Abstract
We report on the identification of active novel antimicrobials determined by screening both the genomic information and the mRNA transcripts from a number of different flatfish for sequences encoding antimicrobial peptides, predicting the sequences of active peptides from the genetic information, producing the predicted peptides chemically, and testing them for their activities. We amplified 35 sequences from various species of flatfish using primers whose sequences are based on conserved flanking regions of a known antimicrobial peptide from winter flounder, pleurocidin. We analyzed the sequences of the amplified products and predicted which sequences were likely to encode functional antimicrobial peptides on the basis of charge, hydrophobicity, relation to flanking sequences, and similarity to known active peptides. Twenty peptides were then produced synthetically and tested for their activities against gram-positive and gram-negative bacteria and the yeast Candida albicans. The most active peptide (with the carboxy-terminus amidated sequence GWRTLLKKAEVKTVGKLALKHYL, derived from American plaice) showed inhibitory activity over a concentration range of 1 to 8 μg/ml against a test panel of pathogens, including the intrinsically antibiotic-resistant organism Pseudomonas aeruginosa, methicillin-resistant Staphylococcus aureus, and C. albicans. The methods described here will be useful for the identification of novel peptides with good antimicrobial activities.
Bacterial resistance to conventional antibiotics is becoming more prevalent. There have therefore been many recent attempts to find effective replacements for use against infections in the clinic and on the farm. In the course of these efforts, cationic antimicrobial peptides (CAPs) have been identified as major defenses against infections in lower organisms and as important components of the innate defenses of mammals, including humans. While several antimicrobial peptides have been isolated from fish (6, 9, 14, 18, 19, 25), their numbers are modest compared to the numbers of peptides isolated from other species. One of the reasons for this small number may be the difficulty of isolating fish CAPs at the protein level. Indeed, several reports indicate that parts of larger molecules with antimicrobial activities, such as histones (6) or lysozyme, may be isolated by purification protocols designed for ribosomally produced peptides. This may render isolation and identification of those ribosomally produced peptide species difficult. In fact, in the case of fish-derived CAPs, the number of reports describing peptides derived from larger precursors is unexpectedly large compared to the number of reports describing ribosomally produced peptides.
Nonetheless, many attempts to isolate new peptides from fish are under way, chiefly because fish theoretically should be peptide rich. The rationale behind this is that since fish rely heavily on their innate defenses (of which peptides are a part), especially immediately after hatching (26) or at low temperatures (4), they should possess an extensive repertoire of CAPs. In addition, some of the fish peptides isolated to date, such as pleurocidin, showed a broad spectrum of activity against gram-positive and gram-negative bacteria, thus holding great promise for other fish peptides.
Pleurocidin was isolated from the winter flounder Pleuronectes americanus (Walbaum) in 1997 (9), and its activity has been under investigation by several research groups (13, 20, 21, 23, 28). Also, pleurocidin genes have been cloned, and their genetic organization has been described (8, 10; S. E. Douglas, A. Patrzykat, J. Pytyck, and J. W. Gallant, submitted for publication). An aspect of pleurocidin genetics of particular interest to this research is the fact that all pleurocidin genes described in winter flounder share common flanking sequences: a signal peptide at the N terminus and an acidic peptide at the C terminus (8, 10; Douglas et al., submitted). The C-terminal portion most likely neutralizes the positive charge of the active peptide, allowing secretion and protection of the host from potential peptide toxicity. Given that the flanking sequences for pleurocidins are conserved and that other cationic antimicrobial peptides from different species, such as cathelicidins (29), share pre-pro sequences, we hypothesized that these conserved sequences in winter flounder would also appear in novel antimicrobial peptides in different flatfish species.
While there is a great deal of uncertainty as to the exact mode of action of pleurocidin, previous studies have shown that many antimicrobial peptides need to be positively charged and show good separation of hydrophilic and hydrophobic residues in their tertiary structures for optimal activity. In addition, peptides amidated at the C terminus frequently showed better antimicrobial activities than their parent unamidated peptides. We hypothesized that by taking these and other factors into consideration we could select those genes that would most likely yield active peptides.
This paper describes a successful attempt to examine the genes of several flatfish in order to identify novel cationic antimicrobial peptide genes. The genetic information was combined with accepted dogma of peptide structure-function relationships in order to identify candidates for novel antimicrobials. Testing of these candidates for their antimicrobial activities against a yeast and a variety of bacterial species confirmed that our approach is appropriate for the identification of new CAPs of potential therapeutic value.
MATERIALS AND METHODS
Amplification of novel peptide genes.
Primers whose sequences were based on conserved regions flanking the sequence of winter flounder pleurocidin and templates from genomic DNA or cDNA from winter flounder, yellowtail flounder (Pleuronectes ferruginea Storer), American plaice (Hippoglossoides platessoides Fabricius), witch flounder (Glyptocephalus cynoglossus L.), and Atlantic halibut (Hippoglossus hippoglossus L.) were used in order to amplify novel peptide genes by PCR. Detailed descriptions of the tissues, primers, and PCR protocols have been published previously (10). The amplification products were sequenced with an ABI 377 automated sequencer and the AmpliTaqFS Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer, Foster City, Calif.). Sequence data were analyzed with Sequencher software (Gene Codes, Inc., Ann Arbor, Mich.) and DNA Strider (17). The amino-terminal signal sequence was predicted by using SignalP (http://www.cbs.dtu.dk/services /SignalP).
