Abstract
Three vancomycin-resistant veal calf fecal streptococci, identified as Streptococcus gallolyticus (n = 2) and Streptococcus lutetiensis, were shown to harbor vanB2 Tn5382-like elements earlier described in enterococci. One S. gallolyticus strain had a 1,495-bp IS256-related element inserted in vanSB. The vanB2 Tn5382 element present in the plasmid-free S. lutetiensis strain was transferable to Enterococcus faecium BM4105-RF, Enterococcus faecalis JH2-2, and its recombination-deficient derivative, UV202. The transfer frequencies were comparable between recipient strains (from 1 × 10−7 to 7 × 10−6). All transconjugants acquired a vanB-containing chromosomal insert of approximately 100 kb, apparently by site-specific integration. Secondary transconjugants were not observed in intraspecies retransfer experiments. These observations are consistent with a conjugative, selftransmissible, integrative element that might be involved in the interspecies spread of vanB2 resistance determinants. Two JH2-2-derived transconjugants had also gained additional copies of large vanB-containing chromosomal fragments, a process that involves unexplained mechanisms that seems to require functional host cell-dependent recombination mechanisms.
The vanB gene cluster is a common cause of transferable high-level vancomycin resistance in human clinical enterococci worldwide (3, 5, 6, 19, 31-33, 40). Its origin remains unknown. The host range and mechanisms for intercellular spread are only partially understood. Enterococcus faecium and Enterococcus faecalis are the main enterococcal hosts, but vanB has also been described in Enterococcus gallinarum (16, 35) and Enterococcus durans (18). Nonenterococcal reservoirs have also been reported, such as three fecal veal calf strains of Streptococcus gallolyticus (25) and one human fecal Streptococcus bovis isolate (27). Recently, the vanB2 gene cluster was identified in a human fecal Eggerthella lenta-related strain and in a nonidentified human fecal Clostridium species (37).
The vanB operons described so far seem to have a conserved order of genes (vanRB, vanSB, vanYB, vanW, vanHB, vanB, and vanXB). Due to sequence diversity, they can be divided in at least three distinct subtypes, vanB1 to B3, indicating different origins (7, 8, 11, 24).
Intra- and interspecies transfer of the vanB operon has been linked to the movement of large (90 to 250 kb) chromosomal fragments or conjugative plasmids (3, 5, 6, 31-33, 40). Recently, the 34-kb vanB2-containing Tn1549, which is similar to the Tn916-Tn1545 family of conjugative transposons, was completely sequenced (11). Tn1549 appears to be similar to the earlier described and partially sequenced Tn5382 (3). These putative conjugative transposons are thus designated Tn5382-like. Several studies suggest that the vanB2 operon is the most prevalent vanB subtype and that it is universally present as an integral part of Tn5382-like elements (5, 11, 23). Although molecular evidence for precise chromosomal or plasmid insertions of a vanB2 Tn5382-like element in enterocooci has been presented (3, 6, 11), direct evidence for conjugative transposition of Tn5382-like elements has not been given (6). Rather, transfer of Tn5382-like elements takes place as an integral part of variably sized chromosomal elements or conjugative plasmids (3, 5, 6). In the present study, we describe the genetic characterization and transferability of vanB2 Tn5382-containing elements in three fecal veal calf streptococci phenotypically identified as S. gallolyticus (25).
(This study was presented in part at the 41st Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 22 to 25 September 2001.)
MATERIALS AND METHODS
Bacterial strains and plasmids.
The vanB clusters of the vanB-positive Streptococcus strains 5-F9, 4-C11, and 4-G10 (Table 1), isolated from mixed fecal samples from veal calf herds in The Netherlands (25), were examined. E. faecium BM4105-RF, E. faecium BM4105-Str (30), E. faecalis JH2-2 (17), E. faecalis BM4110, and the recombination-deficient derivative of JH2-2, E. faecalis UV202 (41), were used as recipient strains in the conjugation experiments.
TABLE 1.
