Abstract
A fundamental question in molecular biology is how proteins fold into domains that can serve as assembly modules for building up large macromolecular structures. The biogenesis of pili on the surface of Gram-negative bacteria requires the orchestration of a complex process that includes protein synthesis, folding via small chaperones, secretion, and assembly. The results presented here support the hypothesis that pilus subunit folding and biogenesis proceed via mechanisms termed donor strand complementation and donor strand exchange. Here we show that the steric information necessary for pilus subunit folding is not contained in one polypeptide sequence. Rather, the missing information is transiently donated by a strand of a small chaperone to allow folding. Providing the missing information for folding, via a 13-amino acid peptide extension to the C-terminal end of a pilus subunit, resulted in the production of a protein that no longer required the chaperone to fold. This mechanism of small periplasmic chaperone function described here deviates from classical hsp60 chaperone-assisted folding.
The PapD-like superfamily of periplasmic chaperones directs the assembly of over 30 diverse adhesive surface organelles that mediate the attachment of many different pathogenic bacteria to host tissues, a critical early step in the development of disease (1, 2). PapD, the prototypical chaperone, is necessary for the assembly of P pili (3). P pili contain the adhesin PapG, which mediates the attachment of uropathogenic Escherichia coli to Galα (1–4) Gal receptors present on kidney cells and are critical for the initiation of pyelonephritis (4). FimC, a homologue of PapD, directs the assembly of type 1 pili (5). Genes important in type 1 pilus biogenesis (fimA-fimH) are organized in the fim operon (Fig. 1A) (6). Type 1 pili are composite fibers consisting of a short thin tip fibrillum joined to a thicker rigid pilus rod (7, 8). The pilus fiber is an ordered array of homologous pilus subunits (FimA, FimF, and FimG) with the FimH adhesin at its tip. FimH and FimG have been purified as a complex and comprise the bulk of the tip fibrillum, which may also contain FimF (8). The rod is comprised of repeating FimA monomers arranged in a right-handed helical cylinder (7). FimH mediates adherence to mannosylated receptors on the bladder epithelium and is critical for the ability of uropathogenic E. coli to cause cystitis (9–12).
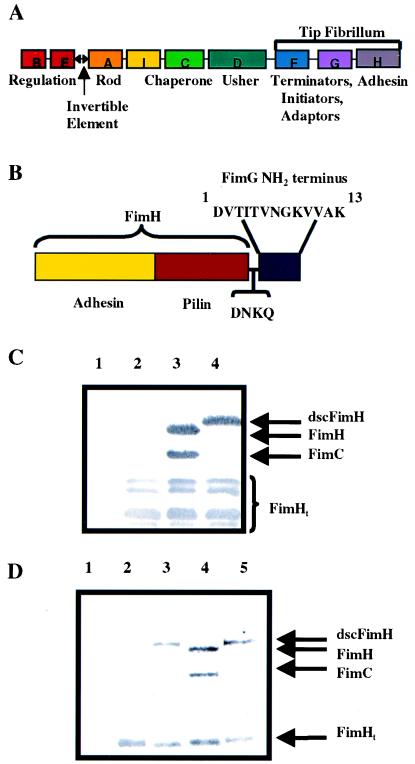
Figure 1.
Donor strand complementation of FimH in cis. Shown are schematic diagrams of the type 1 gene cluster (A) and dscFimH (B). Immunoblots developed with anti-FimCH antiserum of periplasmic extracts (C) after no expression of FimH (lane 1), FimH alone (lane 2), FimH + FimC (lane 3), or dscFimH (lane 4). A proportion of FimH truncation occurred under all conditions and was labeled FimHt. (D) Elution of FimH or dscFimH from mannose-Sepharose after incubation with periplasm containing FimC (lane 1), FimH alone (lane 2), dscFimH (lane 3), FimH + FimC (lane 4), or dscFimH + FimC (lane 5). The elutions were run on a SDS/PAGE gel followed by Western blotting using anti-FimCH antibodies.
