Abstract
The core of intermediary metabolism in autotrophs is the citric acid cycle. In a certain group of chemoautotrophs, the reductive citric acid cycle is an engine of synthesis, taking in CO2 and synthesizing the molecules of the cycle. We have examined the chemistry of a model system of C, H, and O that starts with carbon dioxide and reductants and uses redox couples as the energy source. To inquire into the reaction networks that might emerge, we start with the largest available database of organic molecules, Beilstein on-line, and prune by a set of physical and chemical constraints applicable to the model system. From the 3.5 million entries in Beilstein we emerge with 153 molecules that contain all 11 members of the reductive citric acid cycle. A small number of selection rules generates a very constrained subset, suggesting that this is the type of reaction model that will prove useful in the study of biogenesis. The model indicates that the metabolism shown in the universal chart of pathways may be central to the origin of life, is emergent from organic chemistry, and may be unique.
The chart of metabolic pathways (1) is an expression of the universality of intermediary metabolism. The reaction networks of all extant species of organisms map onto a single chart, the great unity within diversity of the living world. There are a number of possible explanations.
(i) The chart is the reaction network of the universal ancestor, which has survived in all branches of the evolutionary radiation. It is thus a virtual fossil that has persisted because changes deep within the system tend to be lethal, owing to the high degree of connectivity.
(ii) The chart has emerged from a facile interspecific sharing of genes by horizontal transfer across the taxa.
(iii) The chart represents an optimally successful solution to designing biochemical networks.
(iv) Some combination of the above explanations.
All of the possibilities suggest that the metabolic chart or parts thereof can be traced to the earliest organisms and contain information about the chemistry of biogenesis and the prebiotic planet some 4 billion years ago. This period is the preenzymatic domain. A paradox to be faced is that, at present, enzymes are required to define or generate the reaction network, and the network is required to synthesize the enzymes and their component monomers.
In trying to model the beginnings of biochemistry, we assume a vat with the appropriate chemicals, catalytic surfaces, a source of energy, and an energy sink. The source must provide energetic enough quanta to drive reactions involving covalent bond change. In carrying out the modeling, we use generalizations from biochemistry and ecology such as the metabolic chart and the carbon cycle, the notion of fitness, and insights from thermal physics such as the cycling theorem (2) and the notion that the flow of energy from a source to a sink organizes the intermediate system (3).
There has been an ongoing argument as to whether the earliest organisms were autotrophs or heterotrophs. Autotrophy requires metabolic pathways from environmental, one-carbon, minimum free-energy compounds to all intermediates. Heterotrophy in earliest metabolism requires the synthesis of high concentrations of nutrients in an environment free of specific biocatalysts. There is an intermediate case in which a small number of high-probability intermediates arise in the environment and are used by otherwise autotrophic systems. This paper generally assumes autotrophy with the possibility that there may be a preferred reaction network that bridges the gap between environments and cells.
For autotrophs, the metabolic chart has a shell structure (4). The core is the citric acid cycle and related reactions. The first shell is the synthesis of amino acids, which comes from amination of core keto acids. The second shell involves sulfur incorporation into amino acids. The third shell requires the synthesis of dinitrogen heterocycles. We assume that metabolism recapitulates biogenesis; the number of steps from CO2 incorporation to a given biochemical indexes the appearance of that molecule in biogenesis.
At the core of the metabolic chart is the citric acid cycle, which is the pathway to efficient oxidation in aerobic heterotrophs. In autotrophs, the citric acid cycle is the central pathway to all biosynthesis. Lipids come from acetyl CoA, sugars from phosphoenol pyruvate, and amino acids from keto acids and other compounds in the cycle. Nucleic acid components are synthesized from amino acids and sugars. In autotrophs, the citric acid cycle is an engine of synthesis.
Over the past 15 years, a number of chemoautotrophs have been isolated that operate by using the reductive citric acid cycle (5–8). Such organisms gain their energy from environmental redox couples and incorporate CO2 in those steps where CO2 is given off in the oxidative citric acid cycle. These organisms may provide clues as to the origin of metabolism in biogenesis.
The reductive citric acid cycle is found in both eubacteria and archea and in both aerobes and anaerobes. It is found in both mesophiles and thermophiles (9).
