Abstract
The novel antiviral protein cyanovirin-N (CV-N) was initially discovered based on its potent activity against the human immunodeficiency virus (HIV). Subsequent studies identified the HIV envelope glycoproteins gp120 and gp41 as molecular targets of CV-N. More recently, mechanistic studies have shown that certain high-mannose oligosaccharides (oligomannose-8 and oligomannose-9) found on the HIV envelope glycoproteins comprise the specific sites to which CV-N binds. Such selective, carbohydrate-dependent interactions may account, at least in part, for the unusual and unexpected spectrum of antiviral activity of CV-N described herein. We screened CV-N against a broad range of respiratory and enteric viruses, as well as flaviviruses and herpesviruses. CV-N was inactive against rhinoviruses, human parainfluenza virus, respiratory syncytial virus, and enteric viruses but was moderately active against some herpesvirus and hepatitis virus (bovine viral diarrhea virus) strains (50% effective concentration [EC50] = ∼1 μg/ml) while inactive against others. Remarkably, however, CV-N and related homologs showed highly potent antiviral activity against almost all strains of influenza A and B virus, including clinical isolates and a neuraminidase inhibitor-resistant strain (EC50 = 0.004 to 0.04 μg/ml). When influenza virus particles were pretreated with CV-N, viral titers were lowered significantly (>1,000-fold). Further studies identified influenza virus hemagglutinin as a target for CV-N, showed that antiviral activity and hemagglutinin binding were correlated, and indicated that CV-N′s interactions with hemagglutinin involved oligosaccharides. These results further reveal new potential avenues for antiviral therapeutics and prophylaxis targeting specific oligosaccharide-comprised sites on certain enveloped viruses, including HIV, influenza virus, and possibly others.
Though vaccination has been very successful in reducing the human cost of influenza virus infection, the search continues for effective therapeutic or prophylactic agents that could further diminish the threat of, or improve response to, widespread outbreaks (J. C. de Jong, E. C. J. Claas, A. D. M. E. Osterhaus, R. G. Webster, and W. L. Lim, Letter, Nature 389:554, 1997). Toward that end, the recent development of influenza virus neuraminidase inhibitors such as zanamivir and oseltamivir has been noteworthy (14). Though inhibitors of this class have proven clinically effective, further in vitro studies have shown that neuraminidase inhibitor-resistant strains of influenza virus can arise rapidly in the presence of therapeutic agents (19). Unfortunately, in some cases such in vitro resistance has been conferred by only a single amino acid substitution in the viral hemagglutinin (HA) protein (29). Though similar results have not been reported in clinical settings, the influenza virus genome is prone to rapid mutation, and the development of resistance to any agent finely targeted to a viral protein may prove inevitable.
To develop additional active agents against influenza virus, it may therefore be advantageous to target other potential vulnerabilities in the viral life cycle. One potential target is suggested by the apparent propensity of the influenza virus surface glycoproteins to become increasingly glycosylated as they adapt to new susceptible host populations (3). For example, in some strains of influenza virus the HA protein acquires additional glycosylation sites after passaging in humans (25, 30). Though it is clear that the virus can rapidly mutate to add (or delete) glycosylation sites to gain advantage in the human host, the virus itself does not control the structure of the specific oligosaccharides added to those sites. Thus, an agent targeted to the oligosaccharides on the influenza virus glycoproteins might be active against a wide variety of influenza virus strains.
The antiviral protein cyanovirin-N (CV-N) has broad, potent anti-human immunodeficiency virus (HIV) activity (8) and a mechanism of action that is based on the specific targeting of high-mannose oligosaccharides on the HIV envelope glycoproteins gp120 and gp41 (6, 22, 28). As similar oligosaccharides are known to be present on other viruses, including influenza virus, we screened CV-N against a wide array of enteric and respiratory viruses as well as flaviviruses and herpesviruses. Here we demonstrate that CV-N is potently active against almost all tested strains of influenza virus, while having much lower activity against a few select viruses and no activity against the majority of other viruses tested. Additional studies on influenza virus imply a specific molecular target for CV-N. Finally, both a CV-N-sensitive strain and a CV-N-resistant strain of influenza virus were studied to identify the potential basis of resistance.
MATERIALS AND METHODS
Purified recombinant CV-N, a unique 11-kDa cyanobacterial protein, was produced in Escherichia coli as reported previously (21). Functional homologs of CV-N, incorporating an Ala, Gln, or Val substitution at Asn 30 and/or a Gly substitution at Pro 51, were produced as described elsewhere (20). Europium-labeled CV-N (Eu-CV-N) was prepared by addition of chelated europium to free amino groups on CV-N. The labeling was performed by Perkin-Elmer under contract. HA isolated from influenza virus strain A/Beijing/262/95 (H1N1) grown in chicken eggs was kindly provided by Yumi Matsuoka at the Centers for Disease Control and Prevention, Atlanta, Ga. Viral lysates from A/Taiwan/1/86 (H1N1), A/Kiev/301/94 (H3N2), and B/Leningrad/86/93 were purchased from Advanced ImmunoChemical Inc. (Long Beach, Calif.) Anti-CV-N rabbit polyclonal antibodies were provided by Jim McMahon (Molecular Targets Development Program, Center for Cancer Research, National Cancer Institute). Anti-HA antibodies (H3N2) were purchased from Argene Inc. (Massapequa, N.Y.).
Cells and virus.
All cells and viruses were obtained from the American Type Culture Collection (ATCC; Manassas, Va.), unless indicated otherwise. HSB-2 cells were cultured in RPMI 1640; HeLa, Vero, and HEp-2 cells were cultured in Dulbecco's modified Eagle medium (DMEM); MRC-5 cells were cultured in Eagle's minimal essential medium; and BCBL-1, HepG2-2.2.2.15 (AIDS Research and Reference Reagent Program, Bethesda, Md.), and P3HR1 cells were cultured in RPMI 1640. All media were supplemented with 10% fetal bovine serum (FBS), except in assays for coxsackieviruses, enteroviruses, rhinovirus, echovirus, adenovirus, respiratory syncytial virus (RSV), and parainfluenza and influenza viruses, and 2.0 mM l-glutamine, 100 U of penicillin/ml, and 100 mg of streptomycin/ml.
Antiviral bioassays.