Properties of vanB streptococci in this study
| Strain | PFGE pattern | Species, as determined by sodA sequencing | Vancomycin resistance genotypea | vanB localization | Transfer frequencyb
|
|
|---|---|---|---|---|---|---|
| E. faecium BM4105-RFc | E. faecalis JH2-2/isogenic UV202c | |||||
| 5-F9 | I | S. lutetiensis | vanB2 | Chromosomal | 1 × 10−7 | 2 × 10−6/7 × 10−6 |
| 4-C11 | II | S. gallolyticus | vanB2 vanA | Chromosomal | NDd | ND/NTe |
| 4-G10 | III | S. gallolyticus | vanB2 vanA | Chromosomal | ND | ND/NT |
vanA genotype determined by Mevius et al. (25).
Number of transconjugants per donor cell.
Recipient strains.
ND, not detected.
NT, not tested.
Species identification by sodA sequence.
DNA sequence determinations were done by direct sequencing of PCR amplicons (7). The sodA degenerate primers d1 and d2 were used to amplify sodAint, an internal fragment of approximately 480 bp in streptococci, representing 85% of the sodA gene (28).
PCR amplification and nucleotide sequencing.
Amplification of DNA, direct sequencing of PCR amplicons, and restriction analysis of vanB long PCR amplicons by restriction fragment length polymorphism (RFLP) were conducted as described previously (7). A 1,132-bp vanRBSB region of strain 4-C11 was amplified with primers 5′-GTTTGATGCAGAGGCAGACGACT-3′ and 5′-CCTCCAACAGAACGCTTACAG-3′.
Computer analysis.
Editing, initial analysis of sequences, and alignments were performed by using the Sequence Navigator software package (Applied Biosystems, Foster City, Calif.). Nucleotide sequences were compared to those in the GenBank, EMBL, DDBJ, and PDB databases, and protein sequences were compared to nonredundant GenBank CDS translations and sequences in PDB, SwissProt, SPupdate, and PIR by using the BLASTN, BLASTX, and BLASTP local alignment search tools (1). Possible open reading frames (ORFs) were found by using the ORF finder located at the National Center for Biotechnology Information website.
Conjugation experiments.
Filter matings were done with a donor/recipient ratio of 19:1 (5). Various dilutions of resuspended bacteria were spread on brain heart infusion agar containing 8 μg of vancomycin/ml, 20 μg of rifampin/ml, and 10 μg of fusidic acid/ml. Transfer frequencies were expressed as the number of transconjugants per donor cell.
PFGE.
The vanB strains and transconjugants were typed by SmaI digestion and pulsed-field gel electrophoresis (PFGE) of genomic DNA (5) with the run increased to 30 h. A chromosomal vanB location was assessed by sequential vanB, 16S rDNA, and 23S rDNA hybridizations of PFGE-separated I-CeuI (New England Biolabs, Beverly, Mass.)-digested DNA. Agarose plugs were prepared as described previously (5) with 1 mg of proteinase K/ml and digested with the intron-encoded endonuclease I-CeuI, which recognizes a 23-bp sequence specific for 23S rRNA genes (21). Digested DNA was electrophoresed in a 1.2% agarose gel by using a CHEF-DRIII apparatus (Bio-Rad Laboratories, Hercules, Calif.). The pulse time was increased from 60 to 90 s over 22 h at 200 V.
DNA isolation and digestion with restriction enzymes.
Total DNA was isolated by using guanidinium isothiocyanate by a protocol slightly modified from that described by Bickley and Owen (2). Streptococci were grown overnight in brain heart infusion broth, and cells from 1 ml of culture were harvested for lysis. Cold 3 M sodium acetate (60 μl) was added subsequent to cell lysis to salt out proteins. Plasmid DNA was isolated as described by Werner et al. (38). Cleavage of DNA with restriction endonucleases was performed as recommended by the manufacturer (New England Biolabs).
DNA transfer and hybridization.