The cycle may be represented as follows.
 |
1 |
Two features of this cycle should be noted.
(i) It is network autocatalytic (as distinguished from template autocatalytic), and the overall reaction may be represented by,
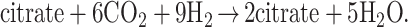 |
2 |
Any of the substrates is autocatalytic for its own synthesis. This type of autocatalysis may be a crucial step on the route to metabolism.
(ii) If the network occurs in a chemical reaction system, then it is a sink for carbon going from CO2 to more complex molecules. It is the simplest extant route for CO2 going to biochemicals.
In chemoautotrophs, the citric acid cycle is the central starting point on the route to all biochemicals. Energy must be supplied from outside the citric acid cycle by reactions going from environmental redox couples to ATP, reduced NAD+, reduced NADP, and reduced FAD. Given this energy, the cycle is the central feature of the metabolic chart.
One approach to the origin of metabolism is therefore a prebiotic nonenzymatic reductive citric acid cycle. In the prebiotic domain, CH3CO-SR can play the role now carried out by acetyl CoA and pyrophosphate can replace ATP. The model we are looking at is a vat of water, CO2, nitrogen, phosphorus, and sulfur and an energy source that will pump the ground state (equilibrium state) to excited states where they will react. We are interested in the occurrence of the reductive tricarboxylic acid (TCA) cycle under possible prebiotic conditions. The vat may contain catalytic surfaces such as pyrite and other metal sulfides (10, 11). The energy source can be photons or environmental redox couples. Carbon is supplied as CO2 and reductants are available.
The task is to find a set of physically motivated selection rules that will lead to a vat with a high concentration of reductive citric acid cycle intermediates and to analyze what the conditions must be for these rules to govern the system in the absence of enzymes. Because at a substrate level the molecules of interest in the citric acid cycle are CxHyOz, this is the universe we deal with first.
The guidance for restricting the domain to CHO comes from certain universal features of present-day metabolism, biochemistry, and chemical ecology. For example, almost all flow of nitrogen into the biosphere involves a series of oxidations and reductions to NH3, followed by the reaction of ammonia with keto acids to form amino acids. This finding strongly suggests the necessity of a network to produce keto acids before nitrogen incorporation and the synthesis of amino acids. Biological phosphorus almost universally occurs in the oxidation states as orthophosphates and pyrophosphates and attaches to intermediates by phosphate ester bonds. The phosphorus is not part of the carbon backbone structure or the small molecules at the core of metabolism. Sulfur also is restricted to the cofactor level in CoA, acetyl CoA, and succinyl CoA.
To study the chemistry within the reaction vessel, the list of all possible compounds can, in theory, be obtained in two ways.
(i) It can be algorithmically generated from the rules of organic chemistry or ultimately from the rules of quantum mechanics.
(ii) It can be extracted from databases of organic chemistry such as the Beilstein handbook (12) or the Dictionary of Organic Compounds (13). Both of these references are now available electronically.
The object of the selection rules is to generate the emergence (14) of the reductive citric acid cycle from the master list of compounds. These rules may be physical, chemical, biological, informational, or a combination of the above.
Because of the difficulty of deriving the network from the fundamental theory of organic chemistry, we have opted to search crossfire (12), an online version of the Beilstein handbook rather than the algorithmic approach. We start by looking at CxHyOz for which:
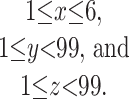 |
3 |
The value 99 is chosen to show that there is no upper limit at this stage. The selection for low-molecular weight compounds embodies an assumption that the beginning of biochemistry starts with C1 compounds and develops into compounds of higher molecular weights. The first cut yielded 2,790 compounds and included all of the intermediates of the reductive TCA cycle.
The next cut came from examining the oil-water partition coefficient and selecting for water solubility. This preference for aqueous solubility deals with phase separation in the original reactions and assumes at some time the capture of the reactions in vesicles of bilayer membranes made of amphiphilic molecules. Partition coefficients are contained in a database maintained by Biobyte Software (Claremont, CA). The quantity p represents the ratio of the concentration in water-saturated octanol divided by the concentration in octanol-saturated water, where those two phases are in equilibrium with each other. Values are available for log p obtained either experimentally or by computation clog p. Negative values of the logarithm are designated as hydrophilic and positive values as lipophilic. Biobyte can be accessed by using the smiles representation of molecules maintained by Daylight Inc.