During pilus biogenesis, the chaperone binds to and forms stable complexes with individual pilus subunits (13, 14). The crystal structures of the FimC-FimH chaperone-adhesin complex and the PapD-PapK chaperone-subunit complex have been solved (15, 16). The chaperones consist of two Ig-like domains oriented toward each other, forming L-shaped molecules (17, 18). The FimH adhesin has both a pilin domain and a receptor-binding domain whereas pilus subunits other than the adhesins contain only a pilin domain. PapK and the pilin domain of FimH have Ig-like folds. However, they lack the seventh C-terminal β-strand (strand G) present in canonical Ig folds (15, 16). The absence of this strand produces a deep groove along the surface of the pilin domain and exposes its hydrophobic core (15, 16), which may account for the instability of pilus subunits when expressed without the chaperone (3, 19). In the chaperone-subunit complex, the G1β strand of the chaperone completes an atypical Ig fold in the subunit by occupying the groove and running parallel to the subunit C-terminal F strand (see Fig. 2A) (15, 16). This donor strand complementation interaction simultaneously stabilizes pilus subunits and caps their interactive surfaces, preventing their premature oligomerization in the periplasm. During pilus biogenesis, the highly conserved N-terminal extension of one subunit has been proposed to displace the chaperone G1β strand from its neighboring subunit in a mechanism termed donor strand exchange (15, 16). The N-terminal strand is thought to insert anti-parallel to the F strand of the neighboring subunit (15, 16) so that the mature pilus would consist of an array of perfectly canonical Ig domains, each of which contributes a strand to the fold of its neighboring subunit.
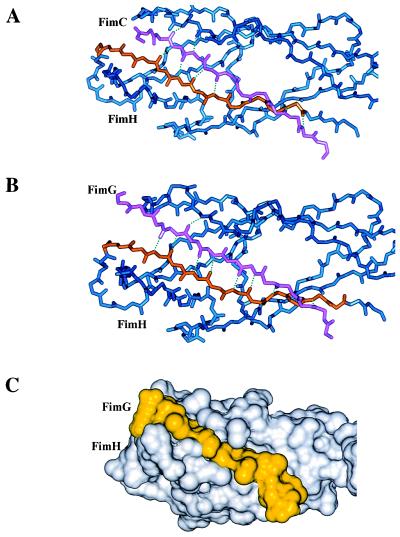
Figure 2.
Modeling of the N terminus of FimG into the FimH pilin domain. (A) Crystal structure of the FimH pilin domain (blue) of the FimCH complex highlighting the interaction between the F strand of FimH (orange) and the G1β strand of FimC (pink). (B) Pilin domain of FimH (blue) modeling the anti-parallel interaction between the F strand of FimH (orange) and the FimG donor strand (pink). (C) Connolly surface representation of the FimH pilin domain (white) with the FimG donor strand (yellow). Note the more extensive hydrogen bonding (dotted lines) with the FimG donor strand.
The hypothesis was tested that pilus subunit proteins are unable to fold independently (or fold inefficiently) because they lack their C-terminal G β-strand and thus require a chaperone to provide this steric information. We investigated whether we could alleviate the need for a chaperone by providing the missing strand in cis, fusing the missing seventh β-strand onto the 3′ end of fimH. The addition of the N-terminal extension of FimG onto the C terminus of FimH (this construct was called donor strand complemented FimH or dscFimH) resulted in the production of a protein that was now stable in the periplasm in the absence of the chaperone. Circular dichroism and fluorescence data show that, unlike FimH, dscFimH was able to fold in the absence of the chaperone.
Materials and Methods
Genetic Constructs.
To construct dscFimH, the following two oligos were annealed together and ligated into the ClaI and BamHI sites of pUC18-FimH (S.J.H. and C. Widberg, unpublished work) to create pUC18-dscFimH: DNKQ top 5′-CGATTATTGGCGTGACTTTTGTTTATCAAGATAACAAACAGGATGTCACCATCACGGTGAACGGTAAGGTCGTCGCCAAATAAG-3′; DNKQ bottom 5′-GATCCT-TATTTGGCGACGACCTTACCGTTCACCGTGATGG-TGACATCCTGTTTGTTATCTTGATAAACAAAAGT-CACGCCAATAAT-3′. pUC18-dscFimH was sequenced followed by subcloning into the EcoRI and BamHI sites of pTrc99A (20) to create pTrc-dscFimH. fimH was subcloned from pUC18-FimH into pTrc99A by using the EcoRI and BamHI sites to create pTrc-FimH. fimH and dscfimH were subcloned from pUC18-FimH and pUC18-dscFimH into pBad18-Kn (21) by using the EcoRI and XbaI sites to create pBad-FimH and pBad-dscFimH.
Periplasmic Preparations and Mannose Binding Assay.
