Abstract
The objective of this study was to further elucidate the role of membrane potential in the mechanism of action of daptomycin, a novel lipopeptide antibiotic. Membrane depolarization was measured by both fluorimetric and flow cytometric assays. Adding daptomycin (5 μg/ml) to Staphylococcus aureus gradually dissipated membrane potential. In both assays, cell viability was reduced by >99% and membrane potential was reduced by >90% within 30 min of adding daptomycin. Cell viability decreased in parallel with changes in membrane potential, demonstrating a temporal correlation between bactericidal activity and membrane depolarization. Decreases in viability and potential also showed a dose-dependent correlation. Depolarization is indicative of ion movement across the cytoplasmic membrane. Fluorescent probes were used to demonstrate Ca2+-dependent, daptomycin-triggered potassium release from S. aureus. Potassium release was also correlated with bactericidal activity. This study demonstrates a clear correlation between dissipation of membrane potential and the bactericidal activity of daptomycin. A multistep model for daptomycin's mechanism of action is proposed.
Daptomycin is a novel lipopeptide antibiotic in late-stage clinical development for the treatment of serious gram-positive infections. Daptomycin exhibits rapid in vitro bactericidal activity against clinically significant strains of gram-positive pathogens including hemolytic streptococci, methicillin-resistant Staphylococcus aureus, and vancomycin-resistant enterococci (4, 10, 12, 14, 19, 22, 23). Daptomycin acts at the cytoplasmic membrane of susceptible bacteria (8), as demonstrated by binding and fractionation studies. Additionally, the activity of daptomycin is dependent on the presence of physiologic levels of free calcium ions (50 mg/liter).
Debate over daptomycin's mechanism of action has continued for more than a decade. One hypothesis suggests that daptomycin bactericidal activity is mediated by inhibition of lipoteichoic acid (LTA) biosynthesis (7, 8). However, a recent investigation has failed to find evidence of a role for LTA in the mechanism of action of daptomycin in S. aureus or Enterococcus faecalis, suggesting that the in vitro bactericidal activity of daptomycin is independent of LTA biosynthesis (V. Laganas, J. Alder, and J. A. Silverman, submitted for publication). A second proposed mechanism of action for daptomycin is that the antibiotic causes dissipation of bacterial membrane potential, resulting in disruption of multiple aspects of cellular function (1, 2). Bactericidal activity via disruption of membrane potential is the proposed mechanism of action for a variety of antimicrobial peptides, including the pore-forming antibiotic nisin (18, 20). We wished to further investigate the role of bacterial membrane potential in the mechanism of action of daptomycin. In this study, we demonstrate a significant correlation between membrane depolarization and bactericidal activity. Furthermore, we demonstrate that one possible mechanism of membrane depolarization involves K+ release by bacteria on daptomycin exposure (J. A. Silverman, N. G. Perlmutter, and H. M. Shapiro, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1800, 2001).
MATERIALS AND METHODS
Strains, media, and antibiotics.
S. aureus ATCC 29213 was grown in Mueller-Hinton broth (Becton Dickinson, Cockeysville, Md.) supplemented with 50 mg of Ca2+/liter (MHBc). The MIC of daptomycin was determined as previously described (21). Daptomycin was provided by Cubist Pharmaceuticals, Inc. (Lexington, Mass.); nisin and valinomycin were obtained from Sigma Chemical Company (St. Louis, Mo.). Using MHBc and Mueller-Hinton agar, bacterial cell viability was determined by following National Committee for Clinical Laboratory Standards guidelines (16).
Fluorimeter assay for membrane potential.