Colony blotting and Southern transfer of digested genomic DNA were performed as previously described (5). Probes were labeled by using a PCR digoxigenin (DIG) probe synthesis kit, and detection was performed by using a DIG luminescent detection kit (Roche Diagnostics, Basel, Switzerland). Total DNAs from the following bacteria were used as a template for probe synthesis. The 23S rDNA probe (669 bp with primers 5′-CGCATGTACAGGATAGGTAGG-3′ and 5′-AGGTGGGCTTCACACTTAGAT-3′) was from E. faecium ATCC19434; the 16S rDNA probe (5) was from E. faecalis DS16C2 (10); the Tn5382 probe was from E. faecium C68 (3); and the vanB probe (7) was from E. faecalis V583 (9).
Nucleotide sequence accession numbers.
Novel nucleotide accession numbers described in the text are AY035703 to AY035715.
RESULTS AND DISCUSSION
Species identification of 5-F9, 4-C11, and 4-G10 by sodA sequence determination.
The three strains have previously been identified as S. gallolyticus by their growth characteristics, biochemical activities, and whole-cell protein analysis (25). The results of Poyart et al. (28) indicate that the superoxid dismutase (sodA) gene has a higher divergence and thus might be a more discriminative target sequence than the 16S rDNA for differentiating closely related streptococcal species. Sequence determination revealed that 4-C11 and 4-G10 had identical sodAint sequences (GenBank accession no. AY035714 and AY035715) that were indistinguishable from those of S. gallolyticus NEM1203 and NEM1204 (GenBank accession no. AJ297196 and AJ297197, respectively). The sodAint region in 5-F9 (GenBank accession no. AY035713) showed only 81% identity to the 4-C11 and 4-G10 sequences but 100% identity to the sequences of NEM760 and NEM1603 (GenBank accession no. AJ297188 and AJ297205, respectively), recently classified as Streptococcus lutetiensis (29).
Molecular characterization of the vanB operons.
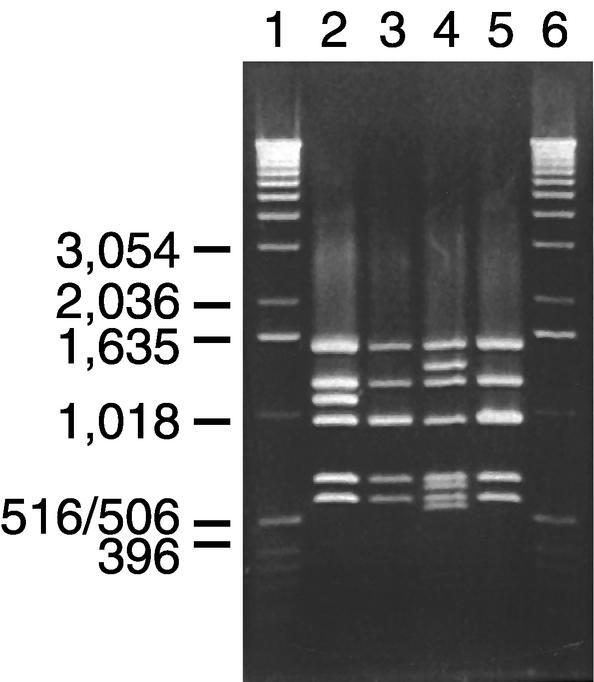
5-F9, 4-C11, and 4-G10 were found to be genomically diverse by PFGE (Table 1). The vanSB to vanYB intergenic regions (GenBank accession no. AY035706 to AY035708) and the vanB2 ligase gene sequences (GenBank accession no. AY035703 to AY035705) were identical to vanB2 operon sequences (7, 12). The RFLP patterns of vanB long PCR amplicons from strains 4-G10 and 5-F9 (Fig. 1, lanes 3 and 5) were also identical to the RFLP-2 pattern of vanB2 clusters found in enterococci (7). The RFLP-2 pattern gives two bands of approximately 1 kb, one in the vanRBSB region and one in the vanYB region. Strain 4-C11 showed a divergent RFLP-2 pattern (RFLP-2#) in which the 1-kb BspHI fragment covering the vanRBSB region in RFLP-2 has been replaced by restriction fragments of approximately 1,350, 600, and 550 bp (Fig. 1, lane 4), indicating the presence of an insertion in this region. The RFLP-2# pattern was accounted for by DNA sequencing, which identified a 1,504-bp insertion starting 103 bp into vanSB2 (GenBank accession number AY035712). Analysis of the inserted DNA revealed a single 1,353-bp ORF oriented in the same direction as the vanB cluster. This ORF encoded a putative 450-amino-acid protein with 29% identity (amino acid 29 to 160) to 139 amino acids found in putative transposases of ISRm3 elements from Rhizobium meliloti (36, 39). ISRm3 belongs to the IS256 family of insertion sequences (ISs), and the vanSB2 insertion shares several characteristics with this IS family (22). Briefly, the insert comprises a potential DDE motif together with a K/R residue, 12 bp at the terminus, which were identical to the vanSB2 sequence at the region of insertion and an imperfect IR of 25 bp located at the terminal ends of the insert, leaving 9 bp external to the repeat, which might be a duplication of the target sequence (Fig. 2) (GenBank accession number AY035712). IS elements have been described earlier in vanB2 Tn5382-like elements (5, 20). However, these IS elements did not interrupt genes involved in expression of the resistance phenotype. Because of the insertion, vanSB2 is likely not expressed in the vanB operon of strain 4-C11. However, this strain also carries a vanA cluster (25), and thus the phenotypical consequences of a nonfunctional VanSB2 or a lack of VanSB2 are not revealed.