The plasmids encoding FimH (pTrc-FimH), dscFimH (pTrc-dscFimH), or FimC (pJP4) (5) were expressed in C600. Overnight cultures were diluted 1:100 into Luria broth and were grown to an OD600 of 0.6 followed by induction with 0.5 mM IPTG for 1 h. Periplasms were prepared, and mannose binding assays were performed as described (5, 22). The presence of FimH, FimC, and dscFimH in the periplasm and in mannose binding assays was monitored by immunoblotting using anti-FimCH antibodies (provided by MedImmune, Gaithersburg, MD).
Hemagglutination Assays.
pBad-Kn, pBad-FimH, and pBad-dscFimH were transformed into ORN103/pETS10. ORN103 does not produce type 1 pili, as it lacks the fim genes (23), and pETS10 encodes a fimH− type 1 gene cluster (S.J.H. and E. T. Saulino, unpublished work). The strains were diluted 1:100 into Luria broth and were grown to an OD600 of 0.8 followed by induction with 0.1 mM IPTG and 0.02% arabinose for 1 h. The cells were harvested, and hemagglutination assays were performed (24).
Protein Purification.
The FimCH complex was purified from the periplasm of C600/pHJ9205/pHJ20 (5, 22). Periplasmic extracts were dialyzed against 20 mM Mes (pH 5.4) and were injected onto a Source 15S column (Pharmacia), and FimCH was eluted at 65 mM NaCl. The eluate was injected onto a Butyl4FF column (Pharmacia) in 0.55 M (NH4)2SO4/20 mM Mes (pH 5.4), and FimCH was eluted at 0.3 M (NH4)2SO4. The FimCH complex was brought to 3 M urea to separate the two proteins. Pure FimH in 3 M urea was collected from the flow through of a Source 15S column (Pharmacia). DscFimH was purified from the periplasm of C600/pTrc-dscFimH (22). The periplasm was dialyzed against 20 mM Tris⋅Cl (pH 8.8), and dscFimH was collected from the flow through of a Source 15Q column (Pharmacia). This flow through was injected onto a Butyl4FF column (Pharmacia) in 0.9 M (NH4)2SO4/20 mM Tris⋅Cl (pH 8.8), and dscFimH was eluted at 0.4 M (NH4)2SO4. The eluate was loaded onto a Source 15S column (Pharmacia) in 20 mM Mes (pH 4.7), and dscFimH was eluted at 55 mM NaCl.
Computer Modeling.
Modeling was performed by using sybyl (Tripos Associates, St. Louis) and insight ii (Molecular Simulations, Waltham, MA) running on a Silicon Graphics (Mountain View, CA) workstation.
Fluorescence and CD.
FimH or dscFimH (22.5 μg) in 20 mM Mes (pH 6.5) + 4 mM DTT was incubated in the appropriate urea concentration. Fluorescence was measured by using an excitation wavelength of 290 nm with emission at 350 nm on an AlphaScan PTI fluorometer (Photon Technologies International, South Brunswick, NJ). CD spectra were measured from 150 μg of protein in 20 mM Mes (pH 6.5) by using a 0.02-cm cell in a JASCO J715 spectropolarimeter. Denatured proteins (in 9 M urea) were diluted to 0.45 M urea to induce refolding.
Results
In Vivo Characterization of dscFimH.
The amino terminal extension of FimG is predicted to complete the Ig fold of the FimH pilin domain in a canonical fashion by interacting anti-parallel to its C-terminal F strand (8, 15). Thus, the DNA sequence encoding the first 13 amino acids of FimG (referred to as the donor strand sequence) was provided to FimH in cis, by fusing it to the 3′ end of fimH to create what will be called donor strand complemented FimH (dscFimH) (Fig. 1B). A hairpin loop region present in PapD consisting of Asp-Asn-Lys-Gln was inserted upstream of the donor strand to allow the donor strand to fold back into the groove of the FimH pilin domain. FimH, FimH + FimC, or dscFimH were expressed separately, and periplasmic extracts were prepared. The presence of FimC and FimH or dscFimH in periplasmic extracts was monitored by immunoblotting using anti-FimCH antibodies. FimH was degraded when expressed alone but was stabilized by the co-expression of the chaperone (Fig. 1C, lanes 2 and 3). In contrast to FimH, dscFimH was stable in the periplasm in the absence of FimC (Fig. 1C, lane 4). Therefore, the addition of the donor strand sequence to FimH resulted in the production of a protein that was now stable in the periplasm without the chaperone.