CV-N and its functional homologs were evaluated against a range of virus isolates by standardized cytopathic effect (CPE) inhibition assays (9) or specific antiviral assays as specified in Tables 1 and 2. In all assays the cytotoxic effects of compounds were measured spectrometrically by formazan dye reduction using either XXT {2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide} or MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt] (CellTiter 96; Promega, Madison, Wis.). Cell-free virus stocks, where appropriate, were produced, clarified by centrifugation, titered in the antiviral assay versus an appropriate control compound, if available, and stored at −80°C until use. For all CPE assays, viruses were titered to provide between 80 and 95% killing of the host cell (approximate multiplicity of infection, 0.01 to 0.1). The varicella-zoster virus (VZV) assay was adjusted to 100 PFU per well. Numbers of chronically or latently infected cells in assays using quantitation of virus DNA were adjusted to provide between 1,000 and 10,000 copies of viral DNA per assay well.
TABLE 1.
Viruses against which CV-N was determined to be inactive (no inhibition of virus-induced cytopathicity at 10 μg/ml)
| Virus | Cells for propagation | Virus strain(s) (if applicable) | Virus assay | End point (Antiviral cytotoxicity) |
|---|---|---|---|---|
| HBV | HepG2-2.2.15 | NAa | Real-time PCR | HBV DNA copy number (MTS) |
| HSV-2 | Vero | MS | CPE | MTS |
| HCMVb | MRC-5 | AD169 | CPE | MTS |
| VZV | MRC-5 | Ellen | Plaque reduction | Visual counting (MTS) |
| HHV-8 | BCBL-1 | NA | Real-time PCR | HH-V8 DNA number copy (MTS) |
| Rhinovirus | MRC-5 | 1A, 14, 26, 1B | CPE | MTS |
| RSV | Vero | Long | CPE | MTS |
| Coxsackie A virus | MRC-5 | A21-Kuykendall, A7-275/58 | CPE | MTS |
| Coxsackie B virus | Vero | B3-Nancy, B1-201468 | CPE | MTS |
| Enterovirus | MC-5 | 70 (J670/71) | CPE | MTS |
| Echovirus | MRC-5 | 21 (HA antigen 27513) | CPE | MTS |
| Adenovirus | HeLa | Type 2 | CPE | MTS |
NA, not applicable.
HCMV, human cytomegalovirus.
TABLE 2.
Moderate in vitro activity of CV-N against select viruses
| Virus (strain) | Cell line | EC50 (μg/ml) | IC50 (μg/ml) | Antiviral index |
|---|---|---|---|---|
| Parainfluenza virus (type 3) | Hep-1 | 15.4 | >10 | >1.9b |
| HSV-1 (ATCC VR-260) | Vero | 0.68 ± 0.12 | 2.3 ± 0.12 | 3.3b |
| EBVa | P3HR1 | 1.2 ± 0.70 | 5.3 ± 0.03 | 4.3c |
| HHV-6 (GS)a | HSB-2 | 1.6 ± 0.06 | 7.1 ± 0.03 | 4.4d |
| BVDV (D/8) | MDBK | 0.54 ± 0.10 | 7.1 ± 0.15 | 13b |
EBV and HHV-6 cell lines are chronically infected with the indicated viruses.
Cytoprotection antiviral assay using XTT dye reduction.
TaqMan antiviral assay.
Cytoprotection-coculture antiviral assay using XTT dye reduction.
CPE assays.
The viruses assayed by determination of CPE were human herpes simplex virus type 1 (HSV-1; strain ATCC VR-260) and HSV-2 (strain MS), human cytomegalovirus (strain AD169), rhinovirus (strains 1A, 1B, 14, and 26), RSV (Long strain), coxsackie A virus (strains A21-Kuykendall and A7-275/58) and B virus (strains B3-Nancy and B1-201468), enterovirus (strain 70([J7670/71]), echovirus (strain 21[HA antigen 27513]), adenovirus (type 2), parainfluenza virus (type 3), and bovine viral diarrhea virus (BVDV; strain D/8). The CPE assay measures the ability of test material to protect the target cells from the CPE of replicating virus. In all assays the number of host cells (Tables 1 to 3) for each virus was optimized to provide exponential cell growth throughout the 5- to 7-day antiviral assay. Antiviral activity (cytoprotection) was measured quantitatively by formazan dye reduction using either XTT or MTS as indicated in Tables 1 to 3. Antiviral and toxicity data were reported as the concentration of compound required to inhibit by 50% virus-induced cell killing (50% effective concentrations [EC50]), the concentrations of compound required to lower cell viability by 50% (cytotoxicity; IC50), and the antiviral index, representing the ratio of IC50 to EC50. All reported data were derived from triplicate determinations.
TABLE 3.
In vitro activity of CV-N against strains of influenza virus
| Virus | Straind | EC50 (μg/ml) | IC50 (μg/ml) | Antiviral index |
|---|---|---|---|---|
| Influenza A | Sydney/05/97 (H3N2)a | 0.04 ± 0.16 | >10 | >228 |
| Victoria/3/75 (H3N2)a | 0.005 ± 0.015 | >1.0 | >187 | |
| Mem/8/99 (H3N2)*a | 0.03 ± 0.55 | >10 | >400 | |
| Mem/2/99 (H3N2)*a | 0.15 ± 0.21 | >10 | >69 | |
| Beijing/262/95 (H1N1)b | 0.11 ± 0.04 | >10 | >90 | |
| PR/8/34 (H1N1)a | >10 | 5.8 ± 0.34 | ||
| NWS/33 (H1N1)b | >10 | >10 | ||
| Shangdong/09/93b | 0.2 ± 0.1 | >10 | >50 | |
| Shangdong/09/93-NIRb,c | 0.028 ± 0.008 | >1.0 | >36 | |
| Influenza B | Hong Kong/5/72b | 0.50 ± 0.2 | >10 | >20 |
| Yamanashi/166/98a | 0.04 ± 0.08 | >1.0 | >270 | |
| Mem/3/99*a | 0.02 ± 0.10 | >10 | >473 | |
| Beijing/184/93b | 0.16 ± 0.06 | >10 | >62 | |
| Sichuan/379/99b | 1.3 ± 0.7 | >10 | >8 | |
| Lee/40b | 0.026 ± 0.004 | 8 | 308 |
Assays performed at the Southern Research Institute, Frederick, Md.
Assays performed at the Institute for Antiviral Research, Utah State University, Logan.
Strain is resistant to neuraminidase inhibitors oseltamivir, zanamivir, and RWJ-270201 in vitro (29).
*, clinical strain of influenza virus.
CPE assays for respiratory and enteric viruses.