Production of active cationic peptide sequences.
The sequences of the peptides used for testing and selected peptide properties are provided in Table 1. All antimicrobial peptides used in this study were synthesized by N-(9-fluorenyl)methoxy carbonyl chemistry at the Nucleic Acid Protein Service unit at the University of British Columbia. The purity of each peptide was confirmed by high-pressure liquid chromatography and mass spectrometry analysis. In the case of NRC-7, the product of chemical synthesis was further purified by reversed-phase fast-protein liquid chromatography. The sequences of peptides NRC-1, -2, -3, -4, -5, -6, -8, -9, and -10 (9, 10, 13; Douglas et al., submitted), as well as the antimicrobial activities of peptides NRC-4 (9, 13, 20; Douglas et al., submitted) and NRC-8, -9, and -10 (Douglas et al., submitted), have been published previously and are included here for reference purposes. The sequence of NRC-4 is sometimes referred to here as original pleurocidin, although the original peptide described by Cole et al. (9) was not reported to be amidated.
TABLE 1.
Origin and one-letter amino acid sequences for flatfish cationic antimicrobial peptides synthesized and tested for activitiesa
| Origin | Amino acid sequence | Code | No. of residues | Mol wt | Charge |
|---|---|---|---|---|---|
| Winter flounder (1) | GKGRWLERIGKAGGIIIGGALDHL-NH2 | NRC-01b,c | 24 | 2,487 | +3.5 |
| Winter flounder (1a) | WLRRIGKGVKIIGGAALDHL-NH2 | NRC-02c | 20 | 2,172 | +4.5 |
| Winter flounder (1a-1) | GRRKRKWLRRIGKGVKIIGGAALDHL-NH2 | NRC-03c | 26 | 2,954 | +9.5 |
| Winter flounder (2) 2.1 | GWGSFFKKAAHVGKHVGKAALTHYL-NH2 | NRC-04b,c | 25 | 2,711 | +6.5 |
| Winter flounder (3) | FLGALIKGAIHGGRFIHGMIQNHH-NH2 | NRC-05c | 24 | 2,625 | +5.0 |
| Winter flounder (4) 1.1 | GWGSIFKHGRHAAKHIGHAAVNHYL-NH2 | NRC-06c | 25 | 2,764 | +6.5 |
| Yellowtail flounder YT2 | RWGKWFKKATHVGKHVGKAALTAYL-NH2 | NRC-07 | 25 | 2,854 | +8.0 |
| Winter flounder X | RSTEDIIKSISGGGFLNAMNA-NH2 | NRC-08b,c | 21 | 2,180 | +2.0 |
| Winter flounder Y | FFRLLFHGVHHGGGYLNAA-NH2 | NRC-09b,c | 19 | 2,112 | +3.5 |
| Winter flounder Z | FFRLLFHGVHHVGKIKPRA-NH2 | NRC-10b,c | 19 | 2,260 | +6.5 |
| American plaice AP1 | GWKSVFRKAKKVGKTVGGLALDHYL-NH2 | NRC-11 | 25 | 2,759 | +6.5 |
| American plaice AP2 | GWKKWFNRAKKVGKTVGGLAVDHYL-NH2 | NRC-12 | 25 | 2,859 | +6.5 |
| American plaice AP3 | GWRTLLKKAEVKTVGKLALKHYL-NH2 | NRC-13 | 23 | 2,653 | +6.5 |
| Witch flounder GcSc4C5 | AGWGSIFKHIFKAGKFIHGAIQAHND-NH2 | NRC-14 | 26 | 2,851 | +4.5 |
| Witch flounder GcSc4B7 | GFWGKLFKLGLHGIGLLHLHL-NH2 | NRC-15 | 21 | 2,356 | +4.5 |
| Witch flounder GC3.8-td | GWKKWLRKGAKHLGQAAIK-NH2 | NRC-16 | 19 | 2,175 | +7.5 |
| Witch flounder GC3.8 | GWKKWLRKGAKHLGQAAIKGLAS | NRC-17 | 23 | 2,505 | +6.5 |
| Witch flounder GC3.2 | GWKKWFTKGERLSQRHFA | NRC-18 | 18 | 2,262 | +4.5 |
| Halibut Hb26 | FLGLLFHGVHHVGKWIHGLIHGHH-NH2 | NRC-19 | 24 | 2,749 | +5.5 |
| Halibut Hb18 | GFLGILFHGVHHGRKKALHMNSERRS | NRC-20 | 26 | 2,985 | +6.0 |
In order to estimate the net charge, K and R were assumed to have values of +1, H was assumed to have a value of of +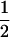 , and D and E of were assumed to have values of −1; C-terminal amidation was counted as an additional +1.
, and D and E of were assumed to have values of −1; C-terminal amidation was counted as an additional +1.
Peptide activities were published previously as described in Materials and Methods.
Clone sequences were published previously, as described in Materials and Methods.
Truncated form of GC3.8.
Bacterial strains and Candida albicans.
