Abstract
It has previously been shown that overexpression of the CdMDR1 gene is a major contributor to resistance in fluconazole-resistant isolates of Candida dubliniensis. However, since CdMdr1p does not mediate transport of other azole drugs such as itraconazole, we investigated the molecular mechanisms of stable resistance to itraconazole obtained in three strains of C. dubliniensis (two with nonfunctional CdCDR1 genes and one with functional CdCDR1 genes) by serial exposure to this antifungal agent in vitro. Seven derivatives that were able to grow on agar medium containing 64 μg of itraconazole per ml were selected for detailed analysis. These derivatives were resistant to itraconazole, fluconazole, and ketoconazole but were not cross resistant to inhibitors. CdMDR1 expression was unchanged in the seven resistant derivatives and their parental isolates; however, all seven derivatives exhibited increased levels of CdERG11 expression, and six of the seven derivatives exhibited increased levels of CdCDR1 expression compared to the levels of expression by their respective parental isolates. Except for one derivative, the level of rhodamine 6G efflux was decreased in the itraconazole-resistant derivatives compared to the level of efflux in their parental isolates, suggesting altered membrane properties in these derivatives. Analysis of their membrane sterol contents was consistent with a defective sterol C5,6-desaturase enzyme (CdErg3p), which was confirmed by the identification of mutations in the alleles (CdERG3) encoding this enzyme and their lack of functional complementation in a Saccharomyces cerevisiae erg3 mutant. The results of this study show that the loss of function of CdErg3p was the primary mechanism of in vitro-generated itraconazole resistance in six of the seven the C. dubliniensis derivatives. However, the mechanism(s) of itraconazole resistance in the remaining seventh derivative has yet to be determined.
Candida dubliniensis is a recently identified yeast species that is phylogenetically closely related to Candida albicans (49) and is frequently misidentified as such (34). Although originally identified in specimens recovered from the oral cavities of human immunodeficiency virus (HIV)-infected individuals in Ireland and Australia, C. dubliniensis has since been identified in a wide variety of clinical settings throughout the world (13, 21, 38, 48). In the most comprehensive study to date, by use of the criteria of the National Committee for Clinical Laboratory Standards (fluconazole MICs, ≤4 μg/ml for susceptible, 8 to 32 μg/ml for susceptible dose dependent, and ≥64 μg/ml for resistant), the majority of C. dubliniensis isolates were found to be susceptible to fluconazole and other commonly used antifungal drugs, including itraconazole, ketoconazole, and amphotericin B (37). However, fluconazole resistance in clinical isolates and the replacement of susceptible strains by resistant derivatives has been documented (21, 31, 36, 41). Interestingly, comparison of the geometric mean MICs of fluconazole, itraconazole, and ketoconazole for 58 isolates each of C. albicans and C. dubliniensis showed that while the fluconazole MICs for C. dubliniensis isolates were below the breakpoint of 64 μg/ml, they were significantly and consistently higher than those for the C. albicans isolates (34). Furthermore, sequential exposure of fluconazole-susceptible clinical isolates of C. dubliniensis to increasing concentrations of fluconazole in agar medium led to the recovery of derivatives expressing stable fluconazole-resistant and susceptible-dose-dependent phenotypes (MIC range, 16 to 64 μg/ml) (29-31). It has been suggested that the ability of C. dubliniensis to develop resistance to fluconazole may contribute to its ability to successfully colonize the oral cavities of HIV-infected individuals receiving prolonged fluconazole treatment (31). Furthermore, this may explain the apparent recent emergence of this species in this patient cohort.
While fluconazole is still very often the drug of choice for the treatment of candidosis, the emergence of fluconazole-resistant strains and recurrent infections recalcitrant to therapy has led to a reappraisal of treatment strategies for these infections. The triazole derivative itraconazole has a broader spectrum of activity than fluconazole and has been shown to be effective against both Candida and Aspergillus species (26). Since itraconazole has been used less extensively than fluconazole, clinical resistance is not yet a major problem.
Azole drugs target an enzyme in Candida responsible for the 14α-demethylation of 24-methylenedihydrolanosterol, a step in the ergosterol biosynthetic pathway. This enzyme, known as 14α-lanosterol demethylase, is encoded by the ERG11 gene. Several mechanisms by which Candida species can develop resistance to azole antifungal agents have been described: cells can fail to accumulate these agents; the affinity of the 14α-demethylase enzyme for these agents can be altered; the cellular content of Erg11p can be elevated; and other enzymes of the ergosterol biosynthetic pathway, such as the sterol C5,6-desaturase, can be inactivated by mutation. A combination of these mechanisms is likely to occur in many clinical azole-resistant isolates, since resistance to azole antifungal agents in C. albicans has been shown to develop in a stepwise process over time (7).
Several studies have demonstrated the importance of the overexpression of multidrug transporters in the development of azole resistance in Candida species (25, 27, 43, 45). In C. albicans, overexpression of the multidrug transporter Cdr1p is associated with resistance to multiple azole drugs, as well as to other unrelated antifungal drugs and particular inhibitors. In contrast, Mdr1p overexpression is associated with resistance to fluconazole and inhibitors but not to itraconazole and ketoconazole (45, 46). In C. albicans, overexpression of Cdr1p is the most frequently reported mechanism of resistance to azole drugs (22, 35). However, in C. dubliniensis the mechanisms of resistance differ from those of C. albicans. Previous studies have shown that in C. dubliniensis the primary mechanism of fluconazole resistance is overexpression of the major facilitator CdMdr1p (30, 54). In this species it has been shown that CdCdr1p is important for mediating reduced susceptibility to itraconazole and ketoconazole but is not essential for fluconazole resistance (29). Moreover, Moran et al. (29) found that 35% of C. dubliniensis isolates harbor mutated alleles of CdCDR1, which encode a truncated nonfunctional CdCdr1p protein. Interestingly, isolates with nonfunctional CdCdr1p all belong to C. dubliniensis genotype 1, a group of very closely related isolates described by Gee et al. (10) that have mainly been recovered from HIV-infected individuals, most of whom have received fluconazole treatment, and that constitute the predominant group of C. dubliniensis isolates.
The aims of the present study were (i) to measure the incidence of itraconazole resistance in C. dubliniensis clinical isolates, (ii) to determine if resistance to itraconazole could be induced by exposing C. dubliniensis isolates to itraconazole in vitro, and (iii) to investigate the molecular mechanisms involved in itraconazole resistance in this species. The last aim was deemed to be of particular interest, given the inability of CdMdr1p to transport itraconazole and the high prevalence of nonsense mutations in the CdCDR1 gene.
MATERIALS AND METHODS
Chemicals, enzymes, radioisotopes, and oligonucleotides.
All chemicals used were of analytical grade or molecular biology grade and were purchased from Sigma-Aldrich Chemical Co. (Tallaght, Dublin, Republic of Ireland), BDH (Poole, United Kingdom), or Roche Diagnostics Ltd. (Lewes, United Kingdom). Enzymes for molecular biology procedures were purchased from Promega Corporation (Madison, Wis.), Roche Diagnostics Ltd., or New England Biolabs Inc. (Beverly, Mass.).
Fluconazole powder was a gift from Pfizer Central Research (Sandwich, United Kingdom), and ketoconazole and itraconazole were both gifts from Janssen Pharmaceuticals (Cork, Republic of Ireland). Amphotericin B was a gift from E. R. Squibb (Swords, Republic of Ireland).
The following drugs were all purchased from Sigma-Aldrich: cerulenin, crystal violet, flucytosine, Geneticin, 4-nitroquinoline-N-oxide, cycloheximide, benomyl, fluphenazine, 1,10-phenanthroline, and rhodamine 6G (R6G).
Strains and media.
The Escherichia coli strain XL2-Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIq ZΔ M15 Tn10 (Tetr) Amy Camr]) (Stratagene, La Jolla, Calif.) was used as the host strain for the plasmid pBluescript IIKS(−) (Stratagene). XL2-Blue and derivatives harboring recombinant plasmids were cultured and maintained as described previously (42).
Saccharomyces cerevisiae ATCC 4002667 Δerg3 was used as the host for the yeast expression vector pYES (4) and its recombinant derivatives. Strain ATCC 4002667 (MATa his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 erg3Δ::kanMX4) is defective in sterol C5,6-desaturation. The S. cerevisiae Δerg3 strain and its transformants harboring CdERG3 were routinely cultured on yeast nitrogen base (Difco Laboratories, Detroit, Mich.) medium supplemented with 50 μg of adenine, histidine, leucine, lysine, uracil, and tryptophan per ml (SC-URA medium) and containing 2% (wt/vol) glucose at 30°C. For heterologous expression of C. dubliniensis open reading frames (ORFs) cloned in pYES, S. cerevisiae Δerg3 transformants were grown on SC-URA medium containing 2% (wt/vol) galactose instead of glucose.
Culture conditions for C. dubliniensis clinical isolates and in vitro-generated derivatives.
The 111 clinical isolates of C. dubliniensis tested for their fluconazole susceptibilities in the present study were recovered from 106 separate patients. The 58 clinical isolates of C. dubliniensis tested for their itraconazole susceptibilities were recovered from 56 separate patients. All isolates were routinely cultured on potato dextrose agar (PDA; Oxoid, Basingstoke, United Kingdom) medium (pH 5.6) at 37°C. For liquid culture, the isolates were grown in yeast extract-peptone-dextrose (YEPD) broth at 37°C in an orbital incubator (Gallenkamp, Leicester, United Kingdom) at 200 rpm.
Susceptibility testing procedures. (i). BMD
Susceptibility testing with fluconazole and itraconazole was carried out by the broth microdilution (BMD) method of Rodriguez-Tudela and Martinez-Suarez (40). All tests were carried out in duplicate. Results were interpreted according to the following MIC breakpoints for itraconazole: susceptible, ≤0.125 μg/ml; susceptible dose dependent, 0.25 to 0.5 μg/ml; resistant, ≥1 μg/ml. For fluconazole the following breakpoints were used: susceptible, ≤4 μg/ml; susceptible dose dependent, 8 to 32 μg/ml; resistant, ≥64 μg/ml.
(ii) Amphotericin B susceptibility testing by E-test.
Determination of susceptibility to amphotericin B by the E-test (AB BIODISK, Solna, Sweden) was carried out according to the instructions of the manufacturer.