CPE assays to quantitate the replication of influenza A and B viruses, coxsackie A and B viruses, enterovirus, rhinovirus, echovirus, adenovirus, RSV, and parainfluenza virus were carried out as identified above for the standard CPE assay with the exception of the media used to infect the host cells. Infections with influenza A and B virus stocks were carried out in DMEM supplemented with 0.3% bovine serum albumin (BSA) and 2.0 mg of TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-trypsin/ml. Infection with the remaining viruses was carried out with virus stocks diluted in DMEM containing 2% FBS.
HHV-6 replication.
Replication of human herpesvirus 6 (HHV-6) was determined by a modified CPE assay. Uninfected HSB-2 (ATCC CL-120.1) cells and HSB-2 cells infected with the GS strain of HSB-2 were cocultivated at a 1:1 ratio for 6 days. Morphology changes indicative of HHV-6 replication (increased size and refractoriness) were determined microscopically at assay completion. The number of infected cells per well was determined microscopically (magnification, ×40) for five fields per well in triplicate. Cytotoxicity of CV-N on HSB-2 cells was determined by MTS dye reduction.
Replication of EBV, HHV-8, and HBV.
The optimal assays for measuring the antiviral effect of compounds on the replication of the Epstein-Barr virus (EBV), HHV-8, and hepatitis B virus (HBV) utilize chronically or latently infected cells. Inhibition of virus replication is measured by real-time PCR. The effect of CV-N on EBV replication was quantitated with P3HR1 cells, which are chronically infected by EBV. EBV replication in the presence of CV-N was quantitated at 5 days. Inhibition of HHV-8 replication was quantitated with phorbol ester (tetradecanoyl phorbol acetate; 10 nM)-induced BCBL-1 cells at 4 days postinduction in cell-free supernatants. HBV replication was quantitated in cell-free supernatants from chronically infected HepG2-2.2.2.15 cells after 6 days of treatment. Cell-free supernatants were collected, clarified by centrifugation, and treated with DNase to degrade unencapsidated viral DNA, followed by proteinase K treatment to release encapsidated DNA. Total viral DNA was then quantitated by real-time PCR (PE Applied Biosystems) using virus-specific primers and probes. Viral DNA copy numbers were estimated from a standard curve of known DNA copy numbers. Cytotoxicity of CV-N in all three assays was determined on replicate cultures using MTS.
VZV antiviral assay.
Inhibition of VZV replication was determined on MRC-5 cells with VZV strain Ellen. MRC-5 cells were seeded at 50% confluence in 24-well plates and allowed to establish overnight. The following day, media were aspirated and the cells were infected with 100 PFU of VZV Ellen per well in media containing 2% serum. Following a 1-h virus adsorption, methylcellulose was added to a final concentration of 0.5% in media with 10% serum, and the cultures were incubated for an addition 4 to 6 days to allow plaque formation. Plaques were counted microscopically after fixing and staining MRC-5 monolayers with crystal violet in 20% methanol. The effect of CV-N on MRC-5 cell viability in the presence of 0.5% methylcellulose was determined on replicate cultures with MTS.
Anti-influenza virus bioassays. (i) Viruses.
The following viruses were provided by H. Regnery of the Influenza Branch of the Centers for Disease Control and Prevention: A/Beijing/262/95 (H1N1), A/Sydney/05/97 (H3N2), A/Shangdong/09/93 (H3N2), and B/Beijing/184/93. A/NWS/33 (H1N1) was provided by K. Cochran of the University of Michigan (Ann Arbor). A/PR/8/34 (H1N1) was obtained from F. Schabel, Jr., Southern Research Institute (Birmingham, Ala.). B/Hong Kong/5/72 and B/Lee/40 were obtained from the ATCC. All were passaged in Madin-Darby canine kidney (MDCK) cells to prepare pools for use in these experiments.
(ii) Cells and media.
MDCK cells (from ATCC) were grown in antibiotic-free minimum essential medium (MEM) with nonessential amino acids (Gibco, Long Island, N.Y.) containing 5% FBS (HyClone Laboratories, Logan, Utah) and 0.1% NaHCO3. Test medium consisted of MEM with 0.18% NaHCO3, 10 U of trypsin/ml, 1 μg of EDTA per ml, and 50 μg of gentamicin/ml.
(iii) Cell culture assays.
Three methods were used to assay antiviral activity in vitro: inhibition of virus-induced CPE determined by visual (microscopic) examination of the cells and confirmed by increase in neutral red (NR) dye uptake into cells, virus yield reduction, and virus neutralization. The CPE inhibition method was reported previously by Smee et al. (29) in which seven concentrations of test compound were evaluated against each virus in 96-well flat-bottom microplates of cells. The compounds were added 5 to 10 min prior to virus, which was used at a concentration of approximately 50 cell culture 50% infectious doses per well. This virus challenge dose equated to a multiplicity of infection of approximately 0.001 infectious particles per cell. The tests were read after incubation at 37°C for 72 h. In the NR uptake assay, dye (0.34% concentration in medium) was added to the same set of plates used to obtain the visual scores. After 2 h, the color intensity of the dye absorbed by and subsequently eluted from the cells was determined by the method of Finter (13) with a computerized EL-309 microplate autoreader (Bio-Tek Instruments, Winooski, Vt.). Antiviral activity was expressed as the EC50 value determined by plotting compound concentration versus percent inhibition on semilogarithmic graph paper. Cytotoxicity of compounds was assessed in parallel with the antiviral determinations in the same microplates, except in the absence of virus. From these, IC50 values were determined.
Virus yield reduction assays were performed by growing the virus in the presence of compound for 3 days as described above; then the plates were frozen at −80°C. Dilutions from the wells containing extracellular virus were later titrated by an end point dilution method (24). In this assay, it was not possible to prevent CV-N carryover from the first part of the experiment. This most likely influenced the recovery of virus from the samples.
The ability of CV-N to neutralize the infectivity of influenza viruses was determined by combining virus-containing medium with an equal part of CV-N (final concentration,10 μg/ml) for 30 min at room temperature. Virus was then diluted to extinction in 96-well plates and titrated (24). By this method the surviving virus titer was quantified.
Preparation of influenza virus for gel analysis.
Five T-75 flasks of MDCK cells were infected with a strain of influenza virus at low multiplicity of infection (∼0.001 infectious particles/cell). The medium used was MEM with 10 U of trypsin/ml and 1 μg of EDTA/ml. Virus was allowed to replicate for 3 days, at which time CPE was 100%. The medium from the flasks was pooled and frozen at −80°C for subsequent use. After being thawed to 4°C, the fluids were centrifuged at 600 × g, the pellet was discarded, and the supernatant was retained. The virus was then purified from the supernatant fluid by a modification of the procedure of Arora et al. (2). Briefly, sucrose solutions of 50 and 22% (wt/wt) in 0.15 M NaCl with 0.08% (wt/vol) NaN3 added were prepared. A step gradient with 2 ml of 50% sucrose and 4 ml of 22% sucrose was prepared. The supernatant was added, followed by centrifugation at 100,000 × g for 1 h in a swinging-bucket rotor. The 22% sucrose layer containing the virus was retained, and the mixture was diluted with cell culture medium and centrifuged again at 100,000 × g for 1 h to pellet the virus. Each pellet was suspended in 0.2 ml of the NaCl-NaN3 buffer containing 0.05% thimerosal. Virus pools were then frozen at −80°C until used.