Two field isolates of the salmonid pathogen Aeromonas salmonicida, isolates 99-1 and 97-4, were from the Institute for Marine Biosciences strain collection. The following strains were supplied by R. E. W. Hancock: Escherichia coli DC2 (outer membrane permeable), E. coli UB1005 (the parent strain of DC2), Pseudomonas aeruginosa K799 (the parent strain of Z61), P. aeruginosa Z61 (antibiotic supersusceptible) (2), Salmonella enterica serovar Typhimurium 14028s (the parent strain of MS7953s), S. enterica serovar Typhimurium MS7953s (defensin supersusceptible) (11), Staphylococcus epidermidis C621, and methicillin-resistant Staphylococcus aureus (MRSA; strain C623; University of British Columbia). The human clinical isolates of S. epidermidis and MRSA were originally isolated by A. Chow, University of British Columbia.
E. coli strain CGSC 4908 (his-67 thyA43 pyr-37), auxotrophic for thymidine, uridine, and l-histidine (7), was provided, free of charge, by the E. coli Genetic Stock Center (Yale University, New Haven, Conn.). Mueller-Hinton broth (Difco Laboratories, Detroit, Mich.) supplemented with 5 mg of thymidine per liter, 10 mg of uridine per liter, and 20 mg of l-histidine per liter (Sigma Chemical Co., St. Louis, Mo.) was used to grow E. coli CGSC 4908 unless specified otherwise.
Most human bacterial pathogens as well as C. albicans were grown at 37°C in Mueller-Hinton broth, while the fish pathogens were maintained at 16°C in tryptic soy broth (Difco) containing 5 g of NaCl per liter. All strains were stored at −70°C until they were thawed for use and were subcultured daily.
MICs.
The activities of the antimicrobial peptides were determined by measurement of the MICs by the broth microdilution method of Amsterdam (1), as modified by Wu and Hancock (27). Serial dilutions of the peptide were made in water or 0.2% bovine serum albumin-0.01% acetic acid solution in 96-well polypropylene microtiter plates (Costar; Corning Incorporated, Corning, N.Y.). The bacterial strains and C. albicans were grown overnight to the mid-logarithmic phase as described above and diluted to give a final inoculum size of 106 CFU/ml. A suspension of bacteria or yeast was added to each well of a 96-well plate, and the plate was incubated overnight at the appropriate temperature. Inhibition was defined as growth less than or equal to one-half of the growth observed in control wells to which no peptide was added. However, for all organisms except P. aeruginosa, for which growth inhibition was indeed gradual, complete inhibition (no growth) was achieved at the lowest inhibitory concentration. Growth was assessed visually. Three replicates of each MIC determination were performed.
Killing assays.
The rates of survival of bacteria and C. albicans upon exposure to selected peptides applied at their MICs and 10 times their MICs were measured by standard methods. One milliliter of each culture containing 106 CFU of the test organism per ml was exposed to the peptides added to the specified final concentrations, and then at defined time intervals, equal aliquots were removed from the cultures and serially diluted and 15 μl was plated onto the appropriate medium. The plates were incubated overnight, and the resulting colonies were counted. Percent killing was calculated as the proportion of live bacteria at a given time point following the addition of the peptide compared to the number of bacteria present prior to the addition of the peptide. Control cultures of each bacterium were incubated without any peptide and were assayed at time points corresponding to the times of assay of the test cultures to ensure that there was no spontaneous loss of viability. Two replicates of each experiment were performed to ensure reproducibility. Maximal percent killing and the time point when maximal killing was reached are reported.
Nucleotide sequence accession numbers.
All new pleurocidin-like sequences reported in this study have been deposited in GenBank (accession numbers AY273172-AY273181).
RESULTS
Amplification and sequencing of cationic peptides from flatfish.
A total of 35 PCR products were obtained by amplification of genomic DNA (21 products) or cDNA from skin or intestine mRNA (14 products) with primers whose sequences are specific for the conserved sequences flanking the winter flounder pleurocidin. In seven cases, a cDNA sequence(s) corresponding to a genomic sequence(s) from the same species was amplified, allowing identification of intron-exon boundaries. All genes encoding pleurocidin-like peptides contained two introns within the coding sequence and one intron just upstream of the initiator methionine, as reported previously (8, 10; Douglas et al., submitted). In 10 cases, several sequences were highly similar, differing by only one or two conservative amino acid replacements. The alignment of the translated sequences from the subset of peptides whose amino acid sequences differed significantly and that were selected for synthesis and further study is shown in Fig. 1. While the high degree of conservation among the flanking regions is evident, the core regions corresponding to the putative mature peptides exhibit a range of amino acid sequences and vary in their compositions, lengths, net charges, and proportions of hydrophobic residues. While some sequences appear to be closely related to the previously reported pleurocidin, most sequences are highly original.
FIG. 1.
Sequences amplified from five different flatfish species with primers whose sequences are based on conserved flanking sequences of the winter flounder pleurocidin. Abbreviations for species are as follows: AP, American plaice; GC, Glyptocephalus cynoglossus; HB, Atlantic halibut; WF, winter flounder; YT, yellowtail flounder. The positions of primers PL5′ and PL3′ are indicated by arrows. The following sequences correspond to the indicated manufactured peptides: AP1, peptide NRC-11; AP2, peptide NRC-12; AP3, peptide NRC-13; WF2, peptide NRC-4; YT2, peptide NRC-7; WF4, peptide NRC-6; GcSc4C5, peptide NRC-14; GC3.8, peptide NRC-17; GC3.2, peptide NRC-18; Wf3, peptide NRC-5; Hb26, peptide NRC-19; Hb18, peptide NRC-20; GcSc4B7, peptide NRC-15; WF1a, peptide NRC-2; Wf1 (NRC-1).