The FimC-FimH complex can be purified from the periplasm by using mannose-Sepharose chromatography (5). Thus, the ability of dscFimH to bind mannose was tested to determine whether it was properly folded in vivo. FimH bound to mannose-Sepharose beads when it was co-expressed with FimC and was eluted as a FimC-FimH complex (Fig. 1D, lane 4). Because FimH is degraded in the absence of FimC, no full length FimH bound to or eluted from the mannose-Sepharose beads when FimH was expressed by itself (Fig. 1D, lane 2). In contrast, dscFimH bound to and specifically eluted from the mannose beads when expressed alone (Fig. 1D, lane 3). When FimC was co-expressed with dscFimH, it did not form a complex with dscFimH and thus did not co-elute with dscFimH from the mannose-Sepharose beads (Fig. 1D, lane 5).
The ability of dscFimH to complement a fimH− type 1 gene cluster was tested (Table 1). Complementation of a fimH− type 1 gene cluster with fimH resulted in the production of hemagglutination positive bacteria. In contrast, complementation of the same strain with dscfimH resulted in hemagglutination-negative bacteria. Thus, dscFimH was not incorporated into the pilus. The added donor strand presumably occupied the groove and completed the Ig fold of the FimH pilin domain, thus shielding the surface that would normally interact first with the chaperone and then with another subunit in the pilus. Hence, dscFimH did not bind FimC nor assemble into the pilus. Thus, the absence of a C-terminal G β-strand in pilus subunits dictates the need for a chaperone to provide this missing steric information, presumably to promote folding and to provide an assembly surface.
Table 1.
DscFimH does not complement a fimH− type 1 gene cluster
| Strain | HA titer |
|---|---|
| ORN103/pUT2002/pBad | 0 |
| ORN103/pUT2002/pBad-FimH | 32 |
| ORN103/pUT2002/pBad-dscFimH | 0 |
Hemagglutination titers were measured as described (24). The results shown are representative of three or more independent experiments.
Modeling of the N-terminal Extension of FimG into the FimH Pilin Domain.
The N-terminal extension of FimG was modeled into the FimH pilin domain to compare this interaction to that of the crystal structure of the G1β strand of FimC with the FimH pilin domain. Because of the twist in the β-sheet formed by strands D, C, and F, the G1β strand of FimC is unable to satisfy all potential backbone hydrogen bonding interactions with the F strand of FimH (Fig. 2A). This may explain why the addition of a sequence of FimC onto the C terminus of FimH did not result in as stable of a protein as dscFimH. In contrast, the antiparallel interaction of the FimG donor strand with the F strand of FimH satisfies all potential backbone hydrogen bonding interactions between the respective strands (Fig. 2 B and C). The canonical Ig folds completed by subunits in the pilus may explain the dramatic increase in stability of subunit-subunit interactions compared with chaperone-subunit interactions (25).
In Vitro Folding of dscFimH.
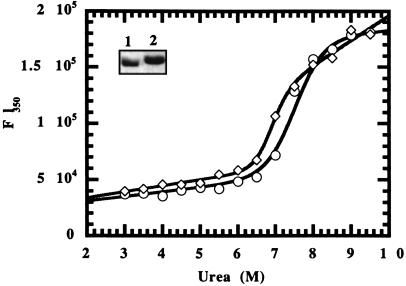
FimH and dscFimH were purified, and urea denaturation was measured (Fig. 3). The presence of 3 M urea was required to stabilize FimH when it was purified away from the FimCH complex, presumably to protect the hydrophobic core of FimH, which is normally occupied by the G1β strand of FimC. FimH retained its native structure in 3 M urea as determined by circular dichroism (CD) (see Fig. 4A) and its ability to bind mannose. FimH and dscFimH had similar denaturation curves, with denaturation complete only above concentrations greater than 8.5 M urea (+ 4 mM DTT) as determined by tyrosine fluorescence spectroscopy emission maxima (350 nm) (Fig. 3). FimH did not begin to denature until 6.5 M urea with the midpoint of the denaturation curve occurring at approximately 7.5 M urea, and thus was not denatured in 4 M urea, as previously assumed (26).
Figure 3.
Urea denaturation curves of FimH and dscFimH. Shown is a Coomassie blue-stained SDS/PAGE gel of purified FimH and dscFimH (inset lanes 1 and 2, respectively). FimH (circles) and dscFimH (diamonds) were incubated with increasing concentrations of urea + 4 mM DTT, and the change in fluorescence at 350 nm was measured to monitor denaturation.