FIG. 1.
Restriction fragment analysis of vanB long PCR amplicons. BspHI/DraI-digested vanB long PCR amplicons were analyzed by agarose gel electrophoresis. Lanes: 1 and 6, 1-kb ladder (Invitrogen Corporation, Carlsbad, Calif.); 2, vanB1 strain V583 with RFLP-1; 3, vanB2 strain 4-G10 with RFLP-2; 4, vanB2 strain 4-C11 with a 1,504-bp insertion in vanSB2 leading to replacement of the 969-bp fragment with restriction fragments of 1,327, 614, and 532 bp (RFLP-2#); 5, vanB2 isolate 5-F9 with RFLP-2. Molecular sizes shown to the left of the gel (in base pairs) refer to the 1-kb ladder.
FIG. 2.
Alignment of the terminal portions of the insertion in the vanSB gene of strain 4-C11. The left end following a 12-bp sequence of vanSB (bold) is aligned to the inverted version of the right end. Vertical lines indicate identical nucleotides. A putative imperfect inverted repeat of 25 bp is underlined. The 12-bp sequence of vanSB (bold) immediately in front of the left end is identical to 12 bp at the right end (bold) of the insertion when this sequence is not inverted.
Presence of Tn5382-like elements.
Southern hybridization of total DNAs showed homology of the Tn5382 probe to an approximately 2.5-kb DdeI restriction fragment in 5-F9 and a 2-kb DdeI restriction fragment in both 4-C11 and 4-G10, consistent with the presence of a single Tn5382-like copy (data not shown). DNA sequencing of the vanXB Tn5382 joint region revealed that the vanB2 clusters are integral parts of Tn5382-like elements in S. gallolyticus and S. lutetiensis. The DNA sequences (GenBank accession no. AY035709 to AY035711) were identical to Tn5382-like elements described in enterococci (5). Thus, the DNA sequence and RFLP analyses show that highly similar vanB2 operons with linkage to Tn5382-like elements are present in enterococci (3, 5, 7, 11) and in streptococci, suggesting a common origin for these operons.
S. gallolyticus strains have been isolated from diverse habitats, including feces of many animals, as well as from animals with bovine mastitis, human clinical sources, and the sheep rumen (26). S. lutetiensis (earlier classified as Streptococcus infantarius subsp. coli) has been isolated from various human specimens, including human feces (34). Thus, these species are able to survive in the gastrointestinal tract and thereby have the opportunity to exchange genetic material with other gastrointestinal commensals. The vanB2 operon has recently been found in human fecal anaerobic bacteria, but possible linkage to Tn5382-like elements was not determined (37). The G + C content of different parts of Tn5382-like elements are 48 (vanB cluster and right extremity, which are implicated in the excision-integration process) to 55 mol% (left extremity, which is probably implicated in conjugative transfer) (GenBank accession no. AF192329). Enterococci (34 to 42 mol%), streptococci (34 to 46 mol%), Clostridium innocuum (41 to 45 mol%), and Eggerthella (30 to 40 mol%) genomes have lower G + C contents, suggesting a different origin of vanB2 Tn5382-like elements. Nevertheless, detection of vanB2 Tn5382-like elements in two different S. gallolyticus strains, in one S. lutetiensis strain, and in genomically unrelated enterococci in both Europe (5, 11, 24) and the United States (3, 5, 14) suggests that these are widespread and successful mobile elements.