Membrane potential was measured by using a fluorescent assay based on the method of Wu and Hancock (24). S. aureus cultures grown to the early exponential phase (optical density at 600 nm [OD600], 0.2 to 0.3) in MHBc were treated with no antibiotic (control), daptomycin (as indicated), nisin (25 μg/ml), or carbonyl cyanide m-chlorophenylhydrazone (CCCP) (10 μM) at 37°C. Samples (2 ml) were transferred to a polystyrene fluorimeter cuvette (VWR International, West Chester, Pa.) containing a stir bar and placed in the heated (37°C) sample chamber of an AMINCO-Bowman series 2 luminescence fluorimeter (Thermo Spectronic, Rochester, N.Y.). The cells were excited at 622 nm, and the fluorescence emission was collected at 670 nm. Background data were collected for 30 s before the addition of DiSC3(5) (3,3′-dipropylthiadicarbocyanine iodide; Molecular Probes, Eugene, Oreg.) to a final concentration of 100 nM. Data were collected for an additional 5 min after the addition of the fluorescent dye.
In a modified form of the assay described above, bacteria were grown to the early exponential phase in culture flasks (OD600, 0.2 to 0.3) and 2.5-ml aliquots were transferred to 14-ml round-bottom polystyrene tubes. The bacterial cells were subsequently treated with no antibiotic (control), daptomycin (as indicated), or nisin (25 μg/ml) at 37°C. At appropriate time points, DiSC3(5) (100 nM final concentration) was added directly to the round-bottom tubes and the samples were incubated for an additional 5 min at 37°C. Samples (2 ml) were transferred to cuvettes without a stir bar, cells were excited at 622 nm, and the fluorescence emission was collected at 670 nm for 10 s.
Flow cytometry.
The membrane potential was determined by flow cytometry by using a laboratory-built dual-laser instrument, as previously described (17), with DiOC2(3) (3,3′-diethyloxacarbocyanine iodide; Molecular Probes). Exponential-phase (OD600 = 0.3) S. aureus cultures grown in MHBc were treated with no antibiotic (control), 5-μg/ml daptomycin, or 25-μg/ml nisin at 37°C. At the indicated time points, samples were stained for 4 min at room temperature with 30 μM DiOC2(3) and 100 nM To-Pro-3 iodide (Molecular Probes). Flow cytometry was performed on 5,000 S. aureus cells per run by using the 488-nm beam from an argon ion laser to excite the DiOC2(3); the green fluorescence was detected through a 530- to 20-nm bandwidth band-pass filter, and the red fluorescence was detected through a 610- to 19-nm bandwidth band-pass filter. The membrane potential was measured as the intensity ratio of the red fluorescence (a membrane potential-dependent signal) and the green fluorescence (a membrane potential-independent signal). The 633-nm beam from a helium-neon laser was used to excite To-Pro-3 iodide; elevated far red fluorescence from this dye (detected through a 695-nm long-pass filter) is indicative of membrane damage.
Potassium ion release assay.
Potassium ion release assays were performed by following the method of Herranz et al. (11). S. aureus was grown to the exponential phase in MHBc (OD600, 0.3) and washed twice with and resuspended in a one-fifth volume of 10 mM HEPES (pH 7.2) and 0.5% glucose. Samples (2 ml) were transferred to a polystyrene fluorimeter cuvette containing a stir bar and placed in the heated (37°C) fluorimeter sample chamber. Bacterial cells were excited at 346 nm, and the fluorescence emission was collected at 505 nm for 30 s before the addition of 1,3-benzenedicarboxylic acid, 4,4′-[1,4,10,13-tetraoxa-7,16-diazacyclooctadecane-7, 16-diylbis(5-methoxy-6,2-benzofurandiyl)]bis (PBFI; Molecular Probes) to a 1 μM final concentration. Data were collected for an additional 2 min to establish a baseline signal before the addition of test compounds. For the control sample, 10 μg of valinomycin/ml was added to the S. aureus cells and followed 15 min later by K+ at a 1 mM final concentration to demonstrate continued indicator responsiveness. For some daptomycin-treated samples, bacterial cells were incubated for 5 min after the addition of daptomycin (1 to 10 μg/ml) and before the addition of Ca2+ (50 mg/liter, final concentration). Data were normalized relative to the fluorescent signal before the addition of calcium (average of the last 10 values prior to addition).