(iii) Plate assays.
The susceptibilities of C. dubliniensis and C. albicans strains to a range of inhibitors and antifungal drugs (R6G, fluphenazine, hygromycin, Geneticin, and cycloheximide) was tested qualitatively by spotting serial dilutions of yeast cultures on complex YEPD medium agar plates, which permit the easy visualization of growth differences between strains. Each plate contained 25 ml of agar in single-vented 90-mm petri dishes. The Candida strains were grown overnight at 30°C with constant shaking in YEPD broth. The cultures were firstly diluted to 2 × 107 cells/ml in sterile deionized water. Four serial 1:10 dilutions of this first dilution were made, 4 μl of each dilution was spotted onto each plate, and the plates were incubated for 48 h at 30°C.
Induction of itraconazole resistance in vitro.
Seven itraconazole-resistant in vitro-generated derivatives of C. dubliniensis isolates CD36, CD570, and CD519-8 were included in this study. These were CD36-C, CD36-F, CD570-A, CD570-B, CD570-C, CD570-D, and CD519-A. Isolates CD36 (type strain C. dubliniensis NCPF 3949) and CD570 belong to C. dubliniensis genotype 1, as described by Gee et al. (10), and harbor the naturally occurring nonfunctional CdCDR1 gene described by Moran et al. (29), while isolate CD519-8 belongs to C. dubliniensis genotype 3 and harbors a functional CdCDR1 gene. A hundred colonies each of the itraconazole-susceptible C. dubliniensis isolates were exposed for 48 to 96 h on 90-mm petri dishes containing 25 ml of Emmons Sabouraud dextrose agar (SDA) medium supplemented with the following increasing concentrations of itraconazole: 0.06, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16, 32, and 64 μg/ml. For each of these concentrations, two successive sequential subcultures were carried out by using toothpicks to transfer a portion of each colony.
DNA isolation and Southern hybridization analysis.
Total genomic DNA of C. dubliniensis clinical isolates and derivatives was prepared from cells grown for 18 h in YEPD broth cultures, as described by Gallagher et al. (8).
Restriction endonuclease-digested DNA fragments were separated by electrophoresis and transferred to MagnaGraph nylon membranes (MSI, Westboro, Mass.), as described by Sullivan et al. (48). Hybridization reactions were carried out under high-stringency conditions with DNA probes labeled with [α-32P]dATP (Perkin Elmer, Life Sciences Inc., Boston, Mass.) by random primer labeling in a rotary hybridization oven (Hybaid, Middlesex, United Kingdom), as described by Sullivan et al. (49).
Yeast chromosomes were prepared for electrophoresis in agarose plugs by the method of Vazquez et al. (50) and separated by electrophoresis as described by Sullivan et al. (49).
PCR amplification and cloning of C. dubliniensis DNA sequences.
PCR amplification was performed in 100-μl reaction volumes containing 100 pmol each of a forward primer and a reverse primer, 10 mM deoxynucleoside triphosphates (2.5 mM each), the high-fidelity enzyme and buffers provided with the Expand PCR System (Roche Diagnostics Ltd.), and 100 ng of C. dubliniensis genomic DNA. PCRs were carried out in a DNA thermal cycler (Perkin-Elmer, Norwalk, Conn.).
For the amplification of CdERG11, the primer set ERG11F-ERG11R (with XhoI and XbaI restriction sites included at the respective 5′ ends; Table 1), which is a modification of the primer set used by Sanglard et al. (44), was used. Reactions were carried out with 30 cycles of denaturation for 1 min at 94°C, primer annealing for 1 min at 55°C, and extension for 1.5 min at 72°C; this was followed by a final incubation at 72°C for 10 min.
TABLE 1.
Nucleotide sequences of primers used to amplify CdERG11 and CdERG3
| Primer | Sequencea (5′-3′) | Restriction site | Nucleotide coordinatesb | Reference |
|---|---|---|---|---|
| ERG11F | CGCTCGAGCGCAATATGGCTATTGTTGAAACTGTC | XhoI | 1 to 21 | 44 |
| ERG11R | GCTCTAGAGCTTAAAACATACAAGTTTCTCTTTT | XbaI | 1587 to 1564 | 44 |
| ERG3F1 | CGCTCGAGCAACCATCCAGAATATTTG | XhoI | 453 to 471 | This study |
| ERG3R1 | GCTCTAGAGTGAATTGACCGTAATTG | XbaI | 986 to 969 | This study |
| NERG3F | CGCTCGAGCTCCAGTCGATGGATTTTTCC | XhoI | 771 to 790 | This study |
| NERG3R | GCTCTAGAGCCACCAGTAGATTCATTGATTT | XbaI | 607 to 587 | This study |
| CAE3F | CGAAGCTTATGGATATCGTACTAGAA | HindIII | 1 to 18 | This study |
| CDE3R | GCGGATCCTTGGAAATATTGAATGGGGG | BamHI | 1202 to 1183 | This study |
The restriction endonuclease recognition sites included in the primer sequences are underlined.
The nucleotide coordinates of the C. dubliniensis gene (where position +1 corresponds to the first base of the ATG translational start codon). The primer pair ERG11F-ERG11R was derived from the primer sequence used for the C. albicans gene by Sanglard et al. (44). The primer pair ERG3F1-ERG3R1 was based on the C. albicans ERG3 sequence (GenBank accession no. AF069752).
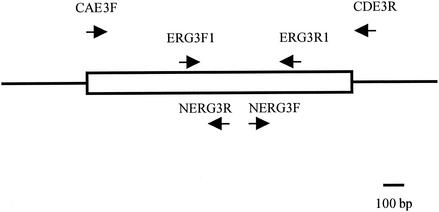
A set of primers (primers ERG3F1 and ERG3R1; Table 1 and Fig. 1) was designed on the basis of the sequence of the C. albicans ERG3 gene spanning a 530-bp region which is most conserved among S. cerevisiae, Candida glabrata, and C. albicans ERG3 sequences and was used in an amplification reaction with C. dubliniensis strain CD36 template DNA. Reactions were carried out with 30 cycles of denaturation for 1 min at 94°C, primer annealing for 1 min at 55°C, and extension for 30 s at 72°C; this was followed by a final incubation at 72°C for 10 min. The amplimer obtained was radiolabeled and used as a probe in Southern hybridization analysis of ClaI-digested CD36 genomic DNA and was found to hybridize to a single 5.5-kb fragment. A second primer set (NERG3F-NERG3R; Table 1 and Fig. 1) was designed on the basis of the sequence of the 530-bp region mentioned above to amplify its flanking sequences by inverse PCR. ClaI-digested CD36 DNA was ligated to itself, and PCR was carried out on religated DNA template with 30 cycles of denaturation for 1 min at 94°C, primer annealing for 1 min at 55°C, and extension for 5 min at 68°C, followed by a 10-min extension at 68°C.
FIG. 1.
Map of the CdERG3 region. The rectangular box represents the CdERG3 ORF. Arrows indicate the positions of the primer sequences used in this study.
On the basis of the sequences obtained by inverse PCR, a set of primers (primers CAE3F and CDE3R; Table 1 and Fig. 1) was designed to amplify the complete CdERG3 ORF. Following restriction enzyme digestion, the amplimers were cloned into pBluescript II KS(−) by conventional methods. These clones were used in sequencing reactions carried out by Lark Inc. (Saffron Walden, United Kingdom). Alignments of the nucleotide and amino acid sequences were carried out by using the CLUSTAL W software program (11). For heterologous expression of CdERG3 in S. cerevisiae, the ORFs cloned in pBluescript were subcloned into the XbaI-digested yeast expression vector pYES.
RNA extraction and Northern analysis.
RNA was extracted from C. dubliniensis cultures grown to the midexponential phase (optical density at 600 nm, approximately 0.6) in 50-ml volumes of YEPD broth at 37°C with shaking in an orbital incubator, as described by Moran et al. (30). Extractions were carried out by the glass bead disruption method described by Hube et al. (12). RNA electrophoresis, transfer, and hybridization were carried out as outlined by Moran et al. (30). Membranes were then exposed to BioMax MS film (Eastman Kodak Company, Rochester, N.Y.) for 24 to 72 h. For normalization purposes, all membranes were hybridized with a probe homologous to the C. dubliniensis gene encoding translation elongation factor 3 (CdTEF3). Hybridization levels were analyzed by scanning densitometry (GelWorks 1D Intermediate; Ultra-Violet Products Ltd., Cambridge, United Kingdom) and normalized with the CdTEF3 expression levels.
Measurement of glucose-induced R6G efflux.
Efflux of R6G was determined by a modification of the method of Maesaki et al. (23). Briefly, yeast cells were grown in YEPD broth at 37°C overnight. A total of 108 cells were transferred to 100 ml of fresh YEPD broth and incubated at 37°C for 4 to 5 h. The cells were harvested by centrifugation at 3,000 × g for 5 min. The pellets were washed twice in phosphate-buffered saline (PBS) and were then resuspended in 10 ml of PBS at a concentration of 108 CFU/ml and incubated at 37°C for 1 h in a shaking incubator. A final concentration of 10 μM R6G was then added to the cell suspension; and the mixture was incubated at 37°C in a shaking incubator for 30 min, after which 1-ml samples were withdrawn and centrifuged at 20,800 × g in 1.5-ml microcentrifuge tubes. The supernatants were collected, and the absorption was measured at 527 nm. To observe the effect of glucose, the remainders of the cell suspensions were divided equally and placed in two 50-ml Falcon tubes, one of which was supplemented with 10 μM glucose, while an equal volume of PBS was added to the other. Both suspensions were incubated at 37°C for a further 30 min. One-milliliter samples were withdrawn following incubation and were processed as described above. The concentration of R6G was determined by using a standard curve of R6G concentrations.
Sterol extraction and analysis. (i) Extraction method for qualitative spectrophotometric analysis.