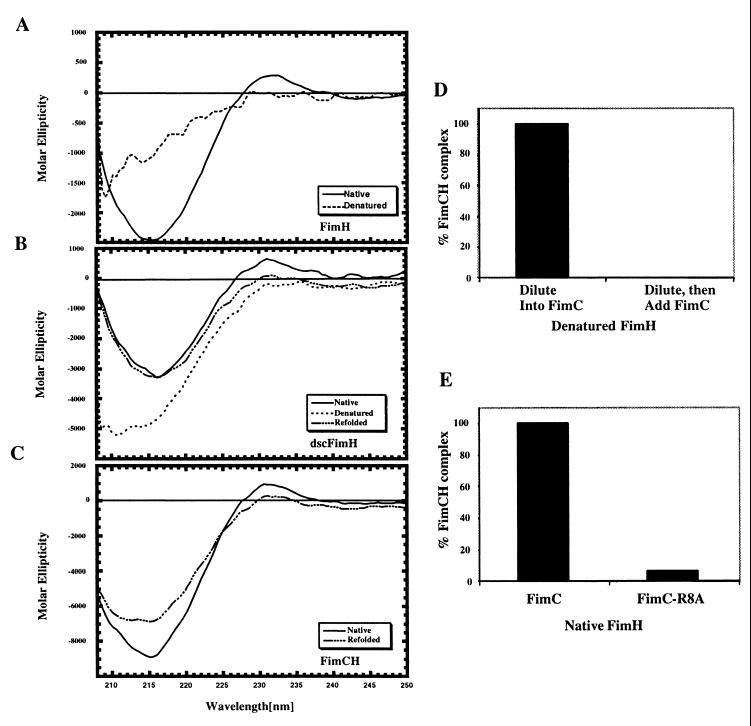
Figure 4.
Refolding of dscFimH and FimH. The CD spectra were measured for native (solid line) and 9 M urea denatured (dashed line) FimH (A), dscFimH (B), and native FimCH (C). The CD spectra of denatured dscFimH after rapid dilution to 0.45 M urea (long and short dashed line) (B) and denatured FimH after rapid dilution to 0.45 M urea in the presence (long and short dashed line) (C) or absence of FimC (elicited no signal attributable to aggregation of FimH) were also determined. The ability of FimC to bind to denatured FimH that was subjected to rapid dilution was measured (D). FimC was either present in the diluent (left side) or added after dilution (right side). The ability of FimH separated from FimCH in 3 M urea to bind to FimC (left) or R8A FimC (right) when diluted to 0.45 M urea was measured (E).
An in vitro folding assay was developed to directly test the hypothesis that the missing steric information in the amino acid sequence of pilus subunits is necessary for folding and can be provided in cis. Attempts to refold urea-denatured FimH (obtained from the FimCH complex) and dscFimH were investigated by examining the CD spectra after rapid dilution of the denatured proteins. Before denaturation, FimH (3 M urea) had a virtually identical β-sheet CD spectrum as native dscFimH (Fig. 4 A and B). After denaturation in 9 M urea (+ 4 mM DTT), the CD spectra of FimH and dscFimH became characteristic of non-native proteins (Fig. 4 A and B). Light scattering of the denatured dscFimH indicated the presence of large aggregates, possibly explaining the somewhat unusual CD spectrum of this species. Rapid dilution of dscFimH led to the refolding of the protein into its native β-sheet structure (Fig. 4B). The refolded dscFimH bound mannose and was monodisperse, as shown by light scattering, indicating that it had refolded into its native structure. In contrast, attempts to refold FimH led to insoluble aggregates and therefore elicited no signal after filtering. FimC was unable to bind to denatured FimH after its rapid dilution (Fig. 4D). However, if FimC was present in the diluent, FimH formed a complex with FimC (Fig. 4D) and folded into its native mannose-binding β-sheet structure (Fig. 4C). Thus, in these assays, dscFimH folds independently and FimH folds in the presence but not in the absence of FimC. FimC was capable of binding to native FimH separated from the FimCH complex by 3 M urea (Fig. 4E), confirming that the chaperone can bind folded subunits. Finally, a mutation in Arg8 of FimC, a residue previously shown to be critical in chaperone-subunit complex formation (22), abolished the ability of the mutant protein to bind to native FimH or facilitate re-folding of denatured FimH (Fig. 4E).