Localization of the vanB-containing elements.
The strains were characterized with regard to plasmid or chromosomal localization of the vanB gene cluster by Southern hybridization of plasmid DNA and I-CeuI PFGE fragments. Hybridization with the vanB probe, and not with the 16S rDNA probe, to plasmid DNA indicates a plasmid localization of the vanB operon, whereas cohybridization of the vanB and either 16S rDNA or 23S rDNA probes to I-CeuI PFGE fragments was interpreted as a chromosomal location of the vanB cluster. A chromosomal location of the vanB operon was shown in all three strains (data not shown).
Mobility of the vanB gene clusters.
Filter mating experiments were performed between the vanB-containing streptococci as donor strains and plasmid-free enterococcal recipients. Five vancomycin-resistant transconjugants from each mating pair were analyzed for the presence of vanB by PFGE and subsequent hybridization. All transconjugants tested harbored vanB and showed PFGE patterns related to that of the recipient.
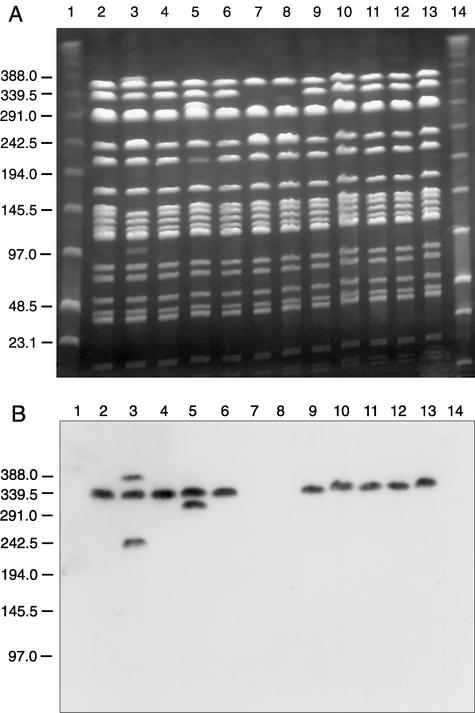
Filter mating experiments using 4-C11 and 4-G10 as donors gave no detectable transconjugants (transfer rates, <1 × 10−9). Successful mating was shown with the 5-F9 strain, consistent with the findings of Mevius et al. (25). Transfer frequencies of about 7 × 10−6 transconjugants per donor were observed with the recombination-deficient recipient E. faecalis UV202. The transfer rates were somewhat lower with E. faecalis JH2-2 (2 × 10−6 transconjugants per donor) or E. faecium BM4105-RF (1 × 10−7 transconjugants per donor) as recipients. SmaI PFGE analysis and subsequent vanB hybridization of the transconjugants (Fig. 3A, lanes 2 to 6 and 9 to 13) derived from the E. faecalis recipients JH2-2 and UV202 (Fig. 3A, lanes 7 and 8, respectively) or the E. faecium recipient BM4105-RF (data not shown) revealed a reproducible insertion event of an approximately 100-kb vanB-containing element which was confirmed by SfiI/NotI PFGE analysis of the JH202- and UV202-derived transconjugants (data not shown). The JH2-2- and UV202-derived transconjugants (Fig. 3A and B, lanes 2 to 6 and 9 to 13, respectively) showed the replacement of an approximately 240-kb fragment (the double band becomes a single band) by a 340-kb vanB-containing fragment. BM4105-RF-derived transconjugants (data not shown) showed the replacement of a 290-kb fragment by an approximately 390-kb vanB-containing fragment.