The next selection rule is for low heats of combustion, to look first for compounds energetically close to CO2 because this represents the initial domain accessed in the energetic pumping of CO2, water, and reductants. Thermodynamic data can be obtained from experimental databases (15, 16) and by calculation from group contributions (17).
After an examination of a number of CxHyOz compounds, we discovered two informatic selection rules that include the oil-water partitions and thermodynamic selection without the necessity of using the other databases.
The two rules are:
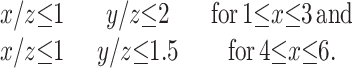 |
4 |
In general for these compounds, the more reduced the molecules, the more hydrophobic and the greater the heat of combustion. Thus, the informatics rules embody the thermodynamic selections and are much easier to apply.
The next selection is to exclude compounds that have no carbonyl groups. The essence of biochemistry of CHO molecules is the domain of carbonyl reactivity, and the set of molecules is restricted to those that can participate in such reactions.
The next selection is to exclude cyclic compounds and compounds with C—O—C on the basis of being difficult to synthesize nonenzymatically in this C, H, O domain.
The next step excludes C C and O—O on the grounds of stability. Radicals and ions are present in the Beilstein list (12) and are not included here. Chiral pairs are treated as single molecules.
The application of the primary rules results in a set of only 153 compounds containing the 11 intermediates of the reductive TCA cycle (see Table 1). Starting with the 3.5 million compounds of Beilstein and applying a small number of pruning rules motivated by physical and chemical considerations, we arrive at a small subset of organic compounds that includes all of the reductive TCA intermediates.
Table 1.
Compounds selected from Beilstein
| No. | Molecular formula | Chemical name | Chemical Abstracts Service registry number |
|---|---|---|---|
| 1 | CH2O | Formaldehyde | 50-00-0 |
| 2 | CH2O2 | Formic acid | 64-18-6 |
| 3 | C2H2O2 | Ethanedial | 107-22-2 |
| 4 | C2H2O3 | Oxo-acetic acid | 298-12-4 |
| 5 | C2H2O4 | Oxalic acid | 144-62-7 |
| 6 | C2H4O2 | Acetic acid | 64-19-7† |
| 7 | C2H4O2 | Hydroxy-acetaldehyde | 141-46-8 |
| 8 | C2H4O3 | Dihydroxy-acetaldehyde | 631-59-4 |
| 9 | C2H4O3 | Hydroxy-acetic acid | 79-14-1 |
| 10 | C2H4O4 | Dihydroxy-acetic acid | 563-96-2 |
| 11 | C3H2O3 | 2-Oxo-malonaldehyde | 497-16-5 |
| 12 | C3H2O4 | 2,3-Dioxo-propionic acid | 815-53-2 |
| 13 | C3H2O5 | 2-Oxo-malonic acid | 473-90-5 |
| 14 | C3H4O3 | 2,3-Dihydroxy-propenal | 636-38-4 |
| 15 | C3H4O3 | 2-Hydroxy-acrylic acid | 19071-34-2 |
| 16 | C3H4O3 | 2-Hydroxy-malonaldehyde | 497-15-4 |