Discussion
In general, cytoplasmic chaperones such as DnaK/DnaJ/GrpE and GroEL/GroES are thought to facilitate protein folding by preventing premature exposure of hydrophobic surfaces to stabilize folding intermediates and prevent nonproductive interactions (reviewed in refs. 27–31). The cytoplasmic chaperones are thought to bind to their substrates in an unfolded or semifolded state. The substrates are then released in a non-native state from the chaperone into the cytoplasm where folding occurs. Cytoplasmic chaperones use ATP hydrolysis for proper function and are not thought to contribute steric information during the folding of their substrate proteins. In contrast, PapD-like periplasmic chaperones function distinctly from these cytoplasmic chaperones; they transiently contribute steric information to pilus subunits to facilitate their folding. In addition, unlike GroEL, which is a large macromolecular complex of two heptameric rings (32), PapD-like chaperones are small proteins that function as monomers, and ATP is not required for PapD function (33).
The addition of the N-terminal extension of FimG onto FimH resulted in the production of a stable mannose binding protein in the periplasm without the co-expression of the chaperone FimC. In vitro folding assays demonstrated that dscFimH was able to fold in the absence of FimC whereas FimH folded only in the presence of FimC. One interpretation of this data is that the amino acid sequence of a pilus subunit is missing steric information necessary for folding and that this missing information is supplied by the chaperone. Thus, the information for folding is contained not in one polypeptide but in two distinct polypeptides. In contrast, in dscFimH, the steric information normally provided by the chaperone is now present in a single polypeptide chain, provided by the sequence corresponding to the N terminus of FimG. The missing sequence provided in cis most likely allows the pilin domain of FimH to fold into a perfectly canonical Ig fold, mimicking the fold that is otherwise completed by FimG in the tip fibrillum.
Alternatively, it is possible that the function of PapD-like chaperones is merely to bind to a subunit after it folds, thereby stabilizing it and simultaneously capping its interactive surface. This could explain the ability of the chaperone to bind to folded subunits and would argue that PapD-like chaperones do not contribute steric information to subunits during folding. However, several lines of evidence argue against this model. First, the native structure of the subunit is unstable. It contains a deep groove on its surface that exposes its hydrophobic core. Thus, the folding of a subunit before an interaction with the chaperone would generate an unstable protein with an exposed hydrophobic core. Pilus subunits are known to spontaneously aggregate into rod-like and fibrillae-like fibers when not bound to a chaperone (34), and such aggregative interactions are toxic in the periplasm (19). Thus, if a subunit were completely folded before binding to the chaperone, there would be a competition between nonproductive subunit-subunit interactions in the periplasm and chaperone binding. Instead, a donor strand complementation mechanism has evolved wherein the folding of a subunit is coupled to the capping of its interactive groove and to its stabilization in the periplasm (15, 16). This elegant mechanism ensures that every folded subunit has its assembly surface simultaneously capped as it forms, eliminating the possibility of nonproductive aggregation reactions in the periplasm.
P pilus subunits are degraded by the DegP protease when expressed in the absence of PapD (19). If PapG did not require the chaperone for folding, then its expression in the absence of both a chaperone and DegP should result in the formation of a native receptor binding protein. This was not the case. In the absence of both DegP and PapD, PapG was unable to fold into a native receptor binding conformation and remained associated with the cytoplasmic membrane (19). In addition, it has been shown by using in vitro protease degradation assays that DegP only degrades proteins in their non-native state and requires the reduction of disulfide bonds in target proteins for degradation (35, 36). Native pilus subunits all have disulfide bonds. Thus, if subunits folded before chaperone recognition, they seemingly would be resistant to DegP degradation because they would possess native structure and disulfide bonds. However, DegP is known to degrade P pilus subunits that are expressed in the absence of PapD.
We suggest the following model for PapD-like chaperone function. The conserved Arg8 and Lys112 residues of the chaperone anchor the very C-terminal carboxyl group of the subunit F strand in the chaperone cleft. Subsequent β-zippering of the F strand along the chaperone G1β strand may facilitate the formation of the F β-strand of the pilus subunit. These interactions would position the strand F hydrophobic side chains of the subunit in register with the G1β strand alternating hydrophobic residues of the chaperone to facilitate the formation of tertiary structure and simultaneous capping of the interactive subunit groove via donor strand complementation. The groove present in the subunit is then used as an assembly surface for the building of the pilus on the bacterial cell. Similar to the pilus biogenesis paradigm, MHC class II molecules have their peptide binding groove occupied during folding by a protein called the invariant chain that prevents nonproductive peptide binding to MHC class II in the endoplasmic reticulum (37, 38).