Several factors were taken into consideration while selecting the peptides to be made chemically and subjected to further testing (listed in Table 1). Overall, peptides which, upon preliminary modeling of their structures, conformed to the characteristics of most CAPs (short and positively charged, with good separation of hydrophobic and hydrophilic residues) were given priority. In addition, those sequences that were amplified from cDNA (and that were therefore known to be expressed in the fish) were also synthesized. While in most cases an attempt was made to accurately predict the sequence, composition, etc., of the natural peptide, in some situations this was difficult and multiple variants were made.
Inhibitory activities.
The peptides produced chemically as described above were tested for their abilities to inhibit the growth of gram-negative and gram-positive bacteria, including the fish pathogen A. salmonicida, and the fungus C. albicans. Activity data for peptides NRC-1, -4, -8, -9, and -10 have been published previously and are included here for reference purposes. All results are shown in Table 2.
TABLE 2.
MICs of cationic peptides synthesized from flatfish
| Peptide code | MIC (μg/ml)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. salmoni- cida 99-1 | A. salmoni- cida 97-4 | S. enterica serovar Typhimurium MS7953s | S. enterica serovar Typhimurium 14028s | P. aeru- ginosa K799 | P. aeru- ginosa Z61 | E. coli CGSC 4908 | E. coli UB1005 | E. coli DC2 | S. epider- midis C621 | MRSA C623 | C. albicans C627 | |
| NRC-1 | 64 | 64 | 16 | >64 | >64 | 32 | 32 | 32 | 32 | >64 | >64 | 64 |
| NRC-2 | >128 | 128 | 64 | >64 | 64 | 32 | 64 | 64 | 64 | >64 | >64 | >64 |
| NRC-3 | 2 | 4 | 2 | 8 | 2 | 1 | 2 | 8 | 2 | 8 | 8 | 4 |
| NRC-4 | 2 | 2 | 2 | 16 | 8 | 4 | 2 | 4 | 2 | 8 | 8 | 8 |
| NRC-5 | >64 | >64 | 64 | >64 | >64 | 32 | 64 | 64 | >64 | 32 | 32 | >64 |
| NRC-6 | 4 | 4 | 4 | 64 | 16 | 4 | 4 | 4 | 2 | >64 | 32 | 32 |
| NRC-7 | 4 | 2 | 2 | 8 | 4 | 2 | 4 | 4 | 4 | 64 | 64 | 16 |
| NRC-8 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 | >64 |
| NRC-9 | >64 | >64 | 64 | >64 | >64 | 64 | 64 | >64 | >64 | >64 | >64 | >64 |
| NRC-10 | >64 | 32 | 16 | >64 | 32 | 8 | 32 | 32 | 32 | 32 | 64 | >64 |
| NRC-11 | 8 | 8 | 4 | 32 | 32 | 4 | 4 | 16 | 4 | 64 | >64 | 32 |
| NRC-12 | 2 | 2 | 2 | 8 | 4 | 1 | 2 | 8 | 2 | 8 | 16 | 4 |
| NRC-13 | 4 | 2 | 2 | 8 | 4 | 1 | 2 | 4 | 2 | 4 | 4 | 4 |
| NRC-14 | 32 | 16 | 16 | >64 | 32 | 8 | 16 | 16 | 16 | 16 | 16 | >64 |
| NRC-15 | 8 | 16 | 4 | 16 | 8 | 4 | 8 | 8 | 8 | 4 | 4 | 16 |
| NRC-16 | 2 | 1 | 0.5 | 16 | 4 | 1 | 1 | 2 | 0.5 | 16 | 32 | 8 |
| NRC-17 | 2 | 1 | 1 | 8 | 4 | 2 | 1 | 4 | 1 | 32 | 16 | 8 |
| NRC-18 | >64 | 128 | 32 | >64 | >64 | 64 | 64 | 64 | 64 | >64 | >64 | >64 |
| NRC-19 | 64 | >64 | 16 | 64 | 32 | 8 | 32 | 16 | 32 | 8 | 8 | 64 |
| NRC-20 | >64 | >64 | >64 | >64 | >64 | 64 | >64 | >64 | >64 | >64 | >64 | >64 |
Following the convention that specifies that a difference in inhibitory activity is defined as a twofold or greater difference in the MICs, an inspection of Table 2 reveals that three peptides, NRC-3, -12, and -13, exhibited inhibitory activities equal to or better than those of the original pleurocidin against all pathogens tested. An additional six peptides, NRC-6, -7, -11, -15, -16, and -17, exhibited inhibitory activities equal to or better than those of the original pleurocidin against most pathogens tested. Overall, of the 20 peptides produced, the 9 listed above, together with pleurocidin, exhibited strong broad-spectrum activities, with 6 others exhibiting a more limited spectrum of activity and only 4 (NRC-8, -9, -18, and -20) being virtually inactive.