Statistical methods.
In the fluorimeter membrane potential experiments, some data were expressed as a percentage of the control, determined by the following: [(max/min)control/(max/min)sample] × 100%, where max was defined as the maximum fluorescent signal in the trace (usually <10 s after dye addition) and min was defined as the minimal signal (the average signal of the last 10 s of the trace) for the control or antimicrobial agent-treated samples. In flow cytometry experiments, some data were expressed as the percent polarized, which was defined as the total number of cells with a fluorescence intensity ratio of >135 arbitrary units divided by the total number of cells counted (n = 5,000).
RESULTS
Membrane depolarization correlates with bactericidal activity.
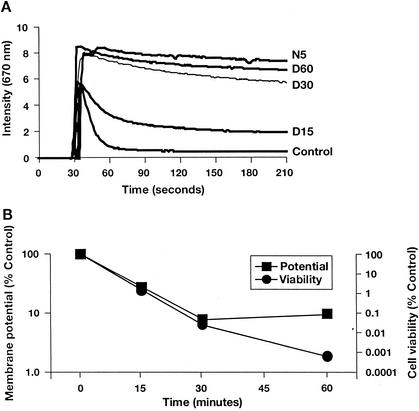
Research has previously demonstrated that daptomycin is capable of reducing the membrane potential of S. aureus (1). However, these studies were performed under conditions in which it was impossible to monitor effects on cell viability and at relatively high (100 μg/ml) drug concentrations. We have reexamined daptomycin's effects on membrane potential with an assay designed to allow simultaneous monitoring of potential and viability under conditions similar to those used for MIC or bactericidal testing. The assay uses DiSC3(5), a membrane potential-sensitive fluorescent probe. The fluorescence of DiSC3(5) decreases as the dye partitions to the surface of polarized cells; depolarization prevents partitioning and can release bound dye into the media. Hence, control cells (no drug treatment) produce a low signal and depolarized cells produce a high signal (24). As shown in Fig. 1A, the addition of daptomycin at 5 μg/ml (approximately eight times the MIC) gradually dissipated the membrane potential in S. aureus. Full depolarization required 30 to 60 min compared with <5 min for the pore-forming antimicrobial nisin (25 μg/ml, four times the MIC) (Fig. 1A) and the proton ionophore CCCP (data not shown). Cell viability decreased in parallel with the changes in membrane potential (Fig. 1B), demonstrating a temporal correlation between these two processes. After 30 min, <0.1% of cells remained viable and membrane potential was reduced to <10% of that of the control. The apparent lack of correlation between membrane potential and cell viability at the 60-min time point probably reflects the limited sensitivity of the depolarization assay.
FIG. 1.
Daptomycin dissipates membrane potential in S. aureus, as measured by fluorimetry. S. aureus cell samples were incubated for 0 (control), 15 (D15), 30 (D30), or 60 (D60) min with 5 μg of daptomycin/ml or for 5 min with 25 μg of nisin (N5)/ml. (A) Individual traces. (B) Changes in membrane potential and cell viability, expressed as percentages of the untreated control.