Intramolecular chaperones (IMCs), which include the pro regions of subtilisin and α lytic protease (reviewed in refs. 39–41), have also been shown to facilitate protein folding by providing steric information (42, 43). IMCs are pro-peptides that are typically N-terminal to the protein they fold and are proteolytically degraded after folding. In the case of subtilisin and α lytic protease, the IMCs mediate folding by binding to the active site of the protease in a substrate-like manner and can then remain bound to the folded protease to act as inhibitors of enzymatic activity (44, 45). Thus, IMCs and PapD-like chaperones function similarly in that they contribute steric information to facilitate folding. However, unlike an IMC, a PapD-like chaperone is a separate polypeptide that transiently completes the fold of the native subunit to stabilize it while simultaneously capping its interactive surface.
The donor strand complementation mechanism presented here could be useful in vaccine development. FimH-based vaccines, including full-length FimH proteins complexed with the FimC chaperone, have been shown to protect mice and monkeys from experimental bladder infections (10, 46). Now, dscFimH vaccines containing the full-length FimH adhesin independent of the FimC chaperone can be produced. Preliminary studies with dscFimH have shown that these proteins induce anti-FimH antibodies that are protective in a murine cystitis model, arguing that this represents promising technology for FimH as well as for any adhesin based vaccine to prevent a multitude of bacterial infections.
Acknowledgments
We thank Stefan Knight for insightful discussions. This work was supported by National Institutes of Health Training Grant 5T32AI07172 (to M.M.B.) and National Institutes of Health Grants RO1DK51406 and RO1AI29549 (to S.J.H.) and R01DK13332 (to C.F.).
Abbreviation
- IMC
intramolecular chaperone
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130183897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130183897
References
- 1.Hung D L, Knight S D, Woods R M, Pinkner J S, Hultgren S J. EMBO J. 1996;15:3792–3805. [PMC free article] [PubMed] [Google Scholar]
- 2.Soto G E, Hultgren S J. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindberg F, Tennent J M, Hultgren S J, Lund B, Normark S. J Bacteriol. 1989;171:6052–6058. doi: 10.1128/jb.171.11.6052-6058.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts J A, Marklund B-I, Ilver D, Haslam D, Kaack M B, Baskin G, Louis M, Mollby R, Winberg J, Normark S. Proc Natl Acad Sci USA. 1994;91:11889–11893. doi: 10.1073/pnas.91.25.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones C H, Pinkner J S, Nicholes A V, Slonim L N, Abraham S N, Hultgren S J. Proc Natl Acad Sci USA. 1993;90:8397–8401. doi: 10.1073/pnas.90.18.8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orndorff P E, Falkow S. J Bacteriol. 1984;159:736–744. doi: 10.1128/jb.159.2.736-744.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinton C C., Jr Trans NY Acad Sci. 1965;27:1003–1165. doi: 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 8.Jones C H, Pinkner J S, Roth R, Heuser J, Nicholes A V, Abraham S N, Hultgren S J. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulvey M A, Lopez-Boado Y S, Wilson C L, Roth R, Parks W C, Heuser J, Hultgren S J. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 10.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner J S, Burlein J, Barren P, Koenig S, Leath S, Jones C H, et al. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 11.Krogfelt K A, Bergmans H, Klemm P. Infect Immun. 1990;58:1995–1999. doi: 10.1128/iai.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connell H, Agace W, Klemm P, Schembri M, Marild S, Svanborg C. Proc Natl Acad Sci USA. 1996;93:9827–9832. doi: 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuehn M J, Normark S, Hultgren S J. Proc Natl Acad Sci USA. 1991;88:10586–10590. doi: 10.1073/pnas.88.23.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hultgren S J, Lindberg F, Magnusson G, Kihlberg J, Tennent J M, Normark S. Proc Natl Acad Sci USA. 1989;86:4357–4361. doi: 10.1073/pnas.86.12.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhury D, Thompson A, Sojanoff V, Langermann S, Pinkner J, Hultgren S J, Knight S. Science. 1999;285:1061–1065. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 16.Sauer F G, Futterer K, Pinkner J S, Dodson K W, Hultgren S J, Waksman G. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 17.Holmgren A, Brändén C. Nature (London) 1989;342:248–251. doi: 10.1038/342248a0. [DOI] [PubMed] [Google Scholar]
