Abstract
The retinoblastoma protein pRB is involved in the transcriptional control of genes essential for cell cycle progression and differentiation. pRB interacts with different transcription factors and thereby modulates their activity by sequestration, corepression, or activation. We report that pRB, but not p107 and p130, binds to and facilitates repression by p120E4F, a ubiquitously expressed GLI-Kruppel-related protein identified as a cellular target of E1A. The interaction involves two distinct regions of p120E4F and the C-terminal part of pRB. In vivo pRB–p120E4F complexes can only be detected in growth-arrested cells, and accordingly contain the hypophosphorylated form of pRB. Repression of an E4F-responsive promoter is strongly increased by combined expression of p120E4F and pRB, which correlates with pRB-dependent enhancement of p120E4F binding activity. Elevated levels of p120E4F have been shown to block growth of mouse fibroblasts in G1. We find this requires pRB, because RB−/− fibroblasts are significantly less sensitive to excess p120E4F.
The members of the retinoblastoma protein family, pRB, p107, and p130 are key signal transducers connecting the cell cycle machinery to the transcriptional control of sets of genes whose products regulate cell growth and differentiation processes. Their cellular function depends on their ability to bind to and modulate the activity of different transcription factors (1–3). Most studies on the pRB family members have focused on their interaction with the E2F/DP family of transcription factors. During G0 and G1 phases of the mammalian cell cycle, the hypophosphorylated form of pRB interacts with E2F/DP bound to DNA, acting either as a sequestrating factor by blocking the transactivating domain of E2Fs or as a corepressor by recruiting histone deacetylases (1–3). Before the G1/S transition, cyclin-associated kinases further phosphorylate pRB, causing its dissociation from E2Fs and thus allowing transcription of E2F target genes. Beside E2Fs, there is also evidence that pRB binds to and promotes the transcriptional activity of many other factors (1–3), including MyoD (4), CCAAT/enhancer binding proteins (C/EBPs, NF-IL6) (5, 6), activator protein-1 (AP-1) factors (7–9), or HBP1 (10). pRB was proposed to enhance their DNA binding, acting as an activator required for setting up the correct assembly, conformation, or concentration of these factors (5, 6, 8, 9).
We now report physical and functional interactions between pRB and the E1A-regulated transcription factor p120E4F. Adenovirus E1A proteins prepare the host cell for viral replication, stimulating cell cycling and viral transcription through the targeting of several critical cellular transcription factors, including E2F complexes and E4F (11, 12). Activation of E4F is essential for the transcription of the adenovirus E4 gene, a product of which (ORF 6/7) associates with E2Fs to drive efficiently the adenovirus E2 promoter (13, 14). Human E4F (13–15) and its mouse homologue, phiAP3 (16), are low abundance and ubiquitously expressed GLI-Kruppel-related zinc finger phosphoproteins synthesized as a predominant 120-kDa species, p120E4F (15–19). In E1A-infected cells, a proteolytic product of 50 kDa, p50E4F, that retained the N-terminal region of p120E4F, was also detected (17). Although both human p50E4F and p120E4F recognize the same DNA motifs (RTGACGTC/AAY) in vitro, they may differentially regulate gene expression in cells. Thus, while p50E4F was shown to transactivate a reporter gene driven by the adenoviral E4 promoter, p120E4F repressed the same reporter (17). In addition, phiAP3, the mouse homologue of p120E4F, was initially identified as a cellular factor binding to and negatively regulating the promoter of the E1A oncogene (16, 20). Cellular genes regulated by p120E4F and/or by p50E4F are currently unknown, but it is likely that they are involved in pathways controlling cell cycle progression and apoptosis because it was reported that overexpression of p120E4F in mouse fibroblasts induced cell cycle arrest in late G1 (21) whereas that of p50E4F in E1A-transformed cells accelerated apoptosis (22).
Here we report that cellular pRB–p120E4F complexes are formed between the hypophosphorylated form of pRB and p120E4F in growth-arrested but not in actively growing cells. pRB enhances both p120E4F DNA binding activity and p120E4F-mediated promoter repression. By using pRB-deficient cells we also present evidence that the antiproliferative effect of p120E4F is, at least in part, dependent on the presence of a functional RB gene. Based on these findings we propose a novel role for pRB as a negative regulator of cell proliferation through its potentiating effect on p120E4F.
Materials and Methods
DNA Plasmids, cDNA Library.
Details of all constructs are available on request. The serum-starved WI38 human fibroblast cDNA library in the multicopy prey plasmid pJG4–5 (23) was described previously (24, 25). Baits for the two-hybrid assays in pEG202 plasmid (23) are as follows: pEG202–RB (aa 2–928 of pRB), pEG202–RB661W [aa 2–894 of pRB pocket mutant RB661W (26)], and pEG202–p107 (aa 1–1068 of p107). Mammalian E4F expression constructs in pcDNA3 (Invitrogen) are as follows: pcDNA–p120E4F (full-length E4F) pcDNA–p50E4F (aa 1–357 of E4F), pcDNA3–E4F–Cter (aa 