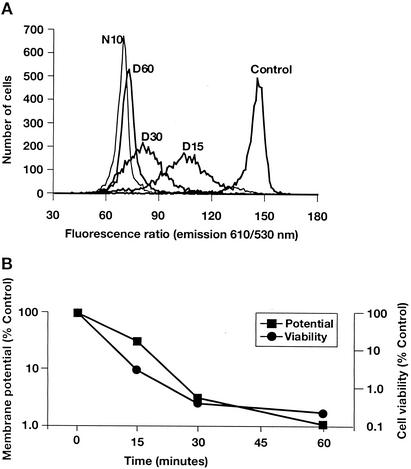
To confirm the results observed with the fluorimetric assay, membrane potential was also studied via flow cytometry with DiOC2(3), a membrane potential-sensitive dye capable of ratiometric measurements of bacterial membrane potential (17). Similar to the fluorometric analysis, flow cytometric assays were performed concurrently with cell viability measurements. Daptomycin gradually dissipated the membrane potential in this assay, with full membrane depolarization observed between 30 and 60 min (Fig. 2A). This timeframe was consistent with the fluorometric analysis results. Further examination of the flow cytometry data suggested that the entire population of S. aureus cells was progressively depolarized, resulting in a wide but unimodal distribution. This distribution suggests that membrane potential in individual bacteria is lost gradually rather than instantaneously. As demonstrated with the fluorometric analysis, there was also a close correlation between the kinetics of membrane depolarization and cell viability (Fig. 2B). Cell viability was reduced to <1% and membrane potential was <10% of that of the control. The sensitivity of the flow cytometry assay allowed the correlation to be demonstrated throughout the 60-min time course. Membrane permeability was monitored simultaneously by measuring the uptake of the membrane-impermeant fluorescent indicator To-Pro-3 iodide. To-Pro-3 iodide fluorescence was not increased in cells treated with daptomycin for as long as 60 min, indicating that membrane depolarization was not caused by rupture or lysis of the membrane (data not shown).
FIG. 2.
Daptomycin dissipates membrane potential in S. aureus, as measured by flow cytometry. S. aureus cell samples were incubated for 0 (control), 15 (D15), 30 (D30), or 60 (D60) min with 5 μg of daptomycin/ml or for 10 min with 25 μg of nisin (N10)/ml. (A) Individual traces. (B) Changes in membrane potential (as measured by the ratio of fluorescence emission intensities at 610 and 530 nm) and cell viability, both expressed as percentages of the untreated control.
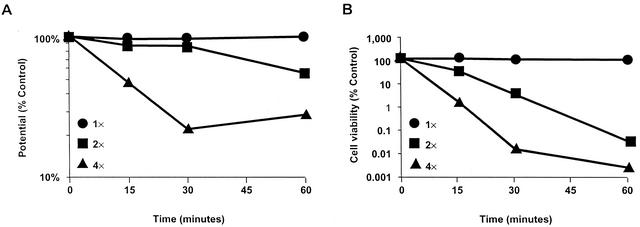
Recently, published studies have demonstrated that some antimicrobial peptides generally believed to act via disruption of membrane function may actually have intracellular targets and that membrane depolarization may only occur in response to artificially high drug concentrations (9). Therefore, we used low doses of daptomycin (one to four times the MIC [MIC = 0.78 μg/ml]) to measure daptomycin bactericidal activity and membrane depolarization in the fluorimeter assay (results are shown in Fig. 3). At daptomycin doses as low as the MIC, bactericidal activity and membrane depolarization were correlated for daptomycin-treated S. aureus cells. The activity of low daptomycin doses was confirmed by using the flow cytometry assay (data not shown).
FIG. 3.
Dose response for membrane potential and cell viability. Membrane potential (A) and cell viability (B) were measured by using the fluorimetry assay after daptomycin treatment at one, two, and four times the MIC. Results from both assays were graphed as percentages of the untreated control.
Daptomycin triggers potassium efflux in S. aureus.