Western blots.
Three commercially available influenza virus lysates (A/Taiwan/1/86 [H1N1], A/Kiev/301/94 [H3N2[, and B/Leningrad/86/93) and two strains of influenza A virus prepared as described above (A/PR/8/34 [H1N1] and A/Sydney/05/97 [H3N2]) were electrophoresed on an 8 to 16% acrylamide gel (Novex) with standard Laemmli buffer systems (18). The gels were electrophoresed at a constant current of 25 mA with cold running buffer at room temperature. After electrophoresis, gels were either stained with Coomassie blue to identify protein bands or blotted onto polyvinylidene difluoride (PVDF) membranes. For the blots, the conditions were a constant voltage of 25 V for 2 h with cold electrode buffer at room temperature. After the blotting, the membranes were blocked overnight at 4°C with 1% BSA (Sigma, St. Louis, Mo.) in phosphate-buffered saline (PBS) and were then washed with PBS-0.05% Tween 20 (TPBS). After being washed, the membranes were incubated with 2 μg of CV-N/ml in PBS overnight at 4°C. After incubation, the membranes were again washed and then incubated with polyclonal rabbit anti-CV-N antibodies for 1 h. After the membranes were washed again with TPBS, they were incubated with goat anti-rabbit secondary antibodies conjugated to horseradish peroxidase (HRP; Amersham) for 1 h. After incubation with the secondary antibodies, the membranes were washed six times with TPBS and then incubated with electrochemiluminescence reagents (Amersham) for 30 s. After being blotted dry, they were exposed to X-ray film (Kodak) for 30 s to 2 min. For visualization of HA with anti-HA antibodies, the blots were blocked and washed as described above and then incubated for 1 h with a 1:1,000 dilution of rabbit polyclonal anti-HA antibodies. After the blot was again washed, the membrane was incubated with goat anti-rabbit secondary antibodies conjugated to HRP and visualized as described above.
CV-N binding studies.
Purified influenza virus HA (A/Beijing 262/95 [H1N1]) at a concentration of 10 pmol/ml in 100-μl aliquots of PBS was bound to individual wells of 96-well protein-binding assay plates (Nunc; Immobilon) by incubation for 2 h at room temperature. Thereafter, the plates were washed three times with TPBS and then blocked by the addition of 200 μl of a solution of 1% BSA in PBS/well, followed by overnight incubation at 4°C and a further wash with TPBS. Eu-CV-N (9.1 pmol/well) was then incubated for 1 h in either PBS alone or increasing concentrations of oligomannose-8 (Man-8). The Eu-CV-N samples were then added to triplicate wells and allowed to incubate for 1 h, followed by six washes with TPBS. After the washes, 100 μl of enhancement solution (Perkin-Elmer) was added to each well, and the plate was monitored for time-resolved fluorescence.
For the binding studies with viral lysates 100 μl of a 1:10,000 dilution of the lysate of either influenza virus A/PR/8/34 (H1N1) or influenza virus A/Sydney/05/97 (H3N2) was bound to the wells of a 96-well plate as described above. The plate was again blocked with BSA, washed with TPBS, and incubated with various concentrations of Eu-CV-N as in the HA binding studies. After a 1-h incubation, the plates were washed thoroughly with TPBS and monitored for time-resolved fluorescence.
RESULTS
In vitro assays to determine the activity of CV-N against a variety of viruses produced results that could be generally separated for three categories of viruses. The first consisted of viruses for which CV-N exhibited no antiviral activity. Included in this group were all of the tested enteric viruses and rhinovirus strains (Table 1). The second category included viruses for which there was moderate antiviral activity (∼1 μg/ml) with low antiviral indices (Table 2). This group comprised herpesviruses and flaviviruses, which showed a level of sensitivity to CV-N similar to that previously reported for the Ebola virus (4). The final category consisted entirely of diverse influenza virus strains (Table 3). These viruses, including laboratory and clinical isolates of both influenza A and B virus and an in vitro-derived neuraminidase inhibitor-resistant strain of influenza A virus, were highly sensitive to CV-N with EC50 values from 0.004 to 0.5 μg/ml. Examples of the antiviral activity of CV-N against strains of influenza A and B virus are shown in Fig. 1. The influenza virus assays were repeated in a second laboratory to confirm the initial results. Interestingly, there were two strains of the influenza virus, A/PR/8/34 (H1N1) and NWS/33 (H1N1), which were insensitive to CV-N. Both of these older, laboratory-adapted strains were not inhibited by CV-N, even at concentrations of 10 μg/ml. Functional homologs of CV-N, engineered to incorporate glycosylation resistance and/or modified stability properties (20), making them potentially more amenable to large-scale production, also showed potent anti-influenza activity (Table 4) essentially indistinguishable from that of the wild-type CV-N.
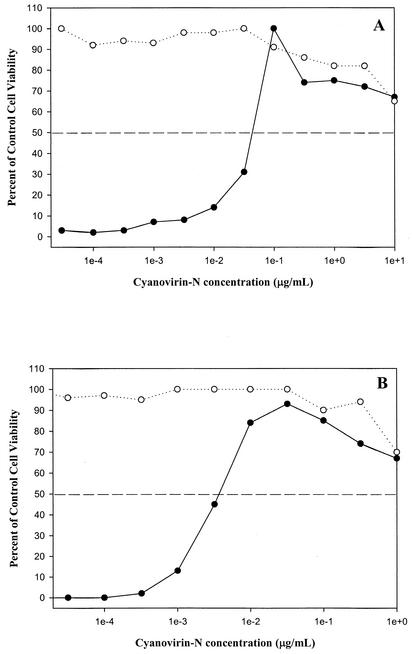
FIG. 1.
Anti-influenza activity of CV-N in MDCK cells infected with influenza virus. Shown is a comparison of the concentration-dependent effects of CV-N on the viability of virus-infected cells (•) and control cells (○) infected with influenza virus A/Sydney/05/97 (H3N2) (A) and influenza virus B/Yamanashi/166/98 (B), as assessed by NR assay.