FIG. 3.
PFGE of SmaI-digested total DNA of the recipients and transconjugants from matings using 5-F9 as donor and JH2-2 and UV202 as recipients (A) and corresponding Southern hybridization with a vanB probe (B). Lanes: 1 and 14, low-range PFG markers (New England Biolabs); 2 to 6, transconjugants obtained after mating with recipient JH2-2; 7, recipient JH2-2; 8, recipient UV202; 9 to 13, transconjugants obtained with UV202 as recipient. JH2-2 and UV202 are isogenic strains and thus have indistinguishable PFGE patterns.
A 100-kb vanB-containing insertion event in the corresponding SmaI fragment, comparable to that demonstrated in this study, has previously been shown in BM4110- and BM4105-RF derived transconjugants obtained by using the vanB2 S. bovis NEM760 as donor (27). BM4110 and JH2-2 have the same PFGE patterns, as they are isogenic strains derived from JH2. Thus, it seems that streptococci might harbor a vanB2-containing transferable 100-kb element that integrates in a site-specific manner in enterococcal recipients. The arrival of the 100-kb vanB Tn5382-containing element in the recombination-deficient recipient strain UV202 as well as its ancestor JH2-2 at comparable transfer frequencies indicates that it does not require a functional host cell recombination system for integration. Thus, the element shares many properties with conjugal self-transmissible, integrating elements (15). Further studies are in progress to characterize this element.
Retransfer experiments using 5-F9-derived transconjugants as donors and BM4105-Str and BM4110 as recipients showed no detectable secondary transconjugants (<1 × 10−9). However, Poyart et al. (27) were able to show successful retransfer of their 100-kb vanB-containing element from S. bovis between enterococcal strains in the presence of the plasmid pIP964. The plasmid contents of 5-F9 and selected transconjugants were therefore analyzed to examine if plasmids could be involved in the observed transfer events. Plasmid DNA was not detected in repeated experiments (data not shown), indicating plasmid-independent chromosomal transfer. Hence, enterococci may not be able to express the transfer functions of the streptococcal 100-kb element, or the factors required for transfer could be retained in the donor after conjugation.
Two of 11 transconjugants obtained by JH2-2 matings revealed the presence of additional vanB-containing fragments (Fig. 3, lanes 3 and 5). These results have been reproduced and confirmed by hybridization of SfiI/NotI PFGE fragments (data not shown). The integration of additional large vanB-containing fragments seems to involve a host cell-encoded recombination mechanism, since this phenomenon was not observed in the 11 UV202-derived transconjugants tested.
The potential spread of vancomycin resistance determinants to other pathogenic bacteria is underlined by the expanded species distribution of the vanB2 Tn5382-containing elements, as seen in this study, by the broad host range demonstrated by the Tn916-Tn1545 family of conjugative transposons in general, as well as by the recent description of vanA gene clusters in clinical Staphylococcus aureus strains (4, 13). Further studies are needed to clarify the reservoirs for transferable vancomycin resistance determinants and address mechanisms for transposition and conjugative mobilization in order to prevent and control their spread.
Acknowledgments
This work was supported by grants from the Norwegian Research Council.