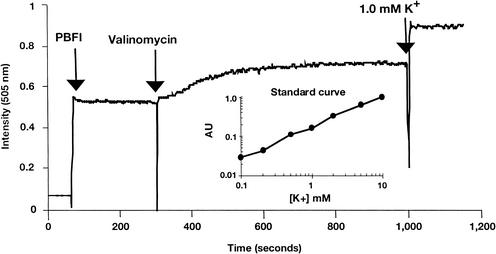
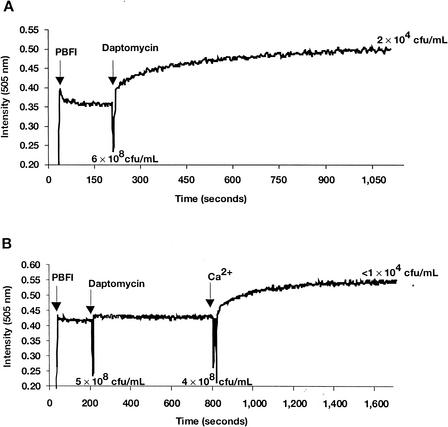
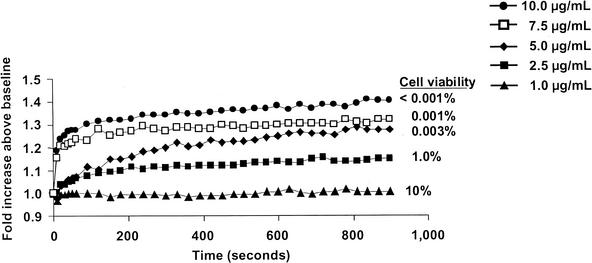
Dissipation of membrane potential requires ion movement across the cytoplasmic membrane. Using the K+-sensitive fluorescent probe PBFI, we examined the ability of daptomycin to trigger potassium release. The addition of PBFI to a suspension of S. aureus cells yielded a stable fluorescent signal (Fig. 4). Adding the potassium ionophore valinomycin resulted in an immediate signal increase consistent with the release of intracellular K+ into the surrounding medium. Adding daptomycin to S. aureus also triggered a similar signal increase (Fig. 5A), accompanied by a cell viability decrease. This result suggests that one component of the mechanism of action of daptomycin is the release of K+. The antibacterial activity of daptomycin is dependent on the physiologic levels of calcium ions. Daptomycin-dependent K+ release was observed only in the presence of Ca2+. As illustrated in Fig. 5B, incubation of S. aureus cells for 10 min with 5-μg/ml daptomycin in the absence of Ca2+ resulted in no significant increase in signal or decrease in cell viability. (The modest change in fluorescence intensity detected upon addition of daptomycin also occurred in the absence of bacterial cells [data not shown].) Adding calcium to the daptomycin-containing sample resulted in an immediate increase in fluorescence, consistent with a rapid release of K+. This was accompanied by a rapid loss of cell viability, as measured 15 min after the addition of calcium. Finally, as demonstrated in the membrane potential experiments, there was a clear correlation between bactericidal activity and the extent and initial rate of K+ release at daptomycin doses between 10 and 1 μg/ml (Fig. 6), although the limited sensitivity of the assay compresses the dose response.
FIG. 4.
Potassium release assay. PBFI (1 μM), valinomycin (10 μg/ml), and KCl (1 mM final concentration) were added at the indicated time points to S. aureus in HEPES-glucose solution. AU, arbitrary unit.
FIG. 5.
Daptomycin triggers potassium release from S. aureus. (A) PBFI (1 μM) and daptomycin (5 μg/ml) were added to S. aureus in HEPES-glucose solution containing 1 mM CaCl2 at the indicated time points. (B) PBFI (1 μM), daptomycin (5 μg/ml), and Ca2+ (50 mg/liter) were added to S. aureus at the indicated time points. For both experiments, viable cell counts were determined at the indicated time points.
FIG. 6.
Bactericidal activity and potassium release. Potassium release was triggered by the addition of 1 mM CaCl2. Data were normalized to the pre-calcium addition baseline values. The cell viability determined 15 min after calcium addition is indicated for each daptomycin dose level.
DISCUSSION
Daptomycin is a novel antibiotic with rapid in vitro bactericidal activity against gram-positive organisms. Although the mechanism of action has not been fully elucidated, previous studies have suggested that daptomycin activity results in the dissipation of cytoplasmic membrane potential (1). In these earlier studies, membrane potential was monitored in S. aureus by using the equilibration of the membrane-permeant anion tetraphenylphosphonium bromide with bacterium and daptomycin concentrations of 1010 CFU/ml and 100 μg/ml, respectively (1). Bacterial viability measurements were not made and no correlation could be established between membrane depolarization and daptomycin bactericidal activity. Therefore, we measured membrane potential under conditions typically used for determining the time-kill kinetics of daptomycin—106 to 107 CFU of mid-exponential-phase cells/ml in MHBc, with daptomycin concentrations of one to eight times the MIC.