| 17 | C3H4O3 | 2-Oxo-propionic acid | 127-17-3† |
| 18 | C3H4O3 | 3-Hydroxy-2-oxo-propionaldehyde | 997-10-4 |
| 19 | C3H4O3 | 3-Hydroxy-acrylic acid | 65034-30-2 |
| 20 | C3H4O3 | 3-Oxo-propionic acid | 926-61-4 |
| 21 | C3H4O4 | 2,2-Dihydroxy-malonaldehyde | 4464-20-4 |
| 22 | C3H4O4 | 2,3-Dihydroxy-acrylic acid | 2702-94-5 |
| 23 | C3H4O4 | 2-Hydroxy-3-oxo-propionic acid | 2480-77-5 |
| 24 | C3H4O4 | 3,3-Dihydroxy-acrylic acid | 177594-62-6 |
| 25 | C3H4O4 | 3-Hydroxy-2-oxo-propionic acid | 1113-60-6 |
| 26 | C3H4O4 | Malonic acid | 141-82-2 |
| 27 | C3H4O5 | 2-Hydroxy-malonic acid | 80-69-3 |
| 28 | C3H4O6 | 2,2-Dihydroxy-malonic acid | 560-27-0 |
| 29 | C3H6O3 | 1,1-Dihydroxy-propan-2-one | 1186-47-6 |
| 30 | C3H6O3 | 1,3-Dihydroxy-propan-2-one | 96-26-4 |
| 31 | C3H6O3 | 2,3-Dihydroxy-propionaldehyde | 453-17-8 |
| 32 | C3H6O3 | 2-Hydroxy-propionic acid | 50-21-5 |
| 33 | C3H6O3 | 3-Hydroxy-propionic acid | 503-66-2 |
| 34 | C3H6O4 | 2,2-Dihydroxy-propionic acid | 1825-45-2 |
| 35 | C3H6O4 | 2,3-Dihydroxy-propionic acid | 473-81-4 |
| 36 | C4H2O4 | 2,3-Dihydroxy-buta-1,3-diene-1,4-dione | 7472724* |
| 37 | C4H2O4 | 2,3-Dioxo-succinaldehyde | 97245-29-9 |
| 38 | C4H2O6 | 2,3-Dioxo-succinic acid | 7580-59-8 |
| 39 | C4H4O4 | 2,4-Dioxo-butyric acid | 1069-50-7 |
| 40 | C4H4O4 | 2-Hydroxy-4-oxo-but-2-enoic acid | 114847-32-4 |
| 41 | C4H4O4 | 2-Methylene-malonic acid | 4442-03-9 |
| 42 | C4H4O4 | 3,4-Dioxo-butyric acid | 20602-39-5 |
| 43 | C4H4O4 | 4-Hydroxy-2-oxo-but-3-enoic acid | 1748936* |
| 44 | C4H4O4 | But-2-enedioic acid | 6915-18-0† |
| 45 | C4H4O5 | 2-Hydroxy-but-2-enedioic acid | 7619-04-7 |
| 46 | C4H4O5 | 2-Oxo-succinic acid | 328-42-7† |
| 47 | C4H4O6 | 2,3-Dihydroxy-but-2-enedioic acid | 13096-38-3 |
| 48 | C4H4O6 | 2-Hydroxy-3-oxo-succinic acid | 5651-05-8 |
| 49 | C4H4O7 | 2-Carboxy-2-hydroxy-malonic acid | 44968-58-3 |
| 50 | C4H6O4 | 1,4-Dihydroxy-butane-2,3-dione | 162369-87-1 |
| 51 | C4H6O4 | 2,3-Dihydroxy-succinaldehyde | 34361-91-6 |
| 52 | C4H6O4 | 2-Hydroxy-3-oxo-butyric acid | 37520-05-1 |
| 53 | C4H6O4 | 2-Hydroxy-4-oxo-butyric acid | 62386-30-5 |
| 54 | C4H6O4 | 2-Methyl-malonic acid | 516-05-2 |
| 55 | C4H6O4 | 3,3-Dihydroxy-2-methyl-acrylic acid | 69858-40-8 |
| 56 | C4H6O4 | 3,4-Dihydroxy-2-oxo-butyraldehyde | 496-56-0 |
| 57 | C4H6O4 | 3-Hydroxy-2-oxo-butyric acid | 1944-42-9 |
| 58 | C4H6O4 | 3-Hydroxy-4-oxo-butyric acid | 10495-18-8 |
| 59 | C4H6O4 | 4-Hydroxy-2-oxo-butyric acid | 22136-38-5 |
| 60 | C4H6O4 | Succinic acid | 110-15-6† |
| 61 | C4H6O5 | 2,3,4-Trihydroxy-but-2-enoic acid | 1928462* |
| 62 | C4H6O5 | 2,3-Dihydroxy-4-oxo-butyric acid | 10385-76-9 |
| 63 | C4H6O5 | 2-Hydroxy-2-methyl-malonic acid | 595-48-2 |
| 64 | C4H6O5 | 2-Hydroxymethyl-malonic acid | 4360-96-7 |