We thank P. Butaye and C. Poyart for providing strains. We also thank M. Birkely, B. C. Haldorsen, B. C. Nygård, and M. S. Wesmajervi for excellent technical assistance.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bickley, J., and R. J. Owen. 1995. Preparation of bacterial genomic DNA. Methods Mol. Biol. 46:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control. 2002. Staphylococcus aureus resistant to vancomycin—United States 2002. Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 5.Dahl, K. H., E. W. Lundblad, T. P. Røkenes, Ø. Olsvik, and A. Sundsfjord. 2000. Genetic linkage of the vanB2 gene cluster to Tn5382 in vancomycin-resistant enterococci and characterization of two novel insertion sequences. Microbiology 146:1469-1479. [DOI] [PubMed] [Google Scholar]
- 6.Dahl, K. H., T. P. Røkenes, E. W. Lundblad, and A. Sundsfjord. 2003. Nonconjugative transposition of the vanB-containing Tn5382-like element in Enterococcus faecium. Antimicrob. Agents Chemother. 47:786-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahl, K. H., G. S. Simonsen, Ø. Olsvik, and A. Sundsfjord. 1999. Heterogeneity in the vanB gene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 43:1105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evers, S., and P. Courvalin. 1996. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J. Bacteriol. 178:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evers, S., P. E. Reynolds, and P. Courvalin. 1994. Sequence of the vanB and ddl genes encoding D-alanine:D-lactate and D-alanine:D-alanine ligases in vancomycin-resistant Enterococcus faecalis V583. Gene 140:97-102. [DOI] [PubMed] [Google Scholar]
- 10.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garnier, F., S. Taourit, P. Glaser, P. Courvalin, and M. Galimand. 2000. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146:1481-1489. [DOI] [PubMed] [Google Scholar]
- 12.Gold, H. S., S. Unal, E. Cercenado, C. Thauvin-Eliopoulos, G. M. Eliopoulos, C. B. Wennersten, and R. C. Moellering, Jr. 1993. A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes of enterococci. Antimicrob. Agents Chemother. 37:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Zorn, B., and P. Courvalin. 2003. VanA-mediated high level glycopeptide resistance in MRSA. Lancet Infect. Dis. 3:67-68. [DOI] [PubMed] [Google Scholar]
- 14.Hanrahan, J., C. Hoyen, and L. B. Rice. 2000. Geographic distribution of a large mobile element that transfers ampicillin and vancomycin resistance between Enterococcus faecium strains. Antimicrob. Agents Chemother. 44:1349-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochut, B., and M. K. Waldor. 1999. Site specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol. Microbiol. 32:99-110. [DOI] [PubMed] [Google Scholar]
- 16.Ishii, Y., A. Ohno, S. Kashitani, M. Iwata, and K. Yamaguchi. 1996. Identification of VanB-type vancomycin resistance in Enterococcus gallinarum from Japan. J. Infect. Chemother. 2:102-105. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenney, A., C. Franklin, L. Liolios, and D. Spelman. 2000. Enterococcus durans VanB. J. Antimicrob. Chemother. 46:515.. [DOI] [PubMed] [Google Scholar]
- 19.Lee, W. G., J. A. Jernigan, J. K. Rasheed, G. J. Anderson, and F. C. Tenover. 2001. Possible horizontal transfer of the vanB2 gene among genetically diverse strains of vancomycin-resistant Enterococcus faecium in a Korean hospital. J. Clin. Microbiol. 39:1165-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, W. G., and W. Kim. 2003. Identification of a novel insertion sequence in vanB2-containing Enterococcus faecium. Lett. Appl. Microbiol. 36:186-190. [DOI] [PubMed] [Google Scholar]
- 21.Liu, S.-L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGregor, K. F., C. Nolan, H. K. Young, M.-F. I. Palepou, L. Tysall, and N. Woodford. 2001. Prevalence of the vanB2 gene cluster in VanB glycopeptide-resistant enterococci in the United Kingdom and the Republic of Ireland and its association with a Tn5382-like element. Antimicrob. Agents Chemother. 45:367-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGregor, K. F., and H. Young. 2000. Identification and characterization of vanB2 glycopeptide resistance elements in enterococci isolated in Scotland. Antimicrob. Agents Chemother. 44:2341-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mevius, D., L. Devriese, P. Butaye, P. Vandamme, M. Verschure, and K. Veldman. 1998. Isolation of glycopeptide resistant Streptococcus gallolyticus strains with vanA, vanB, and both vanA and vanB genotypes from faecal samples of veal calves in The Netherlands. J. Antimicrob. Chemother. 42:275-276. [DOI] [PubMed] [Google Scholar]