Exposure to daptomycin gradually (within 30 to 60 min) dissipated the membrane potential in S. aureus cells. The relatively slow depletion of membrane potential suggests a novel mechanism of action compared with other antimicrobials. For example, the pore-forming peptide nisin and the proton ionophore CCCP disrupted the membrane potential in <5 min. The mechanism of action of daptomycin also differs from that of some antimicrobial peptides (e.g., human neutrophil peptide 1) that are capable of rapid depolarization of the cytoplasmic membrane but do not induce cell death for 1 to 2 h (26, 27). It has been suggested that membrane depolarization is not the primary target of some antimicrobial peptides but that depolarization is required to facilitate antibiotic entry into the bacteria for expression of activity (9). This requirement could account for a lag time between membrane depolarization and bacterial killing. The lack of any kinetic separation between depolarization and killing in daptomycin-treated cells strongly suggests that depolarization is the primary mechanism of action. The correlation between membrane depolarization and bactericidal killing observed even at one to two times the MIC further supports this idea and suggests that membrane depolarization is not the result of artificially high antibiotic concentrations. The consistent correlation between these two activities strongly suggests that membrane depolarization is an inherent part of the mechanism of action of daptomycin.
Establishing the importance of membrane depolarization in daptomycin's bactericidal activity prompted an attempt to begin to discern the detailed mechanism of cell depolarization. Bacteria maintain membrane potential by establishing multiple ion gradients across the cytoplasmic membrane. Because proper maintenance of the K+ gradient is important to bacterial viability, we measured K+ release during exposure to daptomycin in S. aureus. Adding daptomycin resulted in a calcium-dependent release of K+ in S. aureus. This calcium-dependent activity supports previous observations indicating that physiologic concentrations of calcium are required for daptomycin activity (3, 13). In addition, potassium release and viability loss were correlated. In these experiments, bactericidal activity was more rapid compared than in membrane depolarization studies. This increased activity resulted from the use of HEPES-glucose assay media in these experiments as opposed to the rich media (MHBc) used in the membrane potential assays. The analysis of the K+ gradient during daptomycin exposure suggests that K+ release may be responsible for membrane depolarization or that K+ may be one of several ions released during membrane depolarization by daptomycin.
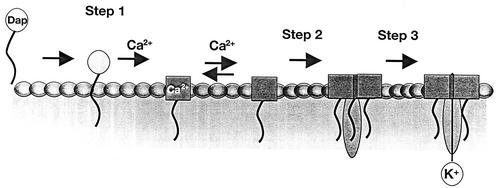
We have demonstrated a clear correlation between daptomycin bactericidal activity and membrane potential dissipation and the calcium-dependent release of potassium from daptomycin-exposed cells. We have proposed a multistep model for the mechanism of action of daptomycin based on the results of these and previously published experiments (Fig. 7). During step 1 of the process, daptomycin inserts into the cytoplasmic membrane of bacteria, as indicated by whole-cell and artificial membrane studies (8, 13). Artificial bilayer studies suggest that insertion can occur in the absence of a bacterium-specific component; however, the possible existence of a gram-positive specific receptor for daptomycin has not been ruled out. Step 2 postulates an as-yet-unproven oligomerization event: daptomycin oligomers could form ion channels, larger nonspecific pores, or irregular aggregate structures (25). The latter have been implicated in the mechanism of action of iturin A, an antifungal membrane-active lipopeptide (15). Alternatively, a cellular component could contribute to this process (5, 6). Formation of any of these structures disrupts the functional integrity of the membrane and triggers a release of intracellular ions (e.g., K+) leading to rapid cell death (step 3). Additional whole-cell and in vitro studies are being conducted to understand the behavior of other ions (e.g., Na+ and H+) and larger molecules (e.g., ATP and proteins) in the presence of daptomycin to determine the nature of the proposed ion-conducting structure.