| 65 | C4H6O5 | 2-Hydroxy-succinic acid | 6915-15-7† |
| 66 | C4H6O5 | 3,4-Dihydroxy-2-oxo-butyric acid | 114579-56-5 |
| 67 | C4H6O6 | 2,2-Dihydroxy-succinic acid | 60047-52-1 |
| 68 | C4H6O6 | 2,3-Dihydroxy-succinic acid | 526-83-0 |
| 69 | C4H6O6 | 2-Hydroxy-2-hydroxymethyl-malonic acid | 54472-64-9 |
| 70 | C4H6O8 | 2,2,3,3-Tetrahydroxy-succinic acid | 76-30-2 |
| 71 | C5H2O5 | 2,3,4-Trioxo-pentanedial | 97245-30-2 |
| 72 | C5H4O5 | 2-Formyl-but-2-enedioic acid | 111598-98-2 |
| 73 | C5H4O5 | 4-Oxo-pent-2-enedioic acid | 6004-32-6 |
| 74 | C5H4O6 | 2-Carboxy-but-2-enedioic acid | 4364-81-2 |
| 75 | C5H4O7 | 2,3-Dihydroxy-4-oxo-pent-2-enedioic acid | 89712-64-1 |
| 76 | C5H4O7 | 2-Carboxy-3-hydroxy-but-2-enedioic acid | 1785338* |
| 77 | C5H4O7 | 2-Carboxy-3-oxo-succinic acid | 4378-81-8 |
| 78 | C5H4O7 | 2-Hydroxy-3,4-dioxo-pentanedioic acid | 89282-33-7 |
| 79 | C5H4O8 | 2,2-Dicarboxy-malonic acid | 193197-67-0 |
| 80 | C5H6O5 | 2-Formyl-succinic acid | 5856-44-0 |
| 81 | C5H6O5 | 2-Hydroxy-3-methyl-but-2-enedioic acid | 148716-85-2 |
| 82 | C5H6O5 | 2-Methyl-3-oxo-succinic acid | 642-93-3 |
| 83 | C5H6O5 | 2-Oxo-pentanedioic acid | 328-50-7† |
| 84 | C5H6O5 | 3-Oxo-pentanedioic acid | 542-05-2 |
| 85 | C5H6O6 | 2,3,5-Trihydroxy-4-oxo-pent-2-enoic acid | 5425275* |
| 86 | C5H6O6 | 2-Carboxy-succinic acid | 922-84-9 |
| 87 | C5H6O6 | 2-Hydroxy-2-methyl-3-oxo-succinic acid | 1777463* |
| 88 | C5H6O6 | 2-Hydroxy-4-oxo-pentanedioic acid | 1187-99-1 |
| 89 | C5H6O6 | 2-Hydroxymethyl-3-oxo-succinic acid | 89323-48-8 |
| 90 | C5H6O7 | 2,3,4-Trihydroxy-pent-2-enedioic acid | 91113-90-5 |
| 91 | C5H6O7 | 2,3-Dihydroxy-4-oxo-pentanedioic acid | 1787046* |
| 92 | C5H6O7 | 2-Carboxy-2-hydroxy-succinic acid | 110863-50-8 |
| 93 | C5H6O7 | 2-Carboxy-3-hydroxy-succinic acid | 80754-80-9 |
| 94 | C5H6O8 | 2-Carboxy-2,3-dihydroxy-succinic acid | 639-51-0 |
| 95 | C5H8O6 | 2,2-Bis-hydroxymethyl-malonic acid | 173783-71-6 |
| 96 | C5H8O6 | 2,2-Dihydroxy-3-methyl-succinic acid | 4980495* |
| 97 | C5H8O6 | 2,2-Dihydroxy-pentanedioic acid | 23788-98-9 |
| 98 | C5H8O6 | 2,3,4-Trihydroxy-5-oxo-pentanoic acid | 114375-57-4 |
| 99 | C5H8O6 | 2,3,5-Trihydroxy-4-oxo-pentanoic acid | 134616-21-0 |
| 100 | C5H8O6 | 2,3-Dihydroxy-2-methyl-succinic acid | 15853-34-6 |
| 101 | C5H8O6 | 2,3-Dihydroxy-pentanedioic acid | 82864-78-6 |
| 102 | C5H8O6 | 2,4-Dihydroxy-pentanedioic acid | 82864-77-5 |
| 103 | C5H8O6 | 2-Hydroxy-2-hydroxymethyl-succinic acid | 2957-09-7 |
| 104 | C5H8O6 | 3,4,5-Trihydroxy-2-oxo-pentanoic acid | 110902-88-0 |
| 105 | C5H8O7 | 2,3,4-Trihydroxy-pentanedioic acid | 608-55-9 |
| 106 | C5H8O7 | 2,3-Dihydroxy-2-hydroxymethyl-succinic acid | 6115630* |
| 107 | C6H4O6 | 4,5-Dioxo-hex-2-enedioic acid | 6123412* |
| 108 | C6H4O8 | 2,3-Dicarboxy-but-2-enedioic acid | 4363-44-4 |