TABLE 4.
Comparison of anti-influenza activities of functional homologs of CV-Na
| Compound | EC50 (μg/ml) against:
|
|
|---|---|---|
| A/Sydney/05/97 (H3N2) virus | B/Yamanashi/166/98 virus | |
| CV-N | 0.026 ± 0.0023 | 0.052 ± 0.042 |
| P51G | 0.015 ± 0.014 | 0.041 ± 0.017 |
| N30A/P51G | 0.011 ± 0.008 | 0.036 ± 0.007 |
| N30Q/P51G | 0.019 ± 0.019 | 0.077 ± 0.029 |
| N30V/P51G | 0.028 ± 0.034 | 0.066 ± 0.019 |
Assays performed at the Southern Research Institute.
To further explore the basis for the anti-influenza activity, virus neutralization assays in which different strains of influenza virus were pretreated with CV-N and then diluted out and tested for infectivity were performed. In these assays, CV-N showed activity (Table 5) against an H1N1 strain and an H3N2 strain of influenza A virus as well as an influenza B virus strain. However, once again CV-N showed no activity against influenza virus strain A/PR/8/34 (H1N1).
TABLE 5.
Virus-neutralizing activity of CV-N for various strains of influenza virus
| Virus strain | Virus titer (log10 PFU/0.1 ml)a
|
Δlog10 | |
|---|---|---|---|
| −CV-N | +CV-N | ||
| A/Beijing/262/95 (H1N1) | 6.3 ± 0.5 | 3.7 ± 0.3 | −2.6 |
| A/PR/8/34 (H1N1) | 5.8 ± 0.4 | 6.2 ± 0.2 | +0.4 |
| A/Sydney/05/97 (H3N2) | 5.3 ± 0.3 | 0.0 | −5.0 |
| B/Beijing/184/93 | 5.4 ± 0.4 | 0.0 | −5.4 |
−CV-N, without CV-N; +CV-N, with CV-N.
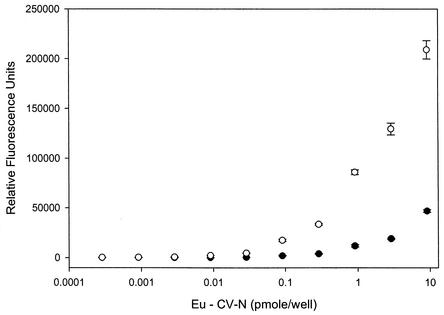
These virus neutralization assays indicated that the likely target for CV-N was present on the influenza virus virion. Accordingly, we set out to establish whether or not CV-N bound to any specific influenza virus proteins and to determine if such binding correlated to the in vitro antiviral data. Initially, lysates from two strains of influenza A virus, the CV-N sensitive A/Sydney/05/97 (H3N2) and the CV-N-resistant A/PR/8/34 (H1N1), were prepared and bound to the wells of a 96-well protein-binding plate. The plate was then incubated with increasing concentrations of Eu-CV-N, and the binding was monitored by time-resolved fluorescence. This experiment indicated that Eu-CV-N bound much more readily to the protein component of the sensitive A/Sydney/05/97 (H3N2) virus than to the resistant A/PR/8/34 (H1N1) viral proteins (Fig. 2). This result correlated well with the whole-cell antiviral assay data and further indicated that CV-N′s antiviral activity might indeed be mediated by specific protein-protein or protein-glycoprotein interactions. To determine which (if any) viral proteins were bound by CV-N, we electrophoresed and blotted three commercially available influenza virus lysates (A/Kiev/301/94 [H3N2], B/Leningrad/86/93, and A/Taiwan/1/86 [H1N1]) onto membranes and incubated them with CV-N. The results (data not shown) indicated that CV-N bound preferentially to a band of protein with a molecular mass of ∼50 kDa.
FIG. 2.
Binding of CV-N to influenza virus lysates. Shown is a comparison of the concentration dependent binding of Eu-CV-N to viral lysates of influenza A/PR/8/34 (H1N1) virus (•) and influenza A/Sydney/05/97 (H3N2) (○), as determined by a time-resolved fluorescence binding assay. Points are the averages ± standard deviations of six replicate determinations.
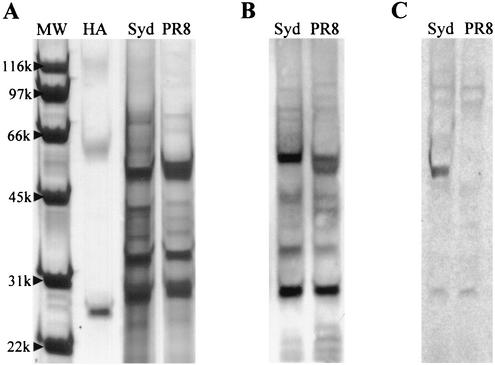
To confirm this result, the A/Sydney/05/97 and A/PR/8/34 sample lysates were electrophoresed on gradient gels and blotted onto PVDF membranes. After the membranes were incubated with CV-N and visualized by anti-CV-N polyclonal antibodies, binding predominantly to a single band of protein was clearly seen with the sensitive A/Sydney/05/97 strain while no significant binding was seen with the resistant A/PR/8/34 strain. Comparison with published studies (16) indicated that the molecular mass of the band binding to CV-N (∼50 kDa) matched that for influenza virus HA1 (Fig. 3). Additional blots for the both the A/Sydney/05/97 and A/PR/8/34 strains with anti-HA antibodies (H3N2) also indicated that the band binding to CV-N was HA1 (Fig. 3). CV-N also appeared to minimally bind to HA2 (molecular mass, ∼28 kDa) in both A/PR/8/34 and A/Sydney/05/97.
FIG. 3.
Analysis by immunoblotting of CV-N binding to influenza A virus lysates. Influenza A virus lysates from either strain A/PR/8/34 (H1N1) (CV-N resistant; PR8) or A/Sydney/05/97 (H3N2) (CV-N sensitive, Syd) were electrophoresed by gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The gels were either stained with Coomassie blue (A) or blotted onto a PVDF membrane and incubated with either anti-HA antibodies (B) or CV-N-specific antibodies (C). Lane MW, molecular weight markers; lane HA, purified HA (A/Beijing/262/95 [H1N1]).