For qualitative spectrophotometric analysis, total sterols were extracted from cells grown overnight in YEPD broth at 37°C (unless stated otherwise) by a modification of the method described by Arthington-Skaggs et al. (1). Briefly, cells from 1 ml of an overnight broth culture were harvested by centrifugation, and the pellet was resuspended in 0.5 ml of a solution containing 20% (wt/vol) KOH and 60% (vol/vol) ethanol. The suspension was incubated at 95°C for 1 h. Following incubation, 0.6 ml of hexane was added and the samples were vortexed for a few seconds before centrifugation at 20,800 × g for 2 min. Following centrifugation, the upper hexane layer containing the sterols was removed and put into fresh tubes. The samples were then diluted by the addition of 0.6 ml of hexane before their absorption between 240 and 330 nm was measured with a spectrophotometer (Genosys 2; ThermoSpectronic, AGB, Dublin, Republic of Ireland).
(ii) Extraction method for GC-MS.
For gas chromatography-mass spectrometry (GC-MS), nonsaponifiable sterols were extracted by a modification of the method described by Venkateswarlu et al. (51). Single colonies from the plate cultures were inoculated into 15 ml of YEPD broth and incubated overnight at 30°C and 200 rpm. Cells were recovered by centrifugation at 1,500 × g for 10 min and washed once in 20 ml of sterile water. Each cell pellet (approximately 400 mg [wet weight]) was resuspended in 3 ml of methanol and transferred to a stoppered glass test tube. Then, 2 ml of 60% (wt/vol) potassium hydroxide and 2 ml of 0.5% (wt/vol) pyrogallol dissolved in methanol were added to each tube, and the tubes were gently vortexed to mix the contents. The tubes were stoppered and placed in a water bath at 90°C for 2 h and then removed and allowed to cool to room temperature. Then, 10 ml of hexane was added to each tube, and the tube was vortexed vigorously for 60 s. Following vortexing, the phases were allowed to separate, the upper hexane layer was removed with a glass capillary pipette and placed in a clean glass test tube, and the hexane was evaporated off under nitrogen. Following silylation for 1 h at 60°C with N,O-bis(trimethylsilyl) trifluoroacetamide (50 μl) in 50 μl of toluene, the sterols were separated and identified by GC-MS (VD 12-250; VG Biotech, Manchester, United Kingdom).
Nucleotide sequence accession number.
The sequence of CdERG3 has been deposited in the EMBL nucleotide sequence database under accession no. AJ421248.
RESULTS
C. dubliniensis fluconazole and itraconazole susceptibility testing.
The susceptibilities of C. dubliniensis clinical isolates were determined in an attempt to determine the levels of itraconazole and fluconazole resistance. The fluconazole susceptibilities of 111 C. dubliniensis isolates from 106 separate patients were assessed by BMD. The vast majority of the isolates (105 of 111; 94.6%) exhibited fluconazole-susceptible phenotypes (MIC range, 0.125 to 4 μg/ml) when tested by BMD. The itraconazole susceptibilities of 58 of the 111 C. dubliniensis isolates from 56 separate patients were also assessed by BMD. The vast majority of these isolates (52 of 58; 89.6%) yielded itraconazole-susceptible phenotypes (MIC range, 0.03 to 0.125 μg/ml). Six isolates were susceptible dose dependent to itraconazole (MIC range, 0.25 to 0.5 μg/ml), but none of the 58 isolates tested were found to be resistant to itraconazole (MIC, >1 μg/ml).
Generation of itraconazole-resistant derivatives by in vitro exposure.
Previously, it was shown that stable fluconazole resistance in C. dubliniensis could be induced in vitro by serial subculture on agar plates containing increasing concentrations of drug (31). In order to determine whether stable itraconazole resistance could be induced in C. dubliniensis by exposure to the drug and to investigate the genetic basis of itraconazole resistance in C. dubliniensis, 100 colonies each of three separate itraconazole-susceptible C. dubliniensis isolates (CD36, CD570, and CD519-8) were exposed to increasing concentrations of itraconazole by subculture on Emmons SDA containing itraconazole. C. dubliniensis isolates CD36 and CD570 were chosen because they harbored a naturally occurring nonfunctional version of the CdCDR1 gene due to the presence of a nonsense mutation described by Moran et al. (29), while the third isolate, CD519-8, was chosen because it harbored a functional CdCDR1 gene. Isolates CD36 and CD570 belong to C. dubliniensis genotype 1, while isolate CD519-8 belongs to genotype 3 (10). All colonies grew well on initial culture on Emmons SDA containing 0.06 μg of itraconazole per ml. Following an additional subculture on Emmons SDA containing 0.06 μg of itraconazole per ml and successive sequential subcultures on Emmons SDA containing increasing concentrations of itraconazole (0.125, 0.25, 0.5, 1, 2, 4, 8, 16, 32, and 64 μg/ml), 4 derivatives of CD36, 10 derivatives of CD570, and 4 derivatives of CD519-8 that could grow on agar containing 64 μg of itraconazole per ml were recovered. After subculture on drug-free PDA medium, all derivatives were assessed for their susceptibilities to itraconazole and fluconazole by BMD. The itraconazole and fluconazole MICs for 2 of the 4 CD36 derivatives (CD36-C and CD36-F), 4 of the 10 CD570 derivatives (CD570-A, CD570-B, CD570-C, and CD570-D), and 1 of the 4 CD519-8 derivatives (CD519-8A) were significantly higher than those for their respective parental isolates (Table 2). In addition, the ketoconazole and amphotericin B MICs for all seven derivatives were higher than those for their respective parental isolates (Table 2). The stabilities of the itraconazole and fluconazole resistance phenotypes of these specific derivatives were assessed by monitoring the MICs for the strains following 15 successive subcultures on drug-free PDA and following prolonged storage and subculture over a period of 6 to 36 months. For each of the seven derivatives examined, no change in MIC was detected by BMD following subculture or storage. The remaining derivatives, which did not show stable resistance to itraconazole, were considered to exhibit transient resistance to the drug.
TABLE 2.
Susceptibilities of parental C. dubliniensis isolates and itraconazole-resistant derivatives to antifungal agents and inhibitors
| Strain name | Parent or derivative | MIC (μg/ml)a
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ITRA | FLU | KETO | AMB | BEN | CER | CRY | CYH | 5FC | NQO | PHE | R6G | ||
| CD36 | Parental isolate | 0.03 | 0.5 | 0.03 | 0.015 | 32 | 1 | 0.06 | 32 | 0.125 | 0.25 | 2 | 0.5 |
| CD36-C | CD36 derivative | 64 | 256 | 2 | 0.125 | 32 | 0.5 | 0.03 | 32 | 0.125 | 0.06 | 2 | 0.25 |
| CD36-F | CD36 derivative | 64 | 256 | 2 | 0.125 | 32 | 0.5 | 0.03 | 32 | 0.125 | 0.06 | 2 | 0.25 |
| CD570 | Parental isolate | 0.03 | 0.5 | 0.03 | 0.03 | 32 | 1 | 0.06 | 32 | 0.125 | 0.125 | 4 | 0.5 |
| CD570-A | CD570 derivative | 64 | 256 | 2 | 0.025 | 32 | 1 | 0.03 | 32 | 0.125 | 0.125 | 2 | 0.25 |
| CD570-B | CD570 derivative | 64 | 256 | 4 | 0.06 | 32 | 0.5 | 0.03 | 16 | 0.125 | 0.03 | 2 | 0.5 |
| CD570-C | CD570 derivative | 64 | 256 | 2 | 0.06 | 16 | 0.5 | 0.03 | 16 | 0.125 | 0.03 | 1 | 0.25 |
| CD570-D | CD570 derivative | 32 | 256 | 1 | 0.06 | 16 | 0.5 | 0.03 | 32 | 0.125 | 0.03 | 1 | 0.25 |
| CD519-8 | Parental isolate | 0.06 | 0.5 | 0.03 | 0.03 | 64 | 0.5 | 0.5 | 128 | 0.125 | 0.06 | 4 | 1 |
| CD519-8A | CD519-8 derivative | 32 | 128 | 4 | 0.25 | 64 | 0.5 | 0.25 | 128 | 0.125 | 0.03 | 4 | 0.5 |
MICs were determined by the BMD method. Abbreviations: ITRA, itraconazole; FLU, fluconazole; KETO, ketoconazole; AMB, amphotericin B; BEN, benomyl; CER, cerulenin; CRY, crystal violet; CYH, cycloheximide; 5FC, flucytosine; NQO, 4-nitroquinoline-N-oxide; PHE, 1,10-phenanthroline; R6G, rhodamine 6G.
Because resistance to azole drugs is often associated with resistance to inhibitors, additional susceptibility tests with a range of drugs and inhibitors were performed following subculture on drug-free PDA and following prolonged storage to further characterize the phenotypes of the derivatives. However, the MICs of the inhibitors cycloheximide, benomyl, flucytosine, 1,10-phenanthroline, 4-nitroquinoline-N-oxide, cerulenin, crystal violet, and R6G for the derivatives were found to be either the same or two to fourfold lower than the corresponding MICs for their respective parental isolates (Table 2).
Phenotypic and molecular analyses of itraconazole-resistant derivatives.
In order to ensure that the derivatives were clonal with their respective parental isolates and not exogenous itraconazole-resistant contaminants, the derivatives and their parental isolates were thoroughly examined by a variety of phenotypic and molecular tests. When cultured on itraconazole-free PDA, the in vitro-generated itraconazole-resistant derivatives exhibited the same smooth, white colony morphology as their parental isolates. All the derivatives yielded C. dubliniensis-specific substrate assimilation profiles with the API ID32C yeast identification system, and all yielded C. dubliniensis-specific amplimers when tested by PCR with the C. dubliniensis-specific primers described by Donnelly et al. (3) (data not shown). Hybridization analysis of EcoRI-digested total cellular DNA from strains CD36, CD570, and CD519-8 and their respective derivatives was carried out with C. dubliniensis-specific fingerprinting probe Cd25, which can be used to discriminate between C. dubliniensis isolates (10, 14). Each of the three itraconazole-susceptible parental isolates, CD36, CD570, and CD519-8, yielded distinct Cd25-generated fingerprint profiles (data not shown). The itraconazole-resistant derivatives of CD36 (CD36-C and CD36-F), CD570 (CD570-C and CD570-D), and CD519-8 (CD519-8A) yielded fingerprint profiles identical to those of their respective parental isolates. Minor band variations in the hybridization patterns of both derivative CD570-A and derivative CD570-B compared to that of their parental isolate, CD570, were detected, suggesting that these derivatives had undergone microevolution in vitro (data not shown). Karyotype analysis by pulsed-field gel electrophoresis confirmed the relatedness between the derivatives and their respective parental isolates (data not shown). All of these results indicated that the itraconazole-resistant derivatives were clonally related to their respective parental isolates.