- 18.Pellecchia M, Guntert P, Glockshuber R, Wuthrich K. Nat Struct Biol. 1998;5:885–890. doi: 10.1038/2325. [DOI] [PubMed] [Google Scholar]
- 19.Jones C H, Danese P N, Pinkner J S, Silhavy T J, Hultgren S J. EMBO J. 1997;16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amann E, Ochs B, Abel K J. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 21.Guzman L-M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slonim L N, Pinkner J S, Branden C I, Hultgren S J. EMBO J. 1992;11:4747–4756. doi: 10.1002/j.1460-2075.1992.tb05580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurer L, Orndorff P E. J Bacteriol. 1987;169:640–645. doi: 10.1128/jb.169.2.640-645.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hultgren S J, Duncan J L, Schaeffer A J, Amundsen S K. Mol Microbiol. 1990;4:1311–1318. doi: 10.1111/j.1365-2958.1990.tb00710.x. [DOI] [PubMed] [Google Scholar]
- 25.Striker R, Jacob-Dubuisson F, Frieden C, Hultgren S J. J Biol Chem. 1994;269:12233–12239. [PubMed] [Google Scholar]
- 26.Pellecchia M, Sebbel P, Hermanns U, Wuthrich K, Glockshuber R. Nat Struct Biol. 1999;6:336–339. doi: 10.1038/7573. [DOI] [PubMed] [Google Scholar]
- 27.Ellis R J. Semin Cell Dev Biol. 2000;11:1–5. doi: 10.1006/scdb.1999.0345. [DOI] [PubMed] [Google Scholar]
- 28.Ellis R J, Hemmingsen S M. Trends Biochem Sci. 1989;14:339–342. doi: 10.1016/0968-0004(89)90168-0. [DOI] [PubMed] [Google Scholar]
- 29.Ellis R J, Hartl F U. Curr Opin Struct Biol. 1999;9:102–110. doi: 10.1016/s0959-440x(99)80013-x. [DOI] [PubMed] [Google Scholar]
- 30.Fink A L. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 31.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 32.Braig K, Otwinowski Z, Hedge R, Boisvert D C, Joachimiak A, Horwich A L, Sigler P B. Nature (London) 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 33.Jacob-Dubuisson F, Striker R, Hultgren S J. J Biol Chem. 1994;269:12447–12455. [PubMed] [Google Scholar]
- 34.Bullitt E, Jones C H, Striker R, Soto G, Jacob-Dubuisson F, Pinkner J, Wick M J, Makowski L, Hultgren S J. Proc Natl Acad Sci USA. 1996;93:12890–12895. doi: 10.1073/pnas.93.23.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim K I, Park S-C, Kang S H, Cheong G-W, Chung C H. J Mol Biol. 1999;294:1363–1374. doi: 10.1006/jmbi.1999.3320. [DOI] [PubMed] [Google Scholar]
- 36.Kolmar H, Waller P R H, Sauer R T. J Bacteriol. 1996;178:5925–5929. doi: 10.1128/jb.178.20.5925-5929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Germain R N. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 38.Romagnoli P, Germain R N. J Exp Med. 1994;180:1107–1113. doi: 10.1084/jem.180.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinde U, Inouye M. Semin Cell Dev Biol. 2000;11:35–44. doi: 10.1006/scdb.1999.0349. [DOI] [PubMed] [Google Scholar]
- 40.Inouye M. Enzyme. 1991;45:314–321. doi: 10.1159/000468904. [DOI] [PubMed] [Google Scholar]
- 41.Eder J, Fersht A R. Mol Microbiol. 1995;16:609–614. doi: 10.1111/j.1365-2958.1995.tb02423.x. [DOI] [PubMed] [Google Scholar]
- 42.Shinde U, Liu J J, Inouye M. Nature (London) 1997;389:520–522. doi: 10.1038/39097. [DOI] [PubMed] [Google Scholar]
- 43.Shinde U, Fu X, Inouye M. J Biol Chem. 1999;22:15615–15621. doi: 10.1074/jbc.274.22.15615. [DOI] [PubMed] [Google Scholar]
- 44.Sauter N K, Mau T, Rader S D, Agard D A. Nat Struct Biol. 1998;5:945–950. doi: 10.1038/2919. [DOI] [PubMed] [Google Scholar]
- 45.Baker D, Silen J L, Agard D A. Proteins. 1992;12:339–344. doi: 10.1002/prot.340120406. [DOI] [PubMed] [Google Scholar]
- 46.Langermann S, Mollby R, Burlein J E, Palaszynski S R, Auguste C G, DeFusco A, Strouse R, Schenerman M A, Hultgren S J, Pinkner J S, et al. J Infect Dis. 2000;181:774–778. doi: 10.1086/315258. [DOI] [PubMed] [Google Scholar]