FIG. 7.
Proposed model for the bactericidal mechanism of action of daptomycin. Daptomycin (Dap) inserts into the bacterial cytoplasmic membrane in a calcium-dependent fashion and is followed by oligomerization and disruption of the functional integrity of the cytoplasmic membrane (as described in the text), triggering a release of intracellular ions and rapid cell death.
These studies support the hypothesis that the in vitro bactericidal activity of daptomycin is a result of cytoplasmic membrane potential dissipation in gram-positive organisms. This mechanism of action is novel compared with classes of antibiotics currently marketed; the relatively gradual rates of depolarization suggest the mechanism of action is also novel compared with other membrane-targeted compounds. Daptomycin may provide another critical line of defense against increasingly antibiotic-resistant gram-positive pathogens.
Acknowledgments
N.G.P. and H.M.S. were supported in part by National Institutes of Health grant AI48354.
We thank Jeff Alder, Skip Shimer, Andrea McCracken, and Julian Davies for helpful discussions.
REFERENCES
- 1.Alborn, W. E., Jr., N. E. Allen, and D. A. Preston. 1991. Daptomycin disrupts membrane potential in growing Staphylococcus aureus. Antimicrob. Agents Chemother. 35:2282-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, N. E., W. E. Alborn, Jr., and J. N. Hobbs, Jr. 1991. Inhibition of membrane potential-dependent amino acid transport by daptomycin. Antimicrob. Agents Chemother. 35:2639-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrew, J. H., M. C. Wale, L. J. Wale, and D. Greenwood. 1987. The effect of cultural conditions on the activity of LY146032 against staphylococci and streptococci. J. Antimicrob. Chemother. 20:213-221. [DOI] [PubMed] [Google Scholar]
- 4.Barry, A. L., P. C. Fuchs, and S. D. Brown. 2001. In vitro activities of daptomycin against 2,789 clinical isolates from 11 North American medical centers. Antimicrob. Agents Chemother. 45:1919-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boaretti, M., and P. Canepari. 1995. Identification of daptomycin-binding proteins in the membrane of Enterococcus hirae. Antimicrob. Agents Chemother. 39:2068-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boaretti, M., and P. Canepari. 2000. Purification of daptomycin binding proteins (DBPs) from the membrane of Enterococcus hirae. New Microbiol. 23:305-317. [PubMed] [Google Scholar]
- 7.Boaretti, M., P. Canepari, M. D. M. Lleo, and G. Satta. 1993. The activity of daptomycin on Enterococcus faecium protoplasts: indirect evidence supporting a novel mode of action on lipoteichoic acid synthesis. J. Antimicrob. Chemother. 31:227-235. [DOI] [PubMed] [Google Scholar]
- 8.Canepari, P., M. Boaretti, M. del Mar Lleo, and G. Satta. 1990. Lipoteichoic acid as a new target for activity of antibiotics: mode of action of daptomycin (LY146032). Antimicrob. Agents Chemother. 34:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedrich, C. L., D. Moyles, T. J. Beveridge, and R. E. Hancock. 2000. Antibacterial action of structurally diverse cationic peptides on gram-positive bacteria. Antimicrob. Agents Chemother. 44:2086-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs, P. C., A. L. Barry, and S. D. Brown. 2002. In vitro bactericidal activity of daptomycin against staphylococci. J. Antimicrob. Chemother. 49:467-470. [DOI] [PubMed] [Google Scholar]
- 11.Herranz, C., L. M. Cintas, P. E. Hernandez, G. N. Moll, and A. J. Driessen. 2001. Enterocin P causes potassium ion efflux from Enterococcus faecium T136 cells. Antimicrob. Agents Chemother. 45:901-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King, A., and I. Phillips. 2001. The in vitro activity of daptomycin against 514 Gram-positive aerobic clinical isolates. J. Antimicrob. Chemother. 48:219-223. [DOI] [PubMed] [Google Scholar]