As there was ambiguity as to what the N terminus of a peptide derived from clone WF1a would be, both a short version (NRC-2) and a longer version with an N-terminal GRRKRK fragment (NRC-3) were made. The longer sequence, NRC-3, with a highly cationic N-terminal fragment, produced a peptide active against virtually all pathogens tested, while the shorter peptide was only marginally active. Based on the previously mentioned convention, the activity of NRC-3 was identical to that of the original pleurocidin against all pathogens tested except P. aeruginosa. In the case of P. aeruginosa, NRC-3 was more active than the original pleurocidin. The results obtained for NRC-2 and NRC-3 led to the suggestion that GRRKRK may be the determinant of activity in NRC-3. It can be further hypothesized that the peptide, as it occurs in nature, possesses N-terminal GRRKRK, although an expression study specific for the GRRKRK-containing peptide would be required to confirm the hypothesis.
Two of three of the most active peptides (excluding the original pleurocidin), NRC-12 and NRC-13, were both derived from American plaice; but they shared only limited sequence identity. In addition, another peptide derived from American plaice, NRC-11, which was highly similar to NRC-12, showed good activity against some bacteria. Interestingly, however, even though NRC-11 and NRC-12 were 80% identical and carried identical net charges, their activities against many pathogens were different. We consider it unlikely, although not impossible, that the substitution of V for L (position 21) or R for K (position 8) had a major impact on activity, thus leaving the remaining three substitutions (at positions 4, 5, and 7) as the possible determinants of activity in NRC-12. The replacement of SV in NRC-11 with the bulky and charged KW in NRC-12 and the replacement of R with N are both likely to have had an impact on the differential activities.
An analysis of the peptide sequences of NRC-4, -7, -10, -11, -12, -13, and -19 reveals the presence of an XVGK motif, where X is H, K, or T, all of which are relatively bulky residues. Indeed, in the case of NRC-4 and NRC-7, the motif is even doubled as HVGKHVGK. We hypothesize that this repeat may be compatible with a highly bactericidal amphipathic helix. This hypothesis was reinforced when we threaded repeats of HVGK or KVGK onto a helical wheel (results not shown). Up to three tandem repeats would indeed form a very amphipathic molecule if they were to form an alpha helix.
The halibut-derived peptide NRC-19 was perhaps the most unique peptide in its spectrum of activity, in that it was active against the hardy species P. aeruginosa and MRSA, while it was less active against usually more susceptible bacteria. What was even less expected was that NRC-19 was virtually inactive against A. salmonicida, which can infect halibut in nature.
The activities of the best peptides against the test strains were comparable to or better than those of gramicidin S and polymyxin B (13).
Bactericidal activity.
On the basis of the results of the MIC assays, several of the most active peptides (NRC-3, -12, -13, -15, -16, and -17), as well as NRC-19, which exhibited good activity against P. aeruginosa and gram-positive bacteria, despite its limited activity against other strains, were selected for bactericidal assays. A summary of the results is shown in Table 3, along with previously reported data for NRC-1 (Douglas et al., submitted). In addition, NRC-13, which was one of the most active peptides against P. aeruginosa in the MIC assays and thus potentially useful for the treatment of infections in cystic fibrosis patients, in which elevated NaCl levels may be a factor, was also tested in the presence of 150 mM NaCl.
TABLE 3.
Bactericidal activities of selected antimicrobial peptidesa
| Peptide code | Concn (fold MIC) | Pathogen | Maximal killing (% bacteria killed) | Time when maximal killing was reached |
|---|---|---|---|---|
| NRC-1 | 1 | 99-1 | 100 | 40 |
| 10 | 99-1 | 100 | 20 | |
| 1 | MS7953s | 100 | 20 | |
| 10 | MS7953s | 100 | 10 | |
| 1 | C. albicans | NK | NA | |
| 10 | C. albicans | 90 | 240 | |
| NRC-3 | 1 | 99-1 | 90 | 20 |
| 10 | 99-1 | 100 | 5 | |
| 1 | MS7953s | 97 | 60 | |
| 10 | MS7953s | 100 | 20 | |
| NRC-12 | 1 | C. albicans | NK | NA |
| 10 | C. albicans | 90 | 240 | |
| NRC-13 | 1 | K799 | 95 | 10 |
| 10 | K799 | 100 | 5 | |
| NRC-13 with NaCl | 1 | K799 | 5 | 60 |
| 10 | K799 | 100 | 20 | |
| NRC-13 | 1 | MRSA | 99 | 10 |
| 10 | MRSA | 100 | 5 | |
| 1 | C. albicans | 25 | 240 | |
| 10 | C. albicans | 75 | 240 | |
| NRC-15 | 1 | K799 | 60 | 60 |
| 10 | K799 | 98 | 10 | |
| 1 | MRSA | 100 | 20 | |
| 10 | MRSA | 100 | 5 | |
| NRC-16 | 1 | C. albicans | 35 | 240 |
| 10 | C. albicans | 92 | 240 | |
| NRC-17 | 1 | C. albicans | 10 | 240 |
| 10 | C. albicans | 95 | 240 | |
| NRC-19 | 1 | MRSA | 33 | 60 |
| 10 | MRSA | 90 | 10 | |
| 1 | K799 | 70 | 60 | |
| 10 | K799 | 80 | 40 |
The bacterial strains used are A. salmonicida (99-1), S. typhimurium (MS7953s), P. aeruginosa (K799), and MRSA. NK, no killing; NA, not available.