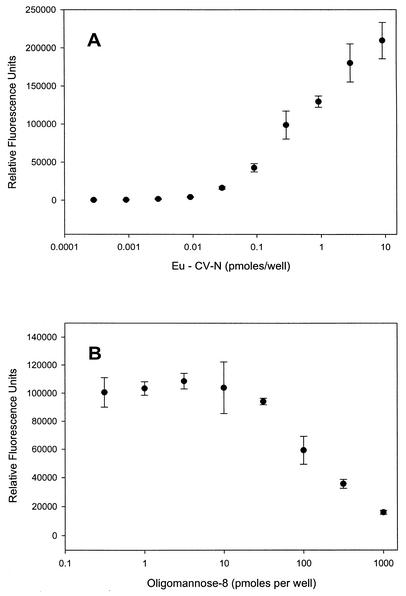
To confirm that influenza virus HA was a target for CV-N, we also performed binding assays to determine if CV-N bound directly to HA purified from influenza virus A/Beijing/262/95 (H1N1) virus. The results of this experiment (Fig. 4A) indicated that CV-N bound to HA in a concentration-dependent manner. Additional experiments were designed to explore whether the binding of CV-N to HA was mediated by protein-carbohydrate interactions. The resulting graph (Fig. 4B) shows a concentration-dependent decrease in the binding of CV-N to HA, reaching a level of 80% inhibition at 1 nmol of Man-8. Taken together, these data indicate that CV-N exerts its antiviral effect on the influenza virus by direct interactions with high-mannose oligosaccharides on influenza virus HA.
FIG. 4.
Binding of CV-N to purified HA isolated from influenza A/Beijing/262/95 (H1N1) virus. (A) Concentration-dependent binding of Eu-CV-N to purified HA as determined by time-resolved fluorescence. One hundred nanograms of CV-N is equivalent to 9.1 pmol of CV-N. (B) Concentration-dependent inhibition of CV-N binding to purified HA by the high-mannose oligosaccharide Man-8. Points are the averages ± standard deviations of four replicate determinations.
DISCUSSION
CV-N has previously been shown to bind high-mannose oligosaccharide structures Man-8 and Man-9 present on the HIV envelope glycoproteins gp120 and gp41 (6, 22, 28). Furthermore, the three-dimensional carbohydrate binding motif of CV-N has been elucidated by both nuclear magnetic resonance titration experiments (5, 27) and X-ray crystallography (7). The unique selectivity of CV-N for high-mannose oligosaccharide structures might explain the activity of CV-N against additional viruses with similar structural constituents. Previous reports had indicated modest in vitro activity of CV-N against both measles virus and HHV-6 but no activity against vaccinia virus, cytomegalovirus, adenovirus type 5, and murine leukemia virus (7, 11). More recently, CV-N has been shown to have moderate in vitro and in vivo activity against the Ebola virus (4). To further elucidate the spectrum of antiviral activity of CV-N, we evaluated in vitro activity of the protein against a wide range of viruses, including those normally transmitted via respiratory, enteric, and sexual routes. For the majority of viruses tested (n = 17), CV-N failed to inhibit replication (Table 1), and it is noteworthy that there are no reports of high-mannose oligosaccharides being present on the CV-N-insensitive rhinoviruses or on the enteric viruses examined in this study. Moderate activity of CV-N against parainfluenza virus, HSV-1, EBV, and HHV-6 was found, and moderate activity was also found in an assay using BVDV as a surrogate for hepatitis C virus (Table 2). In all cases, the antiviral index (IC50/EC50) was very low, similar to that previously reported for the Ebola virus (4) and orders of magnitude lower than that reported for HIV (7). In contrast to the viruses against which CV-N had no activity, all of the viruses exhibiting moderate inhibition by CV-N have been reported to produce envelope glycoproteins bearing high-mannose oligosaccharides (12, 17, 23, 26, 31). These results, taken together with the previous literature on the oligosaccharide composition of the envelope glycoproteins from these viruses, provide further evidence that CV-N′s antiviral activity is indeed mediated through specific protein-carbohydrate interactions. The variation in CV-N′s specific antiviral activity against the different viruses might be due to the density and/or arrangement of these oligosaccharides on the viral glycoproteins.
The most striking revelation in the present study was the potent anti-influenza activity of CV-N. The protein was highly active against a wide spectrum of influenza A and B virus strains, including clinical isolates and an in vitro-derived neuraminidase inhibitor-resistant strain. CV-N was active against influenza virus at concentrations as low as 0.005 μg/ml, with antiviral indices of >473 (Table 3). For the sensitive strains of influenza virus, the antiviral activity demonstrated here (Fig. 1) most closely resembles CV-N′s potent anti-HIV activity (8). Aside from HIV, influenza virus is the virus most sensitive to CV-N so far identified. Also, similar to what was found for CV-N′s activity against HIV (8), there is considerable difference in the EC50s reported for different strains of influenza virus. As CV-N is known to be a surface active agent that binds to specific oligosaccharide structures on viral glycoproteins (22), its inhibition is based on first-order kinetics. This mechanism of action is sensitive to the presence of oligosaccharides on the virus to which CV-N binds that may not be directly associated with its antiviral activity. One possible explanation for these differences in sensitivity could be that, due to the relative amount of ancillary CV-N binding motifs on the various influenza virus strains, it takes greater or lesser amounts of CV-N to effectively inhibit viral infection. Significantly, however, unlike CV-N′s universal activity against all strains of HIV so far tested, certain strains of influenza virus were naturally resistant to CV-N. Two strains used in this study, A/PR/8/34 (H1N1) and A/NWS/33 (H1N1), were not inhibited by CV-N at concentrations up to 9 μM.
The results of viral pretreatment studies indicated that CV-N directly neutralized both influenza A and B viruses, including both H1N1 and H3N2 strains. However, the CV-N-resistant influenza virus strain A/PR/8/34 was completely resistant to any direct neutralizing activity. These results (Table 5) suggested that CV-N bound directly to and inactivated the viral particle, preventing subsequent infection, and that both the likely molecular target for CV-N and the basis of CV-N resistance resided in the viral particle. Binding studies utilizing viral lysates from a CV-N-sensitive strain (A/Sydney/05/97 [H3N2]) and a CV-N-resistant strain (A/PR/8/34 [H1/N1]) clearly confirmed high CV-N binding to the viral lysate of the A/Sydney/05/97 strain, relative to that of the A/PR/8/34 strain (Fig. 2).