| 109 | C6H6O6 | 2,5-Dihydroxy-hexa-2,4-dienedioic acid | 1725831* |
| 110 | C6H6O6 | 2,5-Dioxo-hexanedioic acid | 25466-26-6 |
| 111 | C6H6O6 | 2-Carboxy-3-methyl-but-2-enedioic acid | 1781603* |
| 112 | C6H6O6 | 2-Carboxy-3-methylene-succinic acid | 1779647* |
| 113 | C6H6O6 | 3,4-Dioxo-hexanedioic acid | 533-76-6 |
| 114 | C6H6O6 | 3,6-Dihydroxy-2,5-dioxo-hex-3-enoic acid | 2443471* |
| 115 | C6H6O6 | 3-Carboxy-pent-2-enedioic acid | 499-12-7† |
| 116 | C6H6O7 | 3-Carboxy-2-hydroxy-pent-2-enedioic acid | 1792255* |
| 117 | C6H6O7 | 3-Carboxy-2-oxo-pentanedioic acid | 1948-82-9† |
| 118 | C6H6O8 | 2,3-Dicarboxy-succinic acid | 4378-76-1 |
| 119 | C6H6O8 | 3,4-Dihydroxy-2,5-dioxo-hexanedioic acid | 1794752* |
| 120 | C6H6O8 | 3-Carboxy-2-hydroxy-4-oxo-pentanedioic acid | 3687-15-8 |
| 121 | C6H8O6 | 2-Carboxy-2-methyl-succinic acid | 39994-39-3 |
| 122 | C6H8O6 | 2-Carboxy-3-methyl-succinic acid | 61713-72-2 |
| 123 | C6H8O6 | 2-Carboxy-pentanedioic acid | 4756-09-6 |
| 124 | C6H8O6 | 2-Hydroxy-2-methyl-4-oxo-pentanedioic acid | 19071-44-4 |
| 125 | C6H8O6 | 2-Hydroxy-5-oxo-hexanedioic acid | 13095-45-9 |
| 126 | C6H8O6 | 3-Carboxy-pentanedioic acid | 99-14-9 |
| 127 | C6H8O7 | 2,3,5,6-Tetrahydroxy-4-oxo-hex-2-enoic acid | 5478036* |
| 128 | C6H8O7 | 2,3,5-Trihydroxy-4,6-dioxo-hexanoic acid | 4746-27-4 |
| 129 | C6H8O7 | 2,3-Dihydroxy-5-oxo-hexanedioic acid | 26566-33-6 |
| 130 | C6H8O7 | 3,4,6-Trihydroxy-2,5-dioxo-hexanoic acid | 2595-33-7 |
| 131 | C6H8O7 | 3-Carboxy-2-hydroxy-pentanedioic acid | 320-77-4† |
| 132 | C6H8O7 | 3-Carboxy-3-hydroxy-pentanedioic acid | 77-92-9† |
| 133 | C6H8O7 | 4,5,6-Trihydroxy-2,3-dioxo-hexanoic acid | 7683-53-6 |
| 134 | C6H8O8 | 2,3,4-Trihydroxy-5-oxo-hexanedioic acid | 149250-15-7 |
| 135 | C6H8O8 | 2-Carboxy-2,4-dihydroxy-pentanedioic acid | 82848-19-9 |
| 136 | C6H8O8 | 3-Carboxy-2,3-dihydroxy-pentanedioic acid | 6205-14-7 |
| 137 | C6H8O9 | 2-Carboxy-2,3,4-trihydroxy-pentanedioic acid | 1801017* |
| 138 | C6H10O7 | 2-(1,2-Dihydroxy-ethyl)-2-hydroxy-succinic acid | 1790363* |
| 139 | C6H10O7 | 2,3,4,5,6-Pentahydroxy-hex-2-enoic acid | 113892-19-6 |
| 140 | C6H10O7 | 2,3,4,5-Tetrahydroxy-6-oxo-hexanoic acid | 6814-36-4 |
| 141 | C6H10O7 | 2,3,4,6-Tetrahydroxy-5-oxo-hexanoic acid | 13425-57-5 |
| 142 | C6H10O7 | 2,3,4-Trihydroxy-2-hydroxymethyl-5-oxo-pentanoic acid | 1711202* |
| 143 | C6H10O7 | 2,3,4-Trihydroxy-2-methyl-pentanedioic acid | 469-44-3 |
| 144 | C6H10O7 | 2,3,4-Trihydroxy-hexanedioic acid | 4382-48-3 |
| 145 | C6H10O7 | 2,3,5,6-Tetrahydroxy-4-oxo-hexanoic acid | 54911-28-3 |
| 146 | C6H10O7 | 2,3,5-Trihydroxy-hexanedioic acid | 13427-52-6 |
| 147 | C6H10O7 | 2,3-Dihydroxy-2-(2-hydroxy-ethyl)-succinic acid | 1790420* |