CdERG11 expression in azole-resistant derivatives.
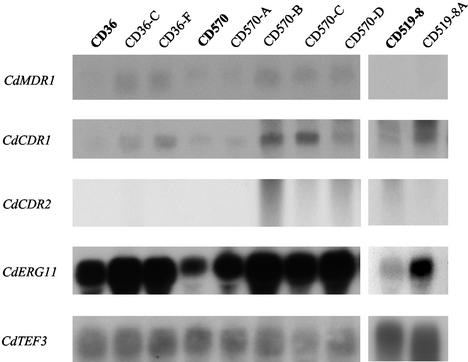
In order to determine if changes in the expression of CdERG11 were involved in the azole resistance phenotype observed in the seven azole-resistant derivatives of the present study, the mRNA levels of the CdERG11 gene were analyzed by Northern blot analysis. CdERG11 mRNA was detected in the three parental strains, CD36, CD570, and CD519-8, and their azole-resistant derivatives, CD36-C, CD36-F, CD570-A, CD570-B, CD570-C, CD570-D, and CD519-8A. However, CdERG11 was expressed four to eight times as much in all seven azole-resistant derivatives as in their respective parental isolates (Fig. 2).
FIG. 2.
Northern blots of total RNA from azole-resistant derivatives CD36-C, CD36-F, CD570-A, CD570-B, CD570-C, CD570-D, and CD519-8A and their azole-susceptible parental isolates, CD36, CD570, and CD519-8. Total RNA was extracted from exponentially growing cultures, and 15 μg was electrophoresed on a denaturing agarose gel. Following transfer to a nylon membrane, the blots were sequentially probed with radiolabeled DNA probes homologous to CdMDR1, CdCDR1, CdCDR2, and CdERG11 and the constitutively expressed CdTEF3 gene. Expression of CdTEF3 was used as an internal control for RNA loading.
CdERG11 sequence analysis of azole-resistant C. dubliniensis derivatives.
Mutations affecting the affinity of Erg11p for azole antifungal drugs that render yeast cells resistant to these drugs have been well documented as a drug resistance mechanism (5, 17, 20, 24, 44). The possibility that such a mechanism contributed to the azole resistance phenotype in some or all of the azole-resistant C. dubliniensis derivatives was therefore investigated. CdERG11 genes from C. dubliniensis isolates CD36, CD570, and CD519-8 and their derivatives, CD36-C, CD36-F, CD570-A, CD570-B, CD570-C, CD570-D, and CD519-8A, were isolated by PCR with primer pair ERG11F-ERG11R (Table 1) and genomic DNA as the template. In each case, fragments of the expected length (1.6 kb) were obtained. Both strands of the coding region of the CdERG11 genes from each derivative were sequenced and compared to the corresponding sequences from their respective parental isolates. To reduce PCR error, a proofreading polymerase (Expand PCR system; Roche Diagnostics Ltd.) was used in each case to obtain amplimers. Furthermore, amplimers from five independent PCRs with template DNA from each parental isolate and derivative were sequenced.
There was no amino acid residue difference between the deduced amino acid sequence of the CdERG11 gene from parental isolate CD36 and the sequence recently published by Perea et al. (36) (GenBank accession no. AY034876). However, there were two heterozygous polymorphisms (R210I and D225Y) in the CdErg11p sequence from parental isolate CD570, and there was one heterozygous polymorphism (V402G) and two homozygous polymorphisms (I188V and R499K) in the amino acid sequence from parental isolate CD519-8. Since strains CD36, CD570, and CD519-8 are susceptible to itraconazole and other azole drugs, the sequence differences found in these strains obviously do not contribute to azole resistance. In each case, the CdERG11 sequences of the itraconazole-resistant derivatives were identical to the corresponding sequences of their respective parental isolates. Therefore, mutation of the CdERG11 gene could not have been responsible for the resistance phenotype observed in the derivatives.
Expression of multidrug resistance genes.
Since overexpression of genes encoding efflux pumps have been implicated in azole resistance in C. albicans and C. dubliniensis, we investigated if the azole resistance of the in vitro-generated derivatives of C. dubliniensis was associated with increased levels of expression of the CdCDR1, CdCDR2, or CdMDR1 gene using Northern blot analysis. For analysis of CdCDR1 and CdCDR2 expression, the cloned 5′ ends of these genes were used as probes, as described by Moran et al. (30). For analysis of CdMDR1 expression, a PCR amplimer spanning the entire ORF was used as a probe. None of the itraconazole-resistant derivatives exhibited significantly elevated levels of CdMDR1 mRNA compared to the levels for their respective azole-susceptible parental isolates (Fig. 2). However, elevated levels of CdCDR1 expression were detected in six of the seven azole-resistant derivatives (Fig. 2). There was no significant elevation in the level of CdCDR1 expression in azole-resistant derivative CD570-A. Large increases in the levels of CdCDR1 expression were observed in derivatives CD570-B (9-fold) and CD570-C (12-fold). Smaller increases were observed in derivatives CD570-D (4-fold), CD36-C (3-fold), CD36-F (5-fold), and CD519-A (2.5-fold). There was no significant elevation of CdCDR2 mRNA levels in any of the azole-resistant derivatives (Fig. 2).
Uptake and glucose-mediated efflux of R6G.
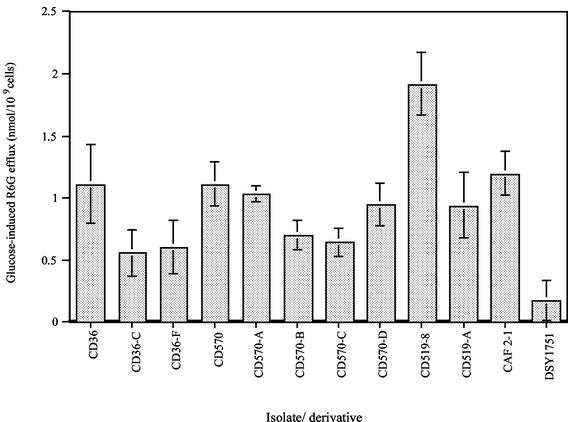
To further investigate the role of energy-dependent efflux mechanisms in the azole-resistant derivatives, the uptake and glucose-mediated efflux of R6G were measured. R6G is a fluorescent compound known to be a substrate of ABC transporters encoded by genes such as CDR1. The method used in this study, as described by Maesaki et al. (23), directly assessed the efflux of R6G from energy-starved cells by measuring the extracellular concentrations of R6G following the addition of glucose. In the absence of glucose, the extracellular concentrations of R6G were roughly similar for the azole-susceptible parental isolates and their respective azole-resistant derivatives (data not shown). In contrast, in the presence of glucose there was an increase in the level of R6G efflux by azole-susceptible parental isolate CD36 (which harbors a defective CdCDR1 gene) compared to those by its azole-resistant derivatives, CD36-C and CD36-F (Fig. 3). Likewise, a notably increased level of R6G efflux was observed for parental isolate CD519-8 (which harbors a functional CdCDR1 gene) compared with that observed for its derivative, CD519-8A. This was also the case for azole-susceptible parental isolate CD570, which showed an increased level R6G efflux compared with those for three of its four azole-resistant derivatives, CD570-B, CD570-C, and CD570-D. However, extracellular R6G levels for azole-resistant derivative CD570-A in the presence of glucose did not show a significant difference compared to those for its azole-susceptible parental isolate, CD570, suggesting no difference in glucose-mediated drug efflux between the parental isolate and its derivative. These results suggest that R6G efflux from the cells mediated by ABC drug transporter extrusion pumps like CDR1 or CDR2 was more efficient in the azole-susceptible parental isolates than in six of their seven azole-resistant derivatives and is therefore unlikely to be involved in resistance. A similar reduction of active transport efficiency has previously been associated with changes in the membranes of erg mutants of S. cerevisiae (15, 32, 47). This led us to investigate whether erg mutations may be present in the itraconazole-resistant derivatives described here, as resistance to azole drugs in Candida species can be caused by alterations in the ergosterol biosynthesis pathway. In particular, alterations resulting from mutations in the ERG3 gene encoding the enzyme sterol C5,6-desaturase have been described in C. albicans and S. cerevisiae (16, 18, 19, 28). In the present study, for comparative purposes, R6G efflux was measured in a C. albicans strain with an altered ergosterol pathway (DSY1751, an erg3 disruptant) (D. Sanglard, unpublished data) and in a wild-type C. albicans strain (CAF 2-1) (6). In glucose-starved cells, the extracellular concentration of R6G was similar for the two strains. However, in the presence of glucose there was a marked increase in the level of R6G efflux in the wild-type strain compared to that in the Δerg3 strain (Fig. 3). The latter finding was similar to the findings for six of the seven C. dubliniensis derivatives, as described above, and suggested that these derivatives could have altered membrane properties.
FIG. 3.
Glucose-induced R6G efflux from C. dubliniensis azole-resistant derivatives CD36-C, CD36-F, CD570-A, CD570-B, CD570-C, CD570-D, and CD519-8A and the azole-susceptible parental isolates CD36, CD570, and CD519-8. For comparative purposes, R6G efflux data for wild-type (CAF2-1) and Δerg3 (DSY1751) C. albicans strains were included. Cells were incubated with 10 μM R6G at 37°C. After 30 min of incubation in glucose-free PBS, glucose was added to a final concentration of 10 μM. After a further 30 min of incubation, with or without glucose, at 37°C, the cells were removed by centrifugation and the absorption of the resulting supernatant was measured at 527 nm. The concentration of R6G was calculated by using a standard curve of the R6G concentration. R6G efflux was calculated by subtracting the extracellular R6G concentration in the absence of glucose from the extracellular R6G concentration in the presence of glucose. Each bar indicates the standard error of the mean of at least two sets of experiments.
Growth in the presence of inhibitors.