The three major proteins present on the surface of the influenza virus particle are HA, neuraminidase, and a transmembrane pore protein (M2). Direct action against any one of these three putative molecular targets could result in anti-influenza activity (e.g., surfactant protein D on HA, zanamivir and oseltamivir on neuraminidase, and amantadine on M2). In experiments with purified viral lysates from A/Sydney/05/97 (CV-N sensitive) and A/PR/8/34 (CV-N resistant), the results clearly showed a single band of protein from the A/Sydney/05/97 lysate, with a molecular mass of ∼50 kDa, bound to CV-N; in contrast, no significant CV-N binding to this band of protein was apparent from the A/PR/8/34 viral lysate (Fig. 3). Both strains of virus appeared to contain a band of protein with a lower molecular mass (∼28 kDa) which minimally interacted with CV-N. The 50-kDa CV-N-binding protein band matched the expected molecular mass for influenza virus HA1 (4) and bound to anti-HA antibodies, thereby suggesting that CV-N exerted its anti-influenza activity by directly binding to HA1 and that resistance to CV-N was the result of some change in HA1. The lower-molecular-mass bands from both strains that bound CV-N were identified as HA2 based on both molecular mass and immunological detection. The low level of CV-N binding to this protein may explain the low level of CV-N binding shown by viral lysates of A/PR/8/34 in the earlier CV-N binding studies (Fig. 2).
The lack of CV-N binding to the HA1 glycoprotein from the A/PR/8/34 strain of influenza virus and its resistance to the antiviral effects of CV-N might be explained by a lack of high-mannose oligosaccharides near the cellular binding region of the HA1 protein in that strain. In fact, previous investigations on this laboratory-adapted strain of influenza virus have shown that it lacks a key oligosaccharide structure at the head of the HA1 protein (10). The absent oligosaccharide might otherwise have comprised a key binding motif for CV-N, and its absence might account for the lack of CV-N binding to HA1 and resistance to the anti-influenza virus effects of CV-N. This resistance is similar to that exhibited by influenza A/PR/8/34 virus for the innate mannose-binding lectins present in the serum of a variety of nonprimate mammals (1, 15, 16).
The broad-spectrum anti-influenza activity of CV-N provides a novel lead for development of new prophylactic or therapeutic treatments for influenza virus infection. Though three surface protein targets are present on the influenza virus virion (HA, neuraminidase, and M2), only two of these proteins (neuraminidase and M2) are currently targeted by commercially available therapeutic agents. The resistance of older laboratory-adapted strains of influenza A virus such as A/PR/8/34 (dating to 1934) and A/NWS/33 (dating to 1933) does not detract from interest in CV-N, especially in light of CV-N′s excellent profile of activity against clinical isolates of both influenza A and B viruses. Furthermore, CV-N′s activity against neuraminidase-resistant strains of influenza virus suggests that CV-N-based agents might be useful in combination with the neuraminidase inhibitor drug class to help prevent or control future pandemics of influenza. The availability of the detailed three-dimensional structure of CV-N and its binding sites with specific oligosaccharide ligands may provide the basis for design of small-molecule mimetics of CV-N. The CV-N protein itself may also have important potential therapeutic and prophylactic utility meriting further investigation. With this in mind, we also tested for efficacy against representative influenza virus strains several glycosylation-resistant and other variants (20) of CV-N potentially more amenable to large-scale production and purification from either prokaryotic or eukaryotic hosts. All these functional homologs of CV-N showed inhibitory activity (Table 4) virtually identical to that of the native protein. Finally, for future consideration, CV-N might provide a new template for vaccine design. Creation of novel immunogens that could stimulate endogenous production of antibodies mimicking CV-N would be of great interest.
Acknowledgments
This work was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400.
We thank Yumi Matsuoka, Centers for Disease Control and Prevention, for the kind gift of purified influenza virus HA, Laura Cartner and ZhaoZhong Han (MTDP) for assistance in providing the CV-N used in this study, and James McMahon (MTDP) for providing anti-CV-N antibodies. We also thank Katherine Tworkoski and Scott Bringans (MTDP) for technical and graphic assistance and Shanta Kodihalli, Jay Wells, Michelle Mathers, and Roger Ptak (Southern Research Institute) for their technical assistance.
REFERENCES
- 1.Anders, E. M., C. A. Hartley, and D. C. Jackson. 1990. Bovine and mouse serum β inhibitors of influenza A viruses are mannose-binding lectins. Proc. Natl. Acad. Sci. USA 87:4485-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora, D. J. S., P. Tremblay, R. Bourgault, and S. Boileau. 1985. Concentration and purification of influenza virus from allantoic fluid. Anal. Biochem. 144:189-192. [DOI] [PubMed] [Google Scholar]
- 3.Banks, J., E. S. Speidel, E. Moore, L. Plowright, A. Piccirillo, I. Capua, P. Cordioli, A. Fioretti, and D. J. Alexander. 2001. Changes in the haemagglutinin and the neuraminidase genes prior to the emergence of highly pathogenic H7N1 avian influenza viruses in Italy. Arch. Virol. 146:963-973. [DOI] [PubMed] [Google Scholar]
- 4.Barrientos, L. G., B. R. O'Keefe, M. Bray, A. Sanchez, A. M. Gronenborn, and M. R. Boyd. 2002. Cyanovirin-N binds to the viral surface glycoprotein GP1,2 and inhibits infectivity of the Ebola virus. Antivir. Res. 58:47-56. [DOI] [PubMed] [Google Scholar]
- 5.Bewley, C. A. 2001. Solution structure of a cyanovirin-N:Man alpha 1-2Man complex: structural basis for high-affinity carbohydrate-mediated binding of gp120. Structure 9:931-940. [DOI] [PubMed] [Google Scholar]
- 6.Bolmstedt, A. J., B. R. O'Keefe, S. R. Shenoy, J. B. McMahon, and M. R. Boyd. 2001. Cyanovirin-N defines a new class of antiviral agent targeting N-linked, high-mannose glycans in an oligosaccharide-specific manner. Mol. Pharmacol. 59:949-954. [DOI] [PubMed] [Google Scholar]