| 148 | C6H10O7 | 2,4-Dihydroxy-2-hydroxymethyl-pentanedioic acid | 98574-40-4 |
| 149 | C6H10O7 | 3,4,5,6-Tetrahydroxy-2-oxo-hexanoic acid | 73803-83-5 |
| 150 | C6H10O8 | 2,3,4,5-Tetrahydroxy-hexanedioic acid | 7558-19-2 |
| 151 | C6H10O8 | 2,3,4-Trihydroxy-2-hydroxymethyl-pentanedioic acid | 1712927* |
| 152 | C6H10O8 | 3,4,5,5,6-Pentahydroxy-2-oxo-hexanoic acid | 7808083* |
| 153 | C6H10O10 | 2,2,3,4,5,5-Hexahydroxy-hexanedioic acid | 1801900* |
Beilstein registry numbers.
Member of the TCA.
One feature of the reductive TCA cycle that immediately attracts attention is that it is network autocatalytic. Any molecule in the cycle is catalytic for its own synthesis. Another feature is that all reactions either are monomolecular or involve substrates interacting with environmental molecules. Because the substrates are at low concentrations, these reactions are kinetically favored over substrate–substrate reactions by an order of magnitude. For these reactions to proceed, therefore, does not require a vesicle to trap the reaction products. The core chemistry can proceed without an envelope. It is a consequence of the type of reactions at the center of the metabolic chart and will no longer apply as soon as a reaction is required that is between two substrates. The dominant reactions are oxidation–reduction, hydration–dehydration, carboxylation–decarboxylation, and splitting; they operate independently of an enclosure.
From the domain of all possible reactions in a reduced world containing H2O and CO2, there emerges through certain physically motivated pruning rules a small set of 153 compounds that includes all of the citric acid cycle intermediates. Another few molecules, such as hydroxypyruvate, occur as intermediates along neighboring metabolic pathways. Thus, the subset of emergent molecules is highly favored as metabolites. In any case, the reductive citric acid cycle is embedded in the emergent subset.
From a point of view of general complexity theory, the Beilstein compendium is a highly structured database of endpoints of reaction networks. The fact that it can be used to generate heuristic approaches to biogenesis indicates a possible approach to the theory of directed database mining. It is facilitated by the rich knowledge of chemistry that accompanies the database.
Efforts have been made to analyze the TCA cycle from the point of view of efficiency (18). They are oriented to acetate oxidation rather than to operating in the reductive direction. We suggest alternative cycles that contain several compounds from our group of 153 and some others that we excluded because they had too high a content of hydrogen. Note that the approach in our study is oriented toward reductive autotrophic metabolism and concentrates on anabolism.