Generally, the bactericidal activities of the peptides against the intrinsically antibiotic-resistant gram-negative pathogen P. aeruginosa K799, the methicillin-resistant gram-positive pathogen S. aureus (MRSA), and the yeast C. albicans were of particular interest, although S. enterica serovar Typhimurium (MS7953s) and A. salmonicida (99-1) were also tested by use of selected peptide-pathogen combinations. The selection was dictated by consideration of the potential future use of the peptides in light of their initial activities in MIC assays.
Upon analysis of the patterns among the peptide-pathogen combinations provided in Table 3, the following trends emerged: (i) in some cases the killing at the MIC and 10 times the MIC were equally efficient but slower at the MIC than at 10 times the MIC (e.g., for NRC-1 with strain 99-1 and NRC-15 with strain MRSA); (ii) in some cases the killing at the MIC was slower and less efficient than that at 10 times the MIC (e.g., for NRC-15 with strain K799 and NRC-19 with MRSA); and (iii) in some cases the killing was as fast at the MIC as it was at 10 times the MIC but it appeared to be less efficient (e.g., for NRC-13, -16, and -17 with C. albicans), although the progression of killing was harder to assess when the maximal killing was only 10% (as in the case of NRC-17 and C. albicans). The particular pattern of killing at the MIC compared to that at 10 times the MIC appeared to be associated with specific peptide-pathogen combinations and did not appear to be dependent on the peptide or the pathogen alone.
The most active peptide based on the MICs, NRC-13, rapidly (5 to 10 min) and efficiently (95 to 100%) killed P. aeruginosa K799 and MRSA when it was applied at the MIC and 10 times the MIC. However, even at 240 min, NRC-13 at 10 times the MIC killed only 75% of the C. albicans cells. The killing of P. aeruginosa K799 by NRC-13 at 10 times the MIC in the presence of 150 mM NaCl was slower that in the absence of salt, but the maximal killing of 100% was nonetheless reached in both cases. Only slight killing (5%) of P. aeruginosa K799 was observed when NRC-13 was used at its MIC in the presence of 150 mM NaCl. It is noteworthy that the MIC of NRC-13 for P. aeruginosa K799 did not change in the presence of 150 mM NaCl and remained 4 μg/ml. These data indicate that although the nature of the activity of NRC-13 against P. aeruginosa K799 changed in the presence of salt and although the killing at the MIC was minimal, the inhibitory properties as well as the bactericidal properties at 10 times the MIC were preserved.
In all cases use of the peptides at 10 times their MICs for the bacteria resulted in a relatively rapid drop in the viable counts, while use of the peptides at 10 times their MICs for C. albicans killed the cells more slowly. Overall, the most active peptides, as determined by the MIC assays, were also efficient killers in bactericidal assays. The various patterns of killing described above may be indicative of distinct modes of peptide-pathogen interaction for the distinct combinations.
DISCUSSION
There are three general approaches to the identification of novel antimicrobial peptides: searching for novel peptides from nature, constructing synthetic peptides by trying to improve the natural ones, and constructing entirely synthetic peptides via combinatorial chemistry. While the first approach has led to the discovery of most truly novel peptides, the technology for isolating them at the protein level is laborious. On the other hand, improvements to existing natural peptides produced many drug candidates, but few of them were truly novel. Finally, the resources required to generate novel peptides via combinatorial chemistry and test them are far too great to efficiently feed the antibiotic discovery pipeline. However, the difficulties described above can be circumvented by applying genetic technologies, which present the opportunity to isolate peptide genes from nature without having to go through a protein purification process.
Identification of over 70 new human and mouse β-defensin sequences by use of a genome-wide computational search strategy (24) and identification of new β-defensins by use of a survey of a defined chromosomal region (12) were reported recently. Although the antimicrobial properties of the sequences were not reported, this work is indicative of the power of genomic approaches.
The most prominent family of peptides exhibiting a common genetic organization and a common N-terminal flanking sequence are the cathelicidins (15, 22, 29). These peptides, derived from humans, monkeys, horses, cattle, goats, pigs, sheep, rabbits, guinea pigs, and mice, not only possess a proregion similar to that in cathelin, but they are all also encoded by four exons separated by three introns. Despite these similarities, mature cathelicidins display a range of amino acid compositions, charges, lengths, and structures.
Since conserved pre- and proregions were identified upon cloning of the pleurocidin genes (10), we hypothesized that pleurocidin may also be a member of a larger family of peptides present in flatfish. This was suggested by a previously published Southern blot analysis (10); and indeed, our data, obtained by PCR amplification of sequences flanked by the conserved pleurocidin regions from diverse species, demonstrate the existence of such a family of peptides. Similar to cathelicidins, the signal and proregions are conserved, but unlike the cathelicidins, in which the pre-pro portion occurs at the N terminus of the mature peptide, in the case of flatfish pleurocidins, the signal is located at the N terminus, while the proregion is located at the C terminus. Although the peptides described here share common flanking regions, they are diverse in their structures and activities.