- 26.Osawa, R., T. Fujisawa, and L. I. Sly. 1995. Streptococcus gallolyticus sp. nov.; Gallate degrading organisms formerly assigned to Streptococcus bovis. Syst. Appl. Microbiol. 18:74-78. [Google Scholar]
- 27.Poyart, C., C. Pierre, G. Quesne, B. Pron, P. Berche, and P. Trieu-Cuot. 1997. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob. Agents Chemother. 41:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poyart, C., G. Quesnes, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poyart, C., G. Quesne, and P. Trieu-Cuot. 2002. Taxonomic dissection of the Streptococcus bovis group by analysis of manganese-dependent superoxide dismutase gene (sodA) sequences: reclassification of ‘Streptococcus infantarius subsp. coli’ as Streptococcus lutetiensis sp. nov. and of Streptococcus bovis biotype 11. 2 as Streptococcus pasteurianus sp. nov. Int. J. Syst. E. Microbiol. 52:1247-1255. [DOI] [PubMed] [Google Scholar]
- 30.Poyart, C., and P. Trieu-Cuot. 1994. Heterogeneric conjugal transfer of the pheromone-responsive plasmid pIP964 (IncHlyI) of Enterococcus faecalis in the apparent absence of pheromone induction. FEMS Microbiol. Lett. 122:173-180. [DOI] [PubMed] [Google Scholar]
- 31.Quintiliani, R., Jr., and P. Courvalin. 1994. Conjugal transfer of the vancomycin resistance determinant vanB between enterococci involves the movement of large genetic elements from chromosome to chromosome. FEMS Microbiol. Lett. 119:359-364. [DOI] [PubMed] [Google Scholar]
- 32.Quintiliani, R., Jr., and P. Courvalin. 1996. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172:1-8. [DOI] [PubMed] [Google Scholar]
- 33.Rice, L. B., L. L. Carias, C. L. Donskey, and S. D. Rudin. 1998. Transferable, plasmid-mediated vanB-type glycopeptide resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 42:963-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlegel, L., F. Grimont, M. D. Collins, B. Renault, P. A. Grimont, and A. Bouvet. 2000. Streptococcus infantarius sp. nov., Streptococcus infantarius subsp. infantarius subsp. nov. and Streptococcus infantarius subsp. coli subsp. nov., isolated from humans and food. Int. J. Syst. E vol. Microbiol. 50:25-34. [DOI] [PubMed] [Google Scholar]
- 35.Simonsen, G. S., B. M. Andersen, A. Digranes, S. Harthug, T. Jacobsen, E. Lingaas, O. B. Natås, Ø. Olsvik, S. H. Ringertz, A. Skulberg, G. Syversen, and A. Sundsfjord. 1998. Low faecal carrier rate of vancomycin resistant enterococci in Norwegian hospital patients. Scand. J. Infect. Dis. 30:465-468. [DOI] [PubMed] [Google Scholar]
- 36.Soto, M. J., A. Zorzano, J. Olivares, and N. Toro. 1992. Nucleotide sequence of Rhizobium meliloti GR4 insertion sequence ISRm3 linked to the nodulation competitiveness locus nfe. Plant Mol. Biol. 20:307-309. [DOI] [PubMed] [Google Scholar]
- 37.Stinear, T. P., D. C. Olden, P. D. R. Johnson, J. K. Davies, and M. L. Grayson. 2001. Enterococcal vanB resistance locus in anaerobic bacteria in human faeces. Lancet 357:855-856. [DOI] [PubMed] [Google Scholar]
- 38.Werner, G., I. Klare, and W. Witte. 1997. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol. Lett. 155:55-61. [DOI] [PubMed] [Google Scholar]
- 39.Wheatcroft, R., and S. Laberge. 1991. Identification and nucleotide sequence of Rhizobium meliloti insertion sequence ISRm3: similarity between the putative transposase encoded by ISRm3 and those encoded by Staphylococcus aureus IS256 and Thiobacillus ferrooxidans IST2. J. Bacteriol. 173:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodford, N., D. Morrison, A. P. Johnson, A. C. Bateman, J. G. M. Hastings, T. S. J. Elliott, and B. Cookson. 1995. Plasmid-mediated vanB glycopeptide resistance in enterococci. Microb. Drug Resist. 1:235-240. [DOI] [PubMed] [Google Scholar]
- 41.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143:966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]