If one wishes to study biogenesis from the bottom up, the first step is to reason from atoms of the periodic table to those molecules that form the core of biochemistry, those molecules central to the chart of intermediary metabolism in chemoautotrophs. We have started with the assumption that the core molecules are made of CHO, possibly supplemented by −SR and polyphosphates. We have assumed that biogenesis moves from simplicity to complexity, from low free energy to high free energy, and from autotrophy to heterotrophy. We convert these assumptions to primary rules as to the kinds of molecules to be selected for and apply this selection to the primary database of organic molecules, Beilstein (12). What emerges is a set of 153 molecules that include the 11 members of the reductive citric acid cycle, as well as some other molecules from the metabolic chart. We argue that there is an enormous simplification as well as indication that the chemistry at the core of the metabolic chart is necessary and deterministic and would likely characterize any aqueous carbon-based life anywhere it is found in this universe. Experiments to find corollaries of these results are in progress.
Acknowledgments
We thank Jennifer Sturgis of the Krasnow Institute, David Weininger of Daylight Inc., Melanie Mitchell and Murray Gell-Mann of the Santa Fe Institute, Albert Leo of Pomona College, Robert Hazen of the Geophysics Laboratory of the Carnegie Institute of Washington, John Holland of the University of Michigan, James Willett of George Mason University, and Sherwood Chang of National Aeronautics and Space Administration Ames Laboratory. Special note should be made of many conversations with Walter Fontana of the Santa Fe Institute. Finally, we want to express our indebtedness to Leo Buss of Yale University for the intensity of his devil's advocacy and the value of his suggestions. This study has been ongoing since 1992, and at various stages many individuals have been most helpful. The work was carried out at the Krasnow Institute of George Mason University, the Santa Fe Institute, and recently as part of a project at the Geophysics Laboratory of the Carnegie Institute of Washington and the National Aeronautics and Space Administration Astrobiology program. The final study was carried out while H.J.M. was on sabbatical at the Department of Ecology and Evolutionary Biology at Yale University. Their welcome is acknowledged.
Abbreviation
- TCA
tricarboxylic acid
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110153997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110153997
References
- 1.Nicholson D E. Metabolic Pathways. St. Louis: Sigma; 1997. [Google Scholar]
- 2.Morowitz H J. J Therm Biol. 1966;13:60–62. [Google Scholar]
- 3.Morowitz H J. Energy Flow in Biology. New York: Academic; 1968. [Google Scholar]
- 4.Morowitz H J. Complexity. 1999;4:39–53. [Google Scholar]
- 5.Evans M C W, Buchanan B B, Arron D I. Proc Natl Acad Sci USA. 1966;55:928–934. doi: 10.1073/pnas.55.4.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanovsky R N, Sintsov N V, Kondratieva E N. Arch Microbiol. 1980;128:239–241. [Google Scholar]
- 7.Shiba H, Kawasumi T, Igarashi Y, Kodoma T, Minoda Y. Arch Microbiol. 1985;142:198–203. [Google Scholar]
- 8.Shima S, Suzuki K I. Int J Syst Bacteriol. 1993;43:703–708. [Google Scholar]
- 9.Danson M J, Hough D W, Lunt G G, editors. The Archaebacteria: Biochemistry and Biotechnology. London: Portland; 1992. pp. 14–20. [Google Scholar]
- 10.Wächterhäuser G. Microbiol Rev. 1988;52:452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wächterhäuser G. Proc Natl Acad Sci USA. 1990;87:200–204. [Google Scholar]
- 12.Beilstein Informationssysteme. Beilstein crossfire. Berlin: Springer; 1998. , update no. BS 9902PR. [Google Scholar]
- 13.Dictionary of Organic Compounds (1999) (Chapman & Hall, London), 6th Ed, CD-ROM.
- 14.Holland J H. Emergence. Reading, MA: Helix; 1998. [Google Scholar]
- 15.Marsh K, editor. TRC Thermodynamic Tables. Texas A & M Univ., College Station, TX: Thermodynamic Research Center; 1999. [Google Scholar]
- 16.Stull D R, Westrum E F, Sinke G C. The Chemical Thermodynamics of Organic Compounds. New York: Wiley; 1969. [Google Scholar]
- 17.Mavrovouniotis M L, Prickett S, Constantinov L. Comput Chem Eng. 1992;16:5353–5360. [Google Scholar]
- 18.Melendez-Hevia E, Waddell T G, Cascanta M. J Mol Evol. 1996;43:393–303. doi: 10.1007/BF02338838. [DOI] [PubMed] [Google Scholar]