The large number of peptide sequences obtained presented a dilemma as to which sequences were likely to encode active peptides. Much of the knowledge regarding sequence-activity relationships has been derived empirically by comparing many naturally occurring peptides, and this knowledge was then used in an attempt to construct improved analogs. Researchers even use their insights about peptide structure-activity relationships to construct entirely synthetic compounds. This knowledge, however, can also be used to select promising candidates from a pool of genes. In this study, we have generally selected peptides which were positively charged and which exhibited likely separation of hydrophilic and hydrophobic residues when they were modeled by using a variety of structure analysis software packages. In addition, whenever the predicted mature sequence was followed by a glycine residue, we decided to amidate the peptide. This was based on empirical knowledge about previously isolated peptide antimicrobials, many of which are naturally amidated. The amidation in nature is an enzymatic modification of an additional C-terminal glycine residue.
Multiple variants of a peptide were made whenever it was difficult to accurately predict the naturally occurring mature peptide. This approach has yielded a score of novel broad-spectrum antimicrobial peptides which will undoubtedly be further developed. The best peptides exhibited inhibitory and bactericidal activities against gram-negative bacteria, gram-positive bacteria, and C. albicans. The peptides with the poorest activities were generally alternative variants of the natural sequences or were based on pseudogenes which had been predicted to be inactive (Douglas et al., submitted). The inability of halibut-derived NRC-19 to kill A. salmonicida could be explained by the fact that the bacteria were isolated from salmon, not halibut, and it is known that A. salmonicida strains naturally infecting halibut are considered atypical (3, 16).
In addition, our approach has elucidated several sequence-activity trends that had not previously been reported in the literature. Among the most important of these is the observation that GRRKRK residues added at the N terminus conferred activity (NRC-3) onto an otherwise identical yet virtually inactive peptide (NRC-2) and the observation that the XVGK motif, in which X was H, K, or T, was present among the most active peptides. While it is not known why GRRKRK confers antimicrobial activity, its presence increases the overall peptide net charge by +5. This increase in charge at the N terminus may aid with the interaction of the peptide with the bacterial membrane. It is, however, equally possible that complex folding characteristics of the peptide are altered by the addition of GRRKRK, thus conferring antimicrobial activity.
Identification of specific determinants of activity among related peptides is not unprecedented. As early as 1994, Casteels et al. (5) delineated constant and variable regions among a family of apidaecins and associated them with the general antibacterial capacity and with the specificity of the antimicrobial spectrum, respectively. As new pleurocidins are identified and studied, more structure-function relationships governing their activity will no doubt become apparent.
The fact that some peptides described here were only moderately bactericidal at their MICs but highly bactericidal at 10 times their MICs suggests that their inhibitory actions against a specific pathogen may be separate from their bactericidal actions against that pathogen. Further studies are required to adequately explain the patterns of bactericidal activity relative to the inhibitory activity.
This study describes a family of flatfish antimicrobial cationic peptides that share common signal and proregion sequences but that differ in their core antimicrobial sequences. The peptide sequences themselves, the methodology used to find their genes, and the principles used to predict active peptides from the multitude of genetic sequences should all be valuable in the search for and the development of novel antimicrobials.
Acknowledgments
We thank the National Research Council of Canada for funding for this project.
We thank Vanya Ewart for kindly reviewing the manuscript. We also thank R. E. W. Hancock for kindly donating many of the bacterial strains used in this study.
Footnotes
National Research Council of Canada publication 42386.
REFERENCES
- 1.Amsterdam, D. 1996. Susceptibility testing for antimicrobials in liquid media, p. 52-111. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. The Williams & Wilkins, Baltimore, Md.
- 2.Angus, B. L., A. M. Carey, D. A. Caron, A. M. Kropinski, and R. E. Hancock. 1982. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild-type with an antibiotic-supersusceptible mutant. Antimicrob. Agents Chemother. 21:299-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergh, O., F. Nilsen, and O. B. Samuelsen. 2001. Diseases, prophylaxis and treatment of the Atlantic halibut Hippoglossus hippoglossus: a review. Dis. Aquat. Organ. 48:57-74. [DOI] [PubMed] [Google Scholar]
- 4.Bly, J. E., and L. W. Clem. 1991. Temperature-mediated processes in teleost immunity: in vitro immunosuppression induced by in vivo low temperature in channel catfish. Vet. Immunol. Immunopathol. 28:365-377. [DOI] [PubMed] [Google Scholar]