- 7.Botos, I., B. R. O'Keefe, S. R. Shenoy, L. K. Cartner, D. M. Ratner, P. H. Seeberger, M. R. Boyd, and A. Wlodawer. 2002. Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high-mannose oligosaccharides. J. Biol. Chem. 277:34336-34342. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, M. R., K. R. Gustafson, J. B. McMahon, R. H. Shoemaker, B. R. O'Keefe, T. Mori, R. J. Gulakowski, L. Wu, M. I. Rivera, C. M. Laurencot, M. J. Currens, J. H. Cardellina II, R. W. Buckheit, Jr., P. L. Nara, L. K. Pannell, R. C. Sowder II, and L. E. Henderson. 1997. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120; potential applications to microbicide development. Antimicrob. Agents Chemother. 41:1521-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckheit, R. W., Jr., T. L. Kinjersky, V. Fliakas-Boltz, J. D. Russell, T. L. Stup, L. A. Panllansch, W. G. Brouwer, D. C. Dao, W. A. Harrison, R. J. Schultz, J. P. Bader, and S. S. Yang. 1995. Structure-activity and cross-resistance evaluations of a series of human immunodeficiency virus type 1-specific compounds related to oxathiin carboxanilide. Antimicrob. Agents Chemother. 39:2718-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caton, A. J., G. G. Brownlee, J. W. Yewdell, and W. Gerhard. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417-427. [DOI] [PubMed] [Google Scholar]
- 11.Dey, B., D. L. Lerner, P. Lusso, M. R. Boyd, J. H. Elder, and E. A. Berger. 2000. Multiple antiviral activities of cyanovirin-N: blocking of gp120 interaction with CD4 and coreceptor, and inhibition of diverse enveloped viruses. J. Virol. 74:4562-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edson, C. M., and D. A. Thorley-Lawson. 1983. Synthesis and processing of the three major envelope glycoproteins of Epstein-Barr virus. J. Virol. 46:547-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finter, N. B. 1969. Dye uptake methods for assessing viral cytopathogenicity and their application to interferon assays. J. Gen. Virol. 5:419-427. [Google Scholar]
- 14.Gubareva, L. V., L. Kaiser, and F. G. Hayden. 2000. Influenza virus neuraminidase inhibitors. Lancet 355:827-835. [DOI] [PubMed] [Google Scholar]
- 15.Hartley, C. A., P. C. Reading, A. C. Ward, and E. M. Anders. 1997. Changes in the hemagglutinin molecule of influenza type A (H3N2) virus associated with increased virulence for mice. Arch. Virol. 142:75-88. [DOI] [PubMed] [Google Scholar]
- 16.Hartshorn, K. L., M. R. White, D. R. Voelker, J. Coburn, K. Zaner, and E. C. Crouch. 2000. Mechanism of binding of surfactant protein D to influenza A viruses: importance of binding to haemagglutinin to antiviral activity. Biochem. J. 351:449-458. [PMC free article] [PubMed] [Google Scholar]
- 17.Inudoh, M., H. Nyunoya, T. Tanaka, M. Hijikata, N. Kato, and K. Shimotohno. 1996. Antigenicity of hepatitis C virus envelope proteins expressed in Chinese hamster ovary cells. Vaccine 17-18:1590-1596. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.McKimm-Breschkin, J. L. 2000. Resistance of influenza viruses to neuraminidase inhibitors—a review. Antivir. Res. 47:1-17. [DOI] [PubMed] [Google Scholar]
- 20.Mori, T., L. G. Barrientos, Z. Han, A. M. Gronenborn, J. A. Turpin, and M. R. Boyd. 2002. Functional homologs of cyanovirin-N amenable to mass production in prokaryotic and eukaryotic hosts. Protein Expr. Purif. 26:42-49. [DOI] [PubMed] [Google Scholar]
- 21.Mori, T., K. R. Gustafson, R. C. Sowder II, L. K. Pannell, J. B. McMahon, R. H. Shoemaker, L. Wu, and M. R. Boyd. 1998. Recombinant production of cyanovirin-N, a potent human immunodeficiency virus-inactivating protein derived from a cultured cyanobacterium. Protein Expr. Purif. 12:151-158. [DOI] [PubMed] [Google Scholar]
- 22.O'Keefe, B. R., S. R. Shenoy, D. Xie, W. Zhang, J. M. Muschik, M. J. Currens, I. Chaiken, and M. R. Boyd. 2000. Analysis of the interaction between the HIV-inactivating protein cyanovirin-N and soluble forms of the envelope glycoproteins gp120 and gp41. Mol. Pharmacol. 58:982-992. [DOI] [PubMed] [Google Scholar]
- 23.Okuno, T., H. Shao, H. Asada, K. Shiraki, M. Takahashi, and K. Yamanishi. 1992. Analysis of human herpesvirus 6 glycoproteins recognized by monoclonal antibody OHV1. J. Gen. Virol. 73:443-447. [DOI] [PubMed] [Google Scholar]
- 24.Reed, L. J., and M. Muench. 1938. A simple method of estimating fifty-percent end points. Am. J. Hyg. 27:493-498. [Google Scholar]
- 25.Seidel, W., F. Kunkel, B. Geisler, W. Garten, B. Hermann, L. Dohner, and H. D. Klenk. 1991. Interepidemic variants of influenza virus H3 hemagglutinin differing in the number of carbohydrate side chains. Arch. Virol. 120:289-296. [DOI] [PubMed] [Google Scholar]
- 26.Serafini-Cessi, F., F. Dall'Olio, L. Pereira, and G. Campadelli-Fiume. 1984. Processing of N-linked oligosaccharides from precursor- to mature-form herpes simplex virus type 1 glycoprotein gC. J. Virol. 51:838-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shenoy, S. R., L. G. Barrientos, D. M. Ratner, B. R. O'Keefe, P. H. Seeberger, A. M. Gronenborn, and M. R. Boyd. 2002. Multi-site and multivalent binding between cyanovirin-N and branched oligomannosides: NMR and calorimetric characterization. Chem. Biol. 9:1109-1118. [DOI] [PubMed] [Google Scholar]
- 28.Shenoy, S. R., B. R. O'Keefe, A. J. Bolmstedt, L. K. Cartner, and M. R. Boyd. 2001. Selective interactions of the human immunodeficiency virus-inactivating protein cyanovirin-N with high-mannose oligosaccharides on gp120 and other glycoproteins. J. Pharmacol. Exp. Ther. 297:704-710. [PubMed] [Google Scholar]
- 29.Smee, D. F., R. W. Sidwell, A. C. Morrison, K. W. Bailey, E. Z. Baum, L. Ly, and P. C. Wagaman. 2001. Characterization of an influenza A (H3N2) virus resistant to the cyclopentane neuraminidase inhibitor RWJ-270201. Antivir. Res. 52:251-259. [DOI] [PubMed] [Google Scholar]
- 30.Tsuchiya, E., K. Sugawara, S. Hongo, Y. Matsuzaki, Y. Muraki, Z.-N. Li, and K. Nakamura. 2001. Antigenic structure of the haemagglutinin of human influenza A/H2N2 virus. J. Gen. Virol. 82:2475-2484. [DOI] [PubMed] [Google Scholar]
- 31.Yoshima, H., M. Nakanishi, Y. Okada, and A. Kobata. 1981. Carbohydrate structures of HVJ (Sendai virus) glycoproteins. J. Biol. Chem. 256:5355-5361. [PubMed] [Google Scholar]