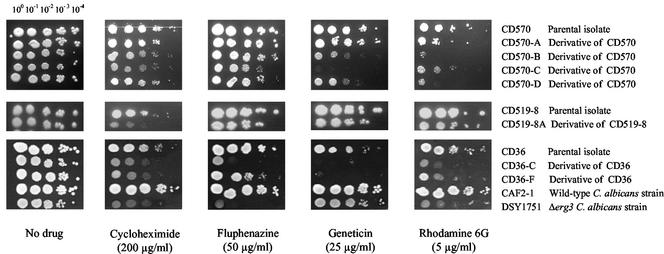
It has previously been shown that yeast cells with altered membrane properties are more susceptible to certain inhibitors. Therefore, the azole-resistant derivatives described above and their respective parental isolates were tested on agar medium for their susceptibilities to a range of inhibitors. C. albicans Δerg3 strain DSY1751 and wild-type C. albicans strain CAF2-1 were also tested for comparison. All the derivatives except CD570-A showed increased susceptibilities to fluphenazine, cycloheximide, and Geneticin (Fig. 4). All the derivatives also exhibited increased susceptibilities to R6G. C. albicans Δerg3 strain DSY1751 also showed increased susceptibilities to these inhibitors compared to those of wild-type C. albicans strain CAF2-1. All of these results strongly suggest that the azole-resistant C. dubliniensis derivatives have altered membrane properties that result in increased susceptibilities to inhibitors that make their phenotypes comparable to the phenotype observed for C. albicans DSY1751 due to the inactivation of the ERG3 gene.
FIG. 4.
Susceptibilities of azole-resistant derivatives CD570-A, CD570-B, CD570-C CD570-D, CD519-8A, CD36-C, and CD36-F and their azole-susceptible parental isolates, CD570, CD519-8, and CD36, to inhibitors. A wild-type C. albicans strain (CAF2-1) and a C. albicans Δerg3 strain (DSY1751) were included for comparison of phenotypes. Each strain was grown to the exponential growth phase to a density of 2 × 107 cells/ml, and 4 μl was spotted in a dilution series on YEPD agar plates containing fixed concentrations of inhibitors, as indicated. The plates were incubated for 48 h at 30°C.
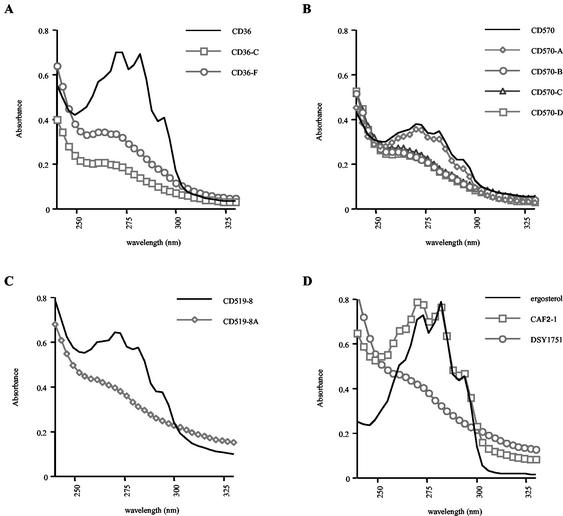
Analysis of sterol contents.
To investigate whether the sterol contents of the azole-resistant C. dubliniensis derivatives were different from those of their respective parental isolates and to determine whether alterations in the ergosterol biosynthetic pathway were responsible for azole resistance, the nonsaponifiable sterol contents of the derivatives and their parental isolates were extracted and examined by UV spectrophotometry. The UV absorption spectra of the wild-type sterol preparations from parental isolates CD36, CD570, and CD519-8 had peaks at 261, 273, 282, and 294 nm, which are typical of ergosterol (Fig. 5). These peaks were absent in the spectra of azole-resistant derivatives CD36-C and CD36-F (Fig. 5A); CD570-B, CD570-C, and CD570-D (Fig. 5B); and CD519-8A (Fig. 5C). The absence of the peaks indicated the absence of ergosterol in these derivatives. However, the UV absorption spectrum of azole-resistant derivative CD570-A exhibited the peaks characteristic of ergosterol (Fig. 5B). The UV absorption spectra of C. albicans Δerg3 strain DSY1751 and wild-type strain CAF2-1 were also examined for comparison. As expected, the spectrum of CAF2-1 exhibited the peaks characteristic of ergosterol, while the spectrum of DSY1751 sterols lacked these peaks (Fig. 5D) and was reminiscent of the profiles of the C. dubliniensis derivatives described above.
FIG. 5.
UV spectrophotometric profiles of nonsaponifiable sterols from C. dubliniensis azole-resistant derivatives and their azole-susceptible parents. (A) CD36 and its azole-resistant derivatives CD36-C and CD36-F; (B) CD570 and its azole-resistant derivatives CD570-A, CD570-B, CD570-C, and CD570-D; (C) CD519-8 and its azole-resistant derivative CD519-8A; (D) UV spectrophotometric profiles for ergosterol and nonsaponifiable sterols from C. albicans wild-type (CAF2-1) and Δerg3 (DSY1751) strains. Sterols were extracted from 5 × 108 cells grown overnight in YEPD broth, as described in Materials and Methods.
The sterols present in all seven derivatives were further analyzed by GC-MS, which allowed their separation and identification. The results of the GC-MS analysis are presented in Table 3. As expected, all of the derivatives except CD570-A lacked ergosterol and mainly accumulated ergosta-7,22-dienol, while the parental isolates accumulated ergosterol (Table 3). Although derivative CD570-A accumulated ergosterol, it accumulated noticeably less ergosta-tetraenol and more unknown sterols than its parental isolate, CD570 (Table 3).
TABLE 3.
Sterols accumulated by C. dubliniensis azole-resistant derivatives and their azole-susceptible parental isolates
| Straina | Accumulated sterolsb |
|---|---|
| CD36 | 9.8% ergosta-tetraenol |
| 64.6% ergosterol | |
| 6.8% fecosterol | |
| 8.4% obtusifoliol | |
| 10.2% unknown sterols | |
| CD36-C | 3.3% 7,22 isomer |
| 73.2% ergosta 7,22 dienol | |
| 9.3% episterol | |
| 13.9% unknown sterols | |
| CD36-F | 4.8% 7,22 isomer |
| 76.4% ergosta 7,22 dienol | |
| 7.0% episterol | |
| 1.3% lanosterol | |
| 10.2% unknown sterols | |
| CD519-8 | 16.8% ergosta-tetraenol |
| 55.8% ergosterol | |
| 6.7% fecosterol | |
| 6.0% episterol | |
| 1.8% obtusifoliol | |
| 12.8% unknown sterols | |
| CD519-8A | 8% ergostadienol |
| 62.4% ergosta 7,22-dienol | |
| 9.2% ergostadienol | |
| 4.4% isomer of ergosterol | |
| 12.5% cholestadienol | |
| 1.6% ergosterol | |
| 1.9% unknown sterols | |
| CD570 | 20% ergosta-tetraenol |
| 60% ergosterol | |
| 4.2% fecosterol | |
| 5.8% obtusifoliol | |
| 9.9% unknown sterols | |
| CD570-A | 11.6% ergosta-tetraenol |
| 55.9% ergosterol | |
| 4.1% fecosterol | |
| 6.2% obtusifoliol | |
| 22.2% unknown sterols | |
| CD570-B | 7.8% 7,22 isomer |
| 61.1% ergosta-7,22-dienol | |
| 2.7% 7,22 isomer | |
| 1.4% fecosterol | |
| 7.3% episterol | |
| 1.9% ergosta-7-enol | |
| 0.8% lanosterol | |
| 16.8% unknown sterols | |
| CD570-C | 11.0% 7,22 isomer |
| 57.0% ergosta-7,22-dienol | |
| 9.4% episterol | |
| 22.5% unknown sterols | |
| CD570-D | 7.4% 7,22 isomer |
| 58.0% ergosta-7,22-dienol | |
| 5.4% 7,22 isomer | |
| 11.8% episterol | |
| 17.4% unknown sterols |
The parental isolates are indicated by boldface type.
Sterols were extracted from cells grown overnight in YEPD broth at 30°C and analyzed by GC-MS, as described in Materials and Methods.
Cloning and sequencing of the C. dubliniensis ERG3 gene.
Since six of seven azole-resistant derivatives investigated had altered membrane properties, lacked ergosterol, and accumulated mainly ergosta-7,22-dienol (a characteristic for defects in ERG3), we hypothesized that they harbored a mutated sterol C5,6-desaturase (Erg3p). The sterol C5,6-desaturase is an enzyme involved in ergosterol biosynthesis that is thought to be located in the endoplasmic reticulum, where it catalyzes the introduction of a double bond between C-5 and C-6 in the B ring of the developing sterol molecule. In order to test this hypothesis, it was necessary to clone and sequence the gene encoding sterol C5,6-desaturase in C. dubliniensis. Initially, a pair of oligonucleotide primers complementary to the C. albicans ERG3 gene sequence, termed ERG3F1 and ERG3R1 (Table 1; Fig. 1), and a proofreading polymerase (Expand PCR system; Roche Diagnostics Ltd.) were used to amplify a 530-bp internal region of the gene with C. dubliniensis CD36 genomic DNA as the template. The 530-bp amplified product obtained was sequenced, and a homology search was carried out with the BLAST computer program. The homology search indicated that the C. dubliniensis sequence is highly homologous to the C. albicans ERG3 gene. In order to clone the remainder of the ERG3 gene, an inverse PCR strategy with the primer pair NERG3F-NERG3R was used (Table 1; Fig. 1). This yielded a fragment of 5.5 kb, both strands of which were sequenced. The reconstructed sequence contained one significant ORF of 1,161 bp. This ORF, termed CdERG3, has the capacity to encode a protein of 387 amino acids with a predicted molecular mass of 45.6 kDa and a pKi of 6.61. The CdERG3 sequence has been deposited in the EMBL nucleotide sequence database (accession no. AJ421248). Alignment of the CdERG3 gene and the C. albicans ERG3 gene by use of the CLUSTAL W sequence alignment computer program demonstrated that the genes are highly similar, being 91% identical at the nucleotide sequence level (data not shown). The predicted amino acid sequence of CdErg3p shows a high degree of amino acid identity (93.3%) to that of CaErg3p (Fig. 6). The predicted CdErg3p contains four histidine-rich motifs (HX3H, HX2HH, HX2H, and HX2HH), three of which are conserved in all known sequences of sterol C5,6-desaturases (Fig. 6). These histidine box domains are thought to contain the active site of the enzyme and are putative iron-binding domains in C. albicans (28).