- 5.Casteels, P., J. Romagnolo, M. Castle, K. Casteels-Josson, H. Erdjument-Bromage, and P. Tempst. 1994. Biodiversity of apidaecin-type peptide antibiotics. Prospects of manipulating the antibacterial spectrum and combating acquired resistance. J. Biol. Chem. 269:26107-26115. [PubMed] [Google Scholar]
- 6.Cho, J. H., I. Y. Park, H. S. Kim, W. T. Lee, M. S. Kim, and S. C. Kim. 2002. Cathepsin D produces antimicrobial peptide parasin I from histone H2A in the skin mucosa of fish. FASEB J. 16:429-431. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, S. 1963. The synthesis of messenger RNA without protein synthesis in normal and phage-infected thymineless strains of Escherichia coli. Proc. Natl. Acad. Sci. USA 49:699.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, A. M., R. O. Darouiche, D. Legarda, N. Connell, and G. Diamond. 2000. Characterization of a fish antimicrobial peptide: gene expression, subcellular localization, and spectrum of activity. Antimicrob. Agents Chemother. 44:2039-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, A. M., P. Weis, and G. Diamond. 1997. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 272:12008-12013. [DOI] [PubMed] [Google Scholar]
- 10.Douglas, S. E., J. W. Gallant, Z. Gong, and C. Hew. 2001. Cloning and developmental expression of a family of pleurocidin-like antimicrobial peptides from winter flounder, Pleuronectes americanus (Walbaum). Dev. Comp. Immunol. 25:137-147. [DOI] [PubMed] [Google Scholar]
- 11.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 12.Jia, H. P., B. C. Schutte, A. Schudy, R. Linzmeier, J. M. Guthmiller, G. K. Johnson, B. F. Tack, J. P. Mitros, A. Rosenthal, T. Ganz, and P. B. McCray. 2001. Discovery of new human beta-defensins using a genomics-based approach. Gene 263:211-218. [DOI] [PubMed] [Google Scholar]
- 13.Jia, X., A. Patrzykat, R. H. Devlin, P. A. Ackerman, G. K. Iwama, and R. E. Hancock. 2000. Antimicrobial peptides protect coho salmon from Vibrio anguillarum infections. Appl. Environ. Microbiol. 66:1928-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauth, X., H. Shike, J. C. Burns, M. E. Westerman, V. E. Ostland, J. M. Carlberg, J. C. Van Olst, V. Nizet, S. W. Taylor, C. Shimizu, and P. Bulet. 2002. Discovery and characterization of two isoforms of moronecidin, a novel antimicrobial peptide from hybrid striped bass. J. Biol. Chem. 277:5030-5039. [DOI] [PubMed] [Google Scholar]
- 15.Lehrer, R. I., and T. Ganz. 2002. Cathelicidins: a family of endogenous antimicrobial peptides. Curr. Opin. Hematol. 9:18-22. [DOI] [PubMed] [Google Scholar]
- 16.Lund, V., L. M. Jenssen, and M. S. Wesmajervi. 2002. Assessment of genetic variability and relatedness among atypical Aeromonas salmonicida from marine fishes, using AFLP-fingerprinting. Dis. Aquat. Organ. 50:119-126. [DOI] [PubMed] [Google Scholar]
- 17.Marck, C. 1992. DNA Strider, version 1.2. Service de Biochimie-Bat 142, Centre d'Etudes Nucleares de Saclay, Gif-sur-Yvette, France.
- 18.Park, C. B., J. H. Lee, I. Y. Park, M. S. Kim, and S. C. Kim. 1997. A novel antimicrobial peptide from the loach, Misgurnus anguillicaudatus. FEBS Lett. 411:173-178. [DOI] [PubMed] [Google Scholar]
- 19.Park, I. Y., C. B. Park, M. S. Kim, and S. C. Kim. 1998. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish. Parasilurus asotus. FEBS Lett. 437:258-262. [DOI] [PubMed] [Google Scholar]
- 20.Patrzykat, A., C. L. Friedrich, L. Zhang, V. Mendoza, and R. E. Hancock. 2002. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 46:605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patrzykat, A., L. Zhang, V. Mendoza, G. K. Iwama, and R. E. Hancock. 2001. Synergy of histone-derived peptides of coho salmon with lysozyme and flounder pleurocidin. Antimicrob. Agents Chemother. 45:1337-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramanathan, B., E. G. Davis, C. R. Ross, and F. Blecha. 2002. Cathelicidins: microbicidal activity, mechanisms of action, and roles in innate immunity. Microbes Infect. 4:361-372. [DOI] [PubMed] [Google Scholar]
- 23.Saint, N., H. Cadiou, Y. Bessin, and G. Molle. 2002. Antibacterial peptide pleurocidin forms ion channels in planar lipid bilayers. Biochim. Biophys. Acta 1564:359.. [DOI] [PubMed] [Google Scholar]
- 24.Schutte, B. C., J. P. Mitros, J. A. Bartlett, J. D. Walters, H. P. Jia, M. J. Welsh, T. L. Casavant, and P. B. McCray. 2002. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc. Natl. Acad. Sci. USA 99:2129-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shike, H., X. Lauth, M. E. Westerman, V. E. Ostland, J. M. Carlberg, J. C. Van Olst, C. Shimizu, P. Bulet, and J. C. Burns. 2002. Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Eur. J. Biochem. 269:2232-2237. [DOI] [PubMed] [Google Scholar]
- 26.Tatner, M. F., and M. T. Horne. 1983. Susceptibility and immunity to Vibrio anguillarum in post-hatching rainbow trout fry, Salmo gairdneri Richardson 1836. Dev. Comp. Immunol. 7:465-472. [DOI] [PubMed] [Google Scholar]
- 27.Wu, M., and R. E. Hancock. 1999. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J. Biol. Chem. 274:29-35. [DOI] [PubMed] [Google Scholar]
- 28.Yoshida, K., Y. Mukai, T. Niidome, C. Takashi, Y. Tokunaga, T. Hatakeyama, and H. Aoyagi. 2001. Interaction of pleurocidin and its analogs with phospholipid membrane and their antibacterial activity. J. Pept. Res. 57:119-126. [DOI] [PubMed] [Google Scholar]
- 29.Zanetti, M., R. Gennaro, and D. Romeo. 1995. Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett. 374:1-5. [DOI] [PubMed] [Google Scholar]