- 12a.Laganas, V., J. Alder, and J. A. Silverman. 2003. In vitro bactericidal activities of daptomycin against Staphylococcus aureus and Enterococcus faecalis are not mediated by inhibition of lipoteichoic acid biosynthesis. Antimicrob. Agents Chemother. 47:2682-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakey, J. H., and M. Ptak. 1988. Fluorescence indicates a calcium-dependent interaction between the lipopeptide antibiotic LY146032 and phospholipid membranes. Biochemistry 27:4639-4645. [DOI] [PubMed] [Google Scholar]
- 14.Lamp, K. C., M. J. Rybak, E. M. Bailey, and G. W. Kaatz. 1992. In vitro pharmacodynamic effects of concentration, pH, and growth phase on serum bactericidal activities of daptomycin and vancomycin. Antimicrob. Agents Chemother. 36:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maget-Dana, R., and F. Peypoux. 1994. Iturins, a special class of pore-forming lipopeptides: biological and physicochemical properties. Toxicology 87:151-174. [DOI] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Novo, D., N. G. Perlmutter, R. H. Hunt, and H. M. Shapiro. 1999. Accurate flow cytometric membrane potential measurement in bacteria using diethyloxacarbocyanine and a ratiometric technique. Cytometry 35:55-63. [DOI] [PubMed] [Google Scholar]
- 18.Ruhr, E., and H. G. Sahl. 1985. Mode of action of the peptide antibiotic nisin and influence on the membrane potential of whole cells and on cytoplasmic and artificial membrane vesicles. Antimicrob. Agents Chemother. 27:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rybak, M. J., E. Hershberger, T. Moldovan, and R. G. Grucz. 2000. In vitro activities of daptomycin, vancomycin, linezolid, and quinupristin-dalfopristin against staphylococci and enterococci, including vancomycin-intermediate and -resistant strains. Antimicrob. Agents Chemother. 44:1062-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahl, H. G., M. Kordel, and R. Benz. 1987. Voltage-dependent depolarization of bacterial membranes and artificial lipid bilayers by the peptide antibiotic nisin. Arch. Microbiol. 149:120-124. [DOI] [PubMed] [Google Scholar]
- 21.Silverman, J. A., N. Oliver, T. Andrew, and T. Li. 2001. Resistance studies with daptomycin. Antimicrob. Agents Chemother. 45:1799-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snydman, D. R., N. V. Jacobus, L. A. McDermott, J. R. Lonks, and J. M. Boyce. 2000. Comparative in vitro activities of daptomycin and vancomycin against resistant gram-positive pathogens. Antimicrob. Agents Chemother. 44:3447-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wise, R., J. M. Andrews, and J. P. Ashby. 2001. Activity of daptomycin against Gram-positive pathogens: a comparison with other agents and the determination of a tentative breakpoint. J. Antimicrob. Chemother. 48:563-567. [DOI] [PubMed] [Google Scholar]
- 24.Wu, M., and R. E. Hancock. 1999. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J. Biol. Chem. 274:29-35. [DOI] [PubMed] [Google Scholar]
- 25.Wu, M., E. Maier, R. Benz, and R. E. Hancock. 1999. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. 38:7235-7242. [DOI] [PubMed]
- 26.Xiong, Y. Q., M. R. Yeaman, and A. S. Bayer. 1999. In vitro antibacterial activities of platelet microbicidal protein and neutrophil defensin against Staphylococcus aureus are influenced by antibiotics differing in mechanism of action. Antimicrob. Agents Chemother. 43:1111-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeaman, M. R., A. S. Bayer, S. P. Koo, W. Foss, and P. M. Sullam. 1998. Platelet microbicidal proteins and neutrophil defensin disrupt the Staphylococcus aureus cytoplasmic membrane by distinct mechanisms of action. J. Clin. Investig. 101:178-187. [DOI] [PMC free article] [PubMed] [Google Scholar]