FIG. 6.
Alignment of deduced amino acid sequences of sterol C5,6-desaturase in C. dubliniensis (EMBL accession no. AJ421248 [this study]) and C. albicans (GenBank accession no. AF069752 [28]) generated with the CLUSTAL W sequence alignment program (11). Asterisks indicate identical residues, colons indicate conservative substitutions, and dots indicate semiconservative substitutions. Four histidine-rich putative iron-binding domains believed to play a role in sterol C5,6-desaturase activity are highlighted in black. Amino acid mutations present in the C. dubliniensis azole-resistant derivatives are designated above the array. Two derivatives, CD570-B and CD519-8A, harbor frameshift mutations introducing stop codons at position 147 and 194, respectively (data not shown).
Sequencing of CdERG3 genes of azole-resistant derivatives.
The primer pair CAE3F-CDE3R (Table 1; Fig. 1) was used to amplify the CdERG3 genes from the azole-susceptible parents and their azole-resistant derivatives by using a proofreading polymerase (Expand PCR system; Roche Diagnostics Ltd.). In each case, an amplimer of 1.1 kb was obtained. All amplimers were cloned into pBluescript and sequenced. There were no differences between the deduced amino acid sequence of parental isolate CD570 and the corresponding amino acid sequence of CD36. There were two amino acid residue differences (Q47H and V119I) between the deduced amino acid sequence obtained from parental isolate CD519-8 and the sequence obtained from C. dubliniensis type strain CD36 described above. Analysis of the sequences obtained for the azole-resistant derivatives revealed that all seven azole-resistant derivatives had alterations in the CdERG3 gene which conferred amino acid changes. Point mutations were found in derivatives CD36-C, CD36-F, CD570-A, CD570-C, CD570-D, and CD519-8A. Derivatives CD570-B and CD519-8A were found to harbor deletions of 2 bp which would introduce frame shifts during translation and therefore result in the synthesis of truncated proteins (Fig. 6).
Derivative CD36-C harbored two mutations, one of which (H318N) is a substitution of a histidine residue in one of the conserved histidine-rich domains which have been shown to be essential for the desaturase activity of the sterol C5,6-desaturase enzyme (39). The point mutations P272L (in CD36-C), Q160K (in CD36-F), H269N (in CD570-A), and Q327K (in CD570-C and CD570-D) affect amino acid residues conserved in fungal and mammalian sterol C5,6-desaturases (data not shown). As well as the frameshift mutation, derivative CD519-8A harbored four heterozygous point mutations in the N-terminal region of CdErg3p, the least conserved region of the protein (Fig. 6). While two independent derivatives of CD570, CD570-C and CD570-D, shared the same Q327K mutation, all other mutations identified were specific to each derivative. In order to determine whether the mutations identified in the derivatives were homozygous or heterozygous or whether they may have been artifacts of PCR amplification, five independent clones were sequenced for each derivative. In addition, when possible (when mutations introduced or removed a restriction endonuclease site) restriction fragment length polymorphism (RFLP) analysis was carried out. Sequence analysis combined with RFLP analysis showed that all mutations identified in the derivatives, with the exception of four heterozygous mutations present in derivative CD519-8A, were homozygous. In this derivative, two distinct alleles were identified, and both of these shared the frameshift mutation.
Expression of CdERG3 alleles in S. cerevisiae.
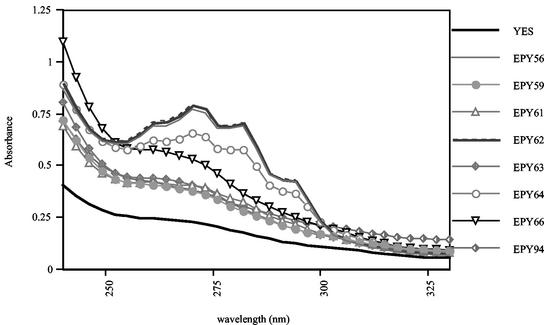
In order to determine if the mutations identified in the azole-resistant derivatives altered the function of the sterol C5,6-desaturase enzyme, the wild-type and mutant alleles of CdERG3 were expressed in a S. cerevisiae Δerg3 strain (ATCC 4002667). First, the CdERG3 alleles were subcloned in the yeast shuttle expression vector pYES and transformed into ATCC 4002667. In the pYES vector, the alleles were placed under the control of the galactose-inducible GAL1 promoter. To verify that the expression system worked, total RNA was extracted from cells growing exponentially in galactose-containing medium, and Northern blot analysis was carried out by using the 530-bp CdERG3 amplimer described above as a probe. All transformants except the control showed high levels of expression of CdERG3 mRNA (data not shown).
Total sterols were extracted from all transformants and were analyzed by spectrophotometry as described above. As expected, the UV absorption profile of the S. cerevisiae Δerg3 strain transformed with control plasmid pYES lacked the peaks characteristic of ergosterol (Fig. 7). The absorption spectra of the transformants expressing wild-type CdERG3 from C. dubliniensis parental isolate CD36, CD570, and CD519-8 alleles showed peaks at 261, 273, 282, and 294 nm, which are typical of ergosterol, indicating that these alleles were able to complement the sterol C5,6-desaturase function lacking in S. cerevisiae Δerg3 since the presence of ergosterol could be detected. The sterol profiles of the transformants expressing the other mutant alleles lacked the ergosterol peaks, which indicated that these mutant alleles are not able to complement the sterol C5,6-desaturase function since ergosterol was not produced by these transformants. The sterol profile of the transformant expressing the CdERG3 allele of CD570-A (EPY64) showed peaks that were typical of those for ergosterol and that were lower than the peaks obtained with the wild-type alleles.
FIG. 7.
UV spectrophotometric profiles of nonsaponifiable sterols extracted from the S. cerevisiae Δerg3 strain expressing CdERG3 alleles from parental isolates CD36 (EPY56) and CD570 (EPY62); derivatives CD36-C (EPY59), CD36-F (EPY61) CD570-A (EPY64), CD570-B (EPY63), CD570-C (EPY66), and CD519-A (EPY94); and a control transformed with the expression vector pYES (YES). Total sterols were extracted from 5 × 108 cells grown overnight in galactose-containing defined broth medium as described in Materials and Methods.
DISCUSSION
C. dubliniensis is now well recognized as a significant human pathogen and is primarily associated with oral carriage and infection in HIV-infected individuals. Previous studies have shown that stable resistance to fluconazole can readily be induced in vitro in this species (10, 30, 31). Moreover, fluconazole-resistant C. dubliniensis isolates have been recovered from HIV-infected and AIDS patients treated with fluconazole for prolonged periods of time and have been shown to replace susceptible strains (36, 41). Recently, Perea and colleagues (36) showed that, as is the case in C. albicans, resistance to fluconazole in C. dubliniensis clinical isolates was associated with combinations of different molecular mechanisms such as upregulation of the multidrug resistance genes CdCDR1 and CdMDR1, upregulation of CdERG11, and the presence of mutations in CdErg11p. In C. albicans, almost all isolates with reduced susceptibilities to azoles analyzed to date exhibit increased levels of CDR1 expression (35). In contrast, increased levels of CdCDR1 expression have been reported in only approximately half of the C. dubliniensis fluconazole-resistant isolates and derivatives analyzed to date, while increased levels of CdMDR1 expression were reported in all C. dubliniensis isolates and derivatives exhibiting high-level resistance to fluconazole (30, 36). In a study that investigated the reasons for the differential regulation of CDR1 expression in C. albicans and C. dubliniensis, Moran et al. (29) cloned and sequenced the CDR1 gene from C. dubliniensis. That study showed that although CdCDR1 is involved in reduced susceptibility to itraconazole and ketoconazole, it is not essential for fluconazole resistance in C. dubliniensis (29). Furthermore, 14 of 40 (35%) C. dubliniensis isolates studied were found to harbor nonfunctional alleles of CdCDR1 encoding a truncated protein (29). Because of the relatively high prevalence of C. dubliniensis isolates harboring nonfunctional CdCDR1 alleles in individuals receiving azole treatment, it was thought to be important to determine if the nonfunctional truncated CdCdr1p expressed in these isolates could affect the development of resistance to itraconazole, a triazole drug that is a substrate of CdCdr1p but not of CdMdr1p (29). Thus, the aims of the present study were to determine the incidence of itraconazole resistance in clinical isolates of C. dubliniensis and to investigate the mechanisms of itraconazole resistance in C. dubliniensis in vitro by using strains harboring either the functional or the nonfunctional alleles of CdCDR1.
In this study, 6 of 58 (10.4%) clinical isolates exhibited reduced susceptibility (MICs, 0.25 to 0.5 μg/ml) to itraconazole. Since the patients from whom these isolates were originally recovered had not been treated with itraconazole, it is likely that the reduced susceptibility observed was a result of fluconazole treatment. While these isolates are not considered clinically resistant according to the breakpoints outlined by the National Committee for Clinical Laboratory Standards (33), they are susceptible dose dependent and the MICs for the isolates are significantly higher than those for susceptible isolates, thus indicating that resistance to itraconazole in C. dubliniensis may be developing clinically.
When investigating molecular mechanisms of resistance, it is preferable to use matched pairs of isolates, i.e., a drug-susceptible derivative and a drug-resistant derivative of the same strain. In the absence of matched pairs of clinical isolates, molecular analysis of parental isolates and in vitro-generated itraconazole-resistant derivatives was used to investigate the mechanisms of itraconazole resistance in C. dubliniensis. In order to induce resistance to itraconazole in C. dubliniensis, 100 colonies each of three itraconazole-susceptible clinical isolates were sequentially exposed to increasing concentrations of the drug in agar medium up to a concentration of 64 μg/ml. The isolates selected for this study included CD36 and CD570, both of which are genotype 1 isolates and both of which harbor a nonfunctional CdCDR1 gene, and genotype 3 isolate CD519-8, which was chosen because it harbored a functional CdCDR1 gene. Exposure to itraconazole resulted in the recovery of seven stable itraconazole-resistant derivatives which exhibited significantly reduced susceptibilities to itraconazole (MIC range, 32 to 64 μg/ml). These derivatives were found to exhibit cross-resistance to fluconazole (MIC range 128 to 264 μg/ml) and ketoconazole (MIC range, 1 to 4 μg/ml). Six of the derivatives exhibited reduced susceptibilities to amphotericin B (MIC range, 0.06 to 0.25 μg/ml). In contrast, no cross-resistance to inhibitors or other antifungal drugs was observed (Table 2). Indeed, the derivatives were slightly more susceptible to these agents than their respective parental isolates.
In order to identify the molecular mechanisms responsible for the derivatives' azole-resistant phenotype, the most common mechanisms of azole resistance in Candida species were investigated. All derivatives exhibited an increased level of CdERG11 expression of at least fourfold. While overexpression of ERG11 has been described in several different azole-resistant clinical isolates of Candida species (36, 46, 53), this has always been accompanied by other modifications associated with azole resistance and the effects of overexpression of ERG11 have not been extensively evaluated. Thus, it was thought to be unlikely that upregulation of CdERG11 alone was responsible for the azole-resistant phenotypes of the derivatives.
Changes in the amino acid sequence of CdErg11p affecting the affinity of this enzyme for azole antifungal agents is a well-described mechanism of azole resistance. However, comparative analysis of the nucleotide sequences of CdERG11 genes from the derivatives and their respective parental isolates did not reveal specific mutations in the derivatives that could be associated with azole resistance.
Because the derivatives exhibited pan-azole resistance but did not exhibit cross-resistance to inhibitors which are normally substrates of multidrug resistance proteins, the involvement of these proteins in resistance seemed unlikely. Nevertheless Northern blot analysis was carried out to investigate levels of expression of the CdCDR and CdMDR1 genes. As expected, overexpression of CdMDR1 did not appear to be involved in the itraconazole-resistant phenotype, as itraconazole is not a substrate of CdMdr1p. There was, however, a noticeable increase (from 2.5- to 12-fold) in the level of expression of CdCDR1 in derivatives CD36-C, CD36-F, CD570-B, CD570-C, CD570-D, and CD519-8A but not in derivative CD570-A. Overexpression of CdCDR2 was not detected in any derivative (Fig. 2). In order to determine if increased levels of expression of multidrug resistance genes resulted in increased drug efflux in the azole-resistant derivatives, the glucose-mediated efflux of R6G was measured. The results obtained suggested that R6G efflux mediated by drug extrusion pumps was less effective in derivatives CD36-C, CD36-F, CD570-B, CD570-C, CD570-D, and CD519-8A than in their parental isolates. This was not the case for derivative CD570-A, for which R6G efflux was similar to that for its parental strain, CD570. These data suggest that the membranes of derivatives CD36-C, CD36-F, CD570-B, CD570-C, CD570-D, and CD519-8A have altered properties, possibly as a result of alterations in the ergosterol biosynthetic pathway. It has previously been demonstrated that altered membrane content can affect the function of efflux pumps (15, 32, 47). Resistance to azole antifungal agents due to alterations of the ergosterol biosynthetic pathway has been documented in S. cerevisiae (16) and in clinical isolates of C. albicans (18, 19). In these studies, a defect in sterol C5,6-desaturase was identified as the mechanism causing resistance to azoles. In C5,6-desaturase-deficient cells, growth in the presence of azoles is supported by the accumulation of 14α-methyl-fecosterol instead of the toxic metabolite 14α-methyl-3,6-diol that normally accumulates during azole treatment, with the accumulation ultimately causing growth arrest. To determine if the derivatives were deficient in sterol C5,6-desaturase, the sterol contents of their membranes were extracted and analyzed. Derivatives CD36-C, CD36-F, CD570-B, CD570-C, CD570-D, and CD519-8A did not produce ergosterol, which was consistent with a lack of sterol C5,6-desaturase activity in these derivatives. This was not the case, however, for derivative CD570-A, which contained ergosterol. In order to identify mutations in the gene encoding sterol C5,6-desaturase (termed ERG3), it was necessary to clone the C. dubliniensis ERG3 gene, which was found to be highly homologous (91.1%) to its C. albicans counterpart. Sequence analysis of the CdERG3 genes of the derivatives revealed that all derivatives (including CD570-A) harbored at least one homozygous mutation which altered the amino acid sequence of CdErg3p.
Heterologous expression studies revealed that these mutations resulted in a loss of Erg3p function for six of the seven azole-resistant derivatives. Mutated alleles of CdERG3 (with the exception of derivative CD570-A) were unable to complement the defective sterol C5,6-desaturase function in an S. cerevisiae Δerg3 mutant, whereas wild-type parental alleles could restore this function.
While loss-of-function mutations in the CdERG3 gene were observed in six derivatives exhibiting azole resistance, this was not the case for derivative CD570-A. This derivative exhibited levels of azole resistance which were similar to those for the other six derivatives and, like them, did not show cross-resistance to inhibitors. Unlike the other six derivatives, CD570-A did not exhibit reduced susceptibility to amphotericin B. Like the other six derivatives, derivative CD570-A exhibited elevated levels of expression of CdERG11, and there were no changes in the predicted amino acid sequence of CdErg11p which could explain its azole resistance phenotype. In contrast to the other six derivatives, the level of CdCDR1 expression was not elevated in CD570-A and R6G efflux did not appear to be altered in this derivative. Although derivative CD570-A accumulated ergosterol in its membranes like its parental isolate, there was a noticeable difference in the overall membrane sterol contents between CD570-A and CD570. The presence of a mutation in CdErg3p seemed to have little effect on the activity of the sterol C5,6-desaturase of CD570-A, as the heterologous expression of the CD570-A CdERG3 gene was able to complement the defective sterol C5,6-desaturase activity of an S. cerevisiae Δerg3 strain. It is unlikely that overexpression of CdERG11 is the only mechanism responsible for azole resistance in derivative CD570-A. While the reasons for this resistance in derivative CD570-A are as yet undetermined, the observed changes in membrane sterol contents could suggest that unidentified alterations of the ergosterol biosynthetic pathway could contribute to the azole-resistant phenotype in this derivative of CD570.
Although the derivatives showed increased levels of CdCDR1 and CdERG11 expression, the data presented in this study are consistent with a loss of function of CdERG3 as the primary mechanism for itraconazole resistance in six of the seven derivatives. The observed overexpression of CdERG11 in all derivatives, although not the primary mechanism, may be a factor contributing to their azole resistance. However, loss-of-function mutations of ERG3 mediate azole resistance by exerting a suppressor effect on yeast cells lacking 14α-lanosterol demethylase activity (52). Therefore, the observed overexpression of the demethylase is likely to be both unnecessary and irrelevant for azole resistance in the derivatives with loss-of-function mutations in CdERG3. Moreover, the observed upregulation of CdERG11 is very likely a consequence of the absence of ergosterol in the derivatives lacking sterol C5,6-desaturase activity. Indeed, such a negative-feedback effect on the regulation of ERG11 expression has previously been observed in a C. glabrata Δerg3 strain (9).
Since two of the parental isolates used in the present study were chosen because they harbored nonfunctional CdCDR1 genes, overexpression of CdCDR1 in their azole-resistant derivatives should have no effect on azole resistance. It is somewhat surprising that derivatives harboring nonfunctional CdCDR1 genes showed increased CdCDR1 mRNA levels. Indeed, data from this study suggest a correlation between a lack of sterol C5,6-desaturase activity and overexpression of CdCDR1 by azole-resistant derivatives. It is conceivable that changes in membrane lipid composition affect factors involved in regulating CdCDR1 expression, as it has been proposed that Cdr1p may be involved in phospholipid translocation (2).
The results from this study show that mutations in sterol C5,6-desaturase are involved in high-level stable azole resistance following exposure to itraconazole in vitro. The C. albicans Darlington strain is an example of a highly resistant clinical strain (itraconazole MIC, >4 μg/ml; fluconazole MIC, 32 μg/ml) harboring, as well as other resistance mechanisms, mutations in sterol C5,6-desaturase (28). This suggests that the levels of azole resistance and the resistance mechanisms observed in the azole-resistant derivatives of C. dubliniensis are of clinical relevance. While it has previously been established that multiple factors contribute to the development of resistance in Candida species, the findings of this study highlight the fact that, if not thoroughly investigated, azole resistance can wrongly be attributed to the apparent overexpression of multidrug resistance genes and of the target enzyme ERG11. Indeed, more studies are needed to evaluate the contribution of sterol C5,6-desaturase loss-of-function mutations in clinical azole resistance.
Acknowledgments
This study was supported by the Dublin Dental School and Hospital and by the Irish Health Research Board (grant 04-97).
REFERENCES
- 1.Arthington-Skaggs, B. A., H. Jradi, T. Desai, and C. J. Morrison. 1999. Quantitation of ergosterol content: novel method for determination of fluconazole susceptibility of Candida albicans. J. Clin. Microbiol. 37:3332-3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dogra, S., S. Krishnamurthy, V. Gupta, B. L. Dixit, C. M. Gupta, D. Sanglard, and R. Prasad. 1999. Asymmetric distribution of phosphatidylethanolamine in C. albicans: possible mediation by CDR1, a multidrug transporter belonging to ATP binding cassette (ABC) superfamily. Yeast 15:111-121. [DOI] [PubMed] [Google Scholar]
- 3.Donnelly, S. M., D. J. Sullivan, D. B. Shanley, and D. C. Coleman. 1999. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology 145:1871-1882. [DOI] [PubMed] [Google Scholar]
- 4.Elledge, S. J., J. T. Mulligan, S. W. Ramer, M. Spottswood, and R. W. Davis. 1991. Lambda YES: A multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc. Natl. Acad. Sci. USA 88:1731-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favre, B., M. Didmon, and N. S. Ryder. 1999. Multiple amino acid substitutions in lanosterol 14α-demethylase contribute to azole resistance in Candida albicans. Microbiology 145:2715-2725. [DOI] [PubMed] [Google Scholar]
- 6.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic gene construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franz, R., S. L. Kelly, D. C. Lamb, D. E. Kelly, M. Ruhnke, and J. Morschhauser. 1998. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob. Agents Chemother. 42:3065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallagher, P. J., D. E. Bennett, M. C. Henman, R. J. Russell, S. R. Flint, D. B. Shanley, and D. C. Coleman. 1992. Reduced azole susceptibility of Candida albicans from HIV-positive patients and a derivative exhibiting colony morphology variation. J. Gen. Microbiol. 138:1901-1911. [DOI] [PubMed] [Google Scholar]
- 9.Geber, A., C. A. Hitchcock, J. E. Swartz, F. S. Pullen, K. E. Marsden, K. J. Kwon-Chung, and J. E. Bennett. 1995. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob. Agents Chemother. 39:2708-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gee, S. F., S. Joly, D. R. Soll, J. F. Meis, P. E. Verweij, I. Polacheck, D. J. Sullivan, and D. C. Coleman. 2002. Identification of four distinct genotypes of Candida dubliniensis and detection of microevolution in vitro and in vivo. J. Clin. Microbiol. 40:556-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 12.Hube, B., M. Monod, D. A. Schofield, A. J. P. Brown, and N. A. R. Gow. 1994. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 13.Jabra-Rizk, M. A., A. A. Baqui, J. I. Kelley, W. A. Falkler, Jr., W. G. Merz, and T. F. Meiller. 1999. Identification of Candida dubliniensis in a prospective study of patients in the United States. J. Clin. Microbiol. 37:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joly, S., C. Pujol, M. Rysz, K. Vargas, and D. R. Soll. 1999. Development and characterization of complex DNA fingerprinting probes for the infectious yeast Candida dubliniensis. J. Clin. Microbiol. 37:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaur, R., and A. K. Bachhawat. 1999. The yeast multidrug resistance pump, Pdr5p, confers reduced drug resistance in erg mutants of Saccharomyces cerevisiae. Microbiology 145:809-818. [DOI] [PubMed] [Google Scholar]
- 16.Kelly, S. L., D. C. Lamb, A. J. Corran, B. C. Baldwin, and D. E. Kelly. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14 alpha-methylergosta-8, 24(28)-dien-3β,6α-diol. Biochem. Biophys. Res. Commun. 207:910-915. [DOI] [PubMed] [Google Scholar]
- 17.Kelly, S. L., D. C. Lamb, and D. E. Kelly. 1999. Y132H substitution in Candida albicans sterol 14α-demethylase confers fluconazole resistance by preventing binding to haem. FEMS Microbiol. Lett. 180:171-175. [DOI] [PubMed] [Google Scholar]
- 18.Kelly, S. L., D. C. Lamb, D. E. Kelly, J. Loeffler, and H. Einsele. 1996. Resistance to fluconazole and amphotericin in Candida albicans from AIDS patients. Lancet 348:1523-1524. [DOI] [PubMed] [Google Scholar]
- 19.Kelly, S. L., D. C. Lamb, D. E. Kelly, N. J. Manning, J. Loeffler, H. Hebart, U. Schumacher, and H. Einsele. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett. 400:80-82. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, S. L., D. C. Lamb, J. Loeffler, H. Einsele, and D. E. Kelly. 1999. The G464S amino acid substitution in Candida albicans sterol 14α-demethylase causes fluconazole resistance in the clinic through reduced affinity. Biochem. Biophys. Res. Commun. 262:174-179. [DOI] [PubMed] [Google Scholar]
- 21.Kirkpatrick, W. R., S. G. Revankar, R. K. McAtee, J. L. Lopez-Ribot, A. W. Fothergill, D. I. McCarthy, S. E. Sanche, R. A. Cantu, M. G. Rinaldi, and T. F. Patterson. 1998. Detection of Candida dubliniensis in oropharyngeal samples from human immunodeficiency virus-infected patients in North America by primary CHROMagar Candida screening and susceptibility testing of isolates. J. Clin. Microbiol. 36:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maebashi, K., M. Niimi, M. Kudoh, F. J. Fischer, K. Makimura, K. Niimi, R. J. Piper, K. Uchida, M. Arisawa, R. D. Cannon, and H. Yamaguchi. 2001. Mechanisms of fluconazole resistance in Candida albicans isolates from Japanese AIDS patients. J. Antimicrob. Chemother. 47:527-536. [DOI] [PubMed] [Google Scholar]
- 23.Maesaki, S., P. Marichal, H. Vanden Bossche, D. Sanglard, and S. Kohno. 1999. Rhodamine 6G efflux for the detection of CDR1-overexpressing azole-resistant Candida albicans strains. J. Antimicrob. Chemother. 44:27-31. [DOI] [PubMed] [Google Scholar]
- 24.Marichal, P., L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. Ramaekers, F. C. Odds, and H. Vanden Bossche. 1999. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145:2701-2713. [DOI] [PubMed] [Google Scholar]
- 25.Marr, K., C. N. Lyons, T. Rustad, R. Bowden, and T. White. 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob. Agents Chemother. 42:2584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meis, J. F., and P. E. Verweij. 2001. Current management of fungal infections. Drugs 61(Suppl. 1):13-25. [DOI] [PubMed] [Google Scholar]
- 27.Miyazaki, H., Y. Miyazaki, A. Geber, T. Parkinson, C. Hitchcock, D. Falconer, D. Ward, K. Marsden, and J. Bennett. 1998. Fluconazole resistance associated with drug efflux and increased expression of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob. Agents Chemother. 42:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyazaki, Y., A. Geber, H. Miyazaki, D. Falconer, T. Parkinson, C. Hitchcock, B. Grimberg, K. Nyswaner, and J. E. Bennett. 1999. Cloning, sequencing, expression and allelic sequence diversity of ERG3 (C-5 sterol desaturase gene) in Candida albicans. Gene 236:43-51. [DOI] [PubMed] [Google Scholar]
- 29.Moran, G., D. Sullivan, J. Morschhauser, and D. Coleman. 2002. The Candida dubliniensis CdCDR1 gene is not essential for fluconazole resistance. Antimicrob. Agents Chemother. 46:2829-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran, G. P., D. Sanglard, S. M. Donnelly, D. B. Shanley, D. J. Sullivan, and D. C. Coleman. 1998. Identification and expression of multidrug transporters responsible for fluconazole resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 42:1819-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran, G. P., D. J. Sullivan, M. C. Henman, C. E. McCreary, B. J. Harrington, D. B. Shanley, and D. C. Coleman. 1997. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob. Agents Chemother. 41:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukhopadhyay, K., A. Kohli, and R. Prasad. 2002. Drug susceptibilities of yeast cells are affected by membrane lipid composition. Antimicrob. Agents Chemother. 46:3695-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth microdilution antifungal susceptibility testing of yeasts: document M27A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 34.Odds, F. C., L. Van Nuffel, and G. Dams. 1998. Prevalence of Candida dubliniensis isolates in a yeast stock collection. J. Clin. Microbiol. 36:2869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perea, S., J. L. Lopez-Ribot, B. L. Wickes, W. R. Kirkpatrick, O. P. Dib, S. P. Bachmann, S. M. Keller, M. Martinez, and T. F. Patterson. 2002. Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 46:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfaller, M. A., S. A. Messer, S. Gee, S. Joly, C. Pujol, D. J. Sullivan, D. C. Coleman, and D. R. Soll. 1999. In vitro susceptibilities of Candida dubliniensis isolates tested against the new triazole and echinocandin antifungal agents. J. Clin. Microbiol. 37:870-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polacheck, I., J. Strahilevitz, D. Sullivan, S. Donnelly, I. F. Salkin, and D. C. Coleman. 2000. Recovery of Candida dubliniensis from non-human immunodeficiency virus-infected patients in Israel. J. Clin. Microbiol. 38:170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahier, A., P. Benveniste, T. Husselstein, and M. Taton. 2000. Biochemistry and site-directed mutational analysis of Δ7-sterol-C5(6)-desaturase. Biochem. Soc. Trans. 28:799-803. [PubMed] [Google Scholar]
- 40.Rodriguez-Tudela, J. L., and J. V. Martinez-Suarez. 1995. Defining conditions for microbroth antifungal susceptibility tests: influence of RPMI and RPMI-2% glucose on the selection of endpoint criteria. J. Antimicrob. Chemother. 35:739-749. [DOI] [PubMed] [Google Scholar]
- 41.Ruhnke, M., A. Schmidt-Westhausen, and J. Morschhauser. 2000. Development of simultaneous resistance to fluconazole in Candida albicans and Candida dubliniensis in a patient with AIDS. J. Antimicrob. Chemother. 46:291-295. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Sanglard, D., F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanglard, D., F. Ischer, M. Monod, and J. Bille. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143: 405-416. [DOI] [PubMed] [Google Scholar]
- 46.Sanglard, D., K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smriti, S. S. Krishnamurthy, and R. Prasad. 1999. Membrane fluidity affects functions of Cdr1p, a multidrug ABC transporter of Candida albicans. FEMS Microbiol. Lett. 173:475-481. [DOI] [PubMed] [Google Scholar]
- 48.Sullivan, D., K. Haynes, J. Bille, P. Boerlin, L. Rodero, S. Lloyd, M. Henman, and D. Coleman. 1997. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J. Clin. Microbiol. 35:960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sullivan, D. J., T. J. Westerneng, K. A. Haynes, D. E. Bennett, and D. C. Coleman. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141:1507-1521. [DOI] [PubMed] [Google Scholar]
- 50.Vazquez, J. A., A. Beckley, S. Donabedian, J. D. Sobel, and M. J. Zervos. 1993. Comparison of restriction enzyme analysis versus pulsed-field gradient gel electrophoresis as a typing system for Torulopsis glabrata and Candida species other than C. albicans. J. Clin. Microbiol. 31:2021-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkateswarlu, K., D. W. Denning, and S. L. Kelly. 1997. In-vitro activity of D0870, a new triazole antifungal drug, in comparison with fluconazole and itraconazole against Aspergillus fumigatus and Candida krusei. J. Antimicrob. Chemother. 39:731-736. [DOI] [PubMed] [Google Scholar]
- 52.Watson, P. F., M. E. Rose, S. W. Ellis, H. England, and S. L. Kelly. 1989. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem. Biophys. Res. Commun. 164:1170-1175. [DOI] [PubMed] [Google Scholar]
- 53.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirsching, S., G. P. Moran, D. J. Sullivan, D. C. Coleman, and J. Morschhauser. 2001. MDR1-mediated drug resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 45:3416-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]