason for these discrepancies remains unclear. It is possible that the methods used to assess drug susceptibilities were different between the studies, and therefore, a more careful comparison needs to be performed by identical susceptibility tests before further speculations can be made. A more direct comparison could be made between the erg3/erg3 mutant constructed here and other clinical strains in which ERG3 mutations have been constructed. In two separate studies, clinical strains resistant to both azoles and amphotericin B were subjected to sterol analysis. The sterol profiles observed in these strains were typical of those for strains with a mutation in ERG3, as they mainly contained ergosta-7,22-dienol and ergosta-7-enol. These sterol profiles therefore resemble those determined in DSY1751. However, no detailed genetic analyses are available for these resistant strains to confirm the basis of the hypothetical ERG3 mutation (18, 25). Another azole-resistant C. albicans strain (the Darlington strain) has been analyzed carefully by Miyazaki et al. (21) and revealed that ergosta-7,22-dienol is a major sterol. This strain carries no functional ERG3 alleles, thus enabling a link between the presence of this sterol and a defect in ERG3. The susceptibility of the Darlington strain to amphotericin B was almost similar to that established for CAF2-1 and shown in Table 2 (J. Bennett, personal communication). The phenotypes of the Darlington strain and the erg3/erg3 homozygote DSY1751 are therefore similar and are in agreement with the idea that the Darlington strain is an erg3 mutant.
A surprising result of this study was the generation of an erg11/erg11 homozygous mutant in the presence of amphotericin B and under aerobic conditions. As inferred from the frequency of amphotericin B resistance and Southern analysis of erg11/erg11 deletion mutants DSY1431 and DSY1769, mitotic recombination was the likely event linked to the generation of amphotericin B resistance. Several studies have used the ability of C. albicans to undergo mitotic recombination to obtain a particular phenotype (9, 32). However, this method of raising homozygous mutants can be linked to additional compensatory mutations during the completion of mitotic recombination. One way to establish that no mutation other than the ERG11 deletion was generated in DSY1769 or other amphotericin B-resistant isolates in the present study would be to restore wild-type amphotericin B susceptibility by reintroduction of an intact ERG11 allele. This experiment was attempted in DSY1769; however, the transformation method used did not allow the recovery of any transformants. The poor transformation efficiency of strain DSY1769 could be due to its weak growth or its altered sterol profile. If compensatory mutations are possible in the erg11/erg11 homozygous mutants produced in this study, their identification remains to be established. Given that the erg11 mutants are viable only in the background of erg3 mutants as mentioned above, we looked for possible loss-of-function mutations in ERG3 alleles recovered from strains DSY1431 and DSY1769. However, the ERG3 alleles from the parental wild type and these mutants were identical at the level of nucleotide sequence.
Different studies performed with S. cerevisiae have reported that strains with the ERG11 mutation are not viable under aerobic conditions unless another suppression mutation that maps to ERG3 is present (3, 16). An erg11 C. glabrata mutant can also be obtained under anaerobic conditions, and from these mutants, spontaneous revertants able to grow under aerobic conditions can be obtained (8). The sterol profiles of such mutants show that they contain high levels of lanosterol and obtusifoliol. erg11/erg11 mutant DSY1769 had a sterol profile similar to this sterol profile in terms of its lanosterol content, but its sterol profile differed with respect to its content of obtusifoliol, a sterol not detected in DSY1769. Obtusifoliol is mostly related to 4,14-dimethyl-zymosterol, a sterol metabolite found in DSY1769, but differs by the addition of an unsaturated methyl group at C-24. A C. albicans mutant called D10 was reported to lack the function conferred by ERG11, although no molecular evidence was given (2, 3). This mutant could grow aerobically and showed a sterol profile similar to those observed for the erg11/erg11 mutants evaluated in this study, with the difference being that no obtusifoliol was detected in our investigation. A common feature among erg11 mutants of C. albicans, C. glabrata, and S. cerevisiae is that they can be generated in an erg3 deletion background without the use of a specific selection procedure. However, the sterol profiles of the double erg3/erg3 erg11/erg11 mutants and the erg3/erg3 mutants of C. albicans are again different from those of both C. glabrata and S. cerevisiae mutants. In the last two yeast species the dominant sterol metabolites are 14α-methyl-fecosterol, followed by lanosterol, while in C. albicans the major sterol is the lanosterol and eburicol fraction, followed by 4,4-dimethyl zymosterol.
In conclusion, this study clarifies the contribution of ERG3 in azole resistance and also suggests that erg11/erg11 mutants can spontaneously appear upon selective drug pressure in vitro when one ERG11 allele is not functional. Whether this event can occur during treatment of patients with amphotericin B is questionable. Experiments in which ERG11 has been “switched off” have been performed with C. glabrata in animal models and showed that the lack of ERG11 expression has only minor effects in animal models, probably because host sterols can compensate for the absence of C. glabrata sterols (22, 23). Therefore, positive selection of erg11 mutants in the context of animal experiments can be envisaged.
Acknowledgments
This work was supported by grant 3100-055901 from the Swiss National Research Foundation (to D.S.).
We thank J. Bennett for communicating results before publication.
REFERENCES
- 1.Arthington, B. A., L. G. Bennett, P. L. Skatrud, C. J. Guynn, R. J. Barbuch, C. E. Ulbright, and M. Bard. 1991. Cloning, disruption and sequence of the gene encoding yeast C-5 sterol desaturase. Gene 102:39-44. [DOI] [PubMed] [Google Scholar]
- 2.Bard, M., N. D. Lees, R. J. Barbuch, and D. Sanglard. 1987. Characterization of a cytochrome P450 deficient mutant of Candida albicans. Biochem. Biophys. Res. Commun. 147:794-800. [DOI] [PubMed] [Google Scholar]
- 3.Bard, M., N. D. Lees, T. Turi, D. Craft, L. Cofrin, R. Barbuch, C. Koegel, and J. C. Loper. 1993. Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids 28:963-967. [DOI] [PubMed] [Google Scholar]
- 4.De Backer, M. D., T. Ilyina, X. J. Ma, S. Vandoninck, W. H. Luyten, and H. Vanden Bossche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fridkin, S. K., and W. R. Jarvis. 1996. Epidemiology of nosocomial fungal infections. Clin. Microbiol. Rev. 9:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaber, R. F., D. M. Copple, B. K. Kennedy, M. Vidal, and M. Bard. 1989. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 9:3447-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geber, A., C. A. Hitchcock, J. E. Swartz, F. S. Pullen, K. E. Marsden, K. J. Kwon-Chung, and J. E. Bennett. 1995. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob. Agents Chemother. 39:2708-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorman, J. A., W. Chan, and J. W. Gorman. 1991. Repeated use of GAL1 for gene disruption in Candida albicans. Genetics 129:19-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning. A practical approach. IRL Press, Oxford, United Kingdom.
- 11.Henry, K. W., J. T. Nickels, and T. D. Edlind. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 44:2693-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 13.Jensen-Pergakes, K. L., M. A. Kennedy, N. D. Lees, R. Barbuch, C. Koegel, and M. Bard. 1998. Sequencing, disruption, and characterization of the Candida albicans sterol methyltransferase (ERG6) gene: drug susceptibility studies in erg6 mutants. Antimicrob. Agents Chemother. 42:1160-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jia, N., B. Arthington-Skaggs, W. Lee, C. A. Pierson, N. D. Lees, J. Eckstein, R. Barbuch, and M. Bard. 2002. Candida albicans sterol C-14 reductase, encoded by the ERG24 gene, as a potential antifungal target site. Antimicrob. Agents Chemother. 46:947-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph-Horne, T., D. Hollomon, R. S. Loeffler, and S. L. Kelly. 1995. Cross-resistance to polyene and azole drugs in Cryptococcus neoformans. Antimicrob. Agents Chemother. 39:1526-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelly, S. L., D. C. Lamb, A. J. Corran, B. C. Baldwin, and D. E. Kelly. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14 alpha-methylergosta-8,24(28)-dien-3 beta,6 alpha-diol. Biochem. Biophys. Res. Commun. 207:910-915. [DOI] [PubMed] [Google Scholar]
- 17.Kelly, S. L., D. C. Lamb, D. E. Kelly, J. Loeffler, and H. Einsele. 1996. Resistance to fluconazole and amphotericin in Candida albicans from AIDS patients. Lancet 348:1523-1524. [DOI] [PubMed] [Google Scholar]
- 18.Kelly, S. L., D. C. Lamb, D. E. Kelly, N. J. Manning, J. Loeffler, H. Hebart, U. Schumacher, and H. Einsele. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta 5,6-desaturation. FEBS Lett. 400:80-82. [DOI] [PubMed] [Google Scholar]
- 19.Lupetti, A., R. Danesi, M. Campa, M. Del Tacca, and S. Kelly. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8:76-81. [DOI] [PubMed] [Google Scholar]
- 20.Masia Canuto, M., and F. Gutierrez Rodero. 2002. Antifungal drug resistance to azoles and polyenes. Lancet Infect. Dis. 2:550-563. [DOI] [PubMed] [Google Scholar]
- 21.Miyazaki, Y., A. Geber, H. Miyazaki, D. Falconer, T. Parkinson, C. Hitchcock, B. Grimberg, K. Nyswaner, and J. E. Bennett. 1999. Cloning, sequencing, expression and allelic sequence diversity of ERG3 (C-5 sterol desaturase gene) in Candida albicans. Gene 236:43-51. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama, H., M. Izuta, N. Nakayama, M. Arisawa, and Y. Aoki. 2000. Depletion of the squalene synthase (ERG9) gene does not impair growth of Candida glabrata in mice. Antimicrob. Agents Chemother. 44:2411-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama, H., N. Nakayama, M. Arisawa, and Y. Aoki. 2001. In vitro and in vivo effects of 14α-demethylase (ERG11) depletion in Candida glabrata. Antimicrob. Agents Chemother. 45:3037-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 1995. Reference method for broth dilution antifungal susceptibility testing of yeast, vol. 12. Tentative standard M27-A. National Committee for Clinical Laboratory Standards, Villanova, Pa.
- 25.Nolte, F. S., T. Parkinson, D. J. Falconer, S. Dix, J. Williams, C. Gilmore, R. Geller, and J. R. Wingard. 1997. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob. Agents Chemother. 44:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parks, L. W., S. J. Smith, and J. H. Crowley. 1995. Biochemical and physiological effects of sterol alterations in yeast—a review. Lipids 30:227-230. [DOI] [PubMed] [Google Scholar]
- 27.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contributing to the resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC-transporter gene. Microbiology 143:405-416. [DOI] [PubMed] [Google Scholar]
- 30.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1996. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 40:2300-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsang, P. W., B. Cao, P. Y. Siu, and J. Wang. 1999. Loss of heterozygosity, by mitotic gene conversion and crossing over, causes strain-specific adenine mutants in constitutive diploid Candida albicans. Microbiology 145:1623-1629. [DOI] [PubMed] [Google Scholar]
- 33.Watson, P. F., M. E. Rose, S. W. Ellis, H. England, and S. L. Kelly. 1989. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem. Biophys. Res. Commun. 164:1170-1175. [DOI] [PubMed] [Google Scholar]
- 34.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zweytick, D., C. Hrastnik, S. D. Kohlwein, and G. Daum. 2000. Biochemical characterization and subcellular localization of the sterol C-24(28) reductase, Erg4p, from the yeast Saccharomyces cerevisiae. FEBS Lett. 470:83-87. [DOI] [PubMed] [Google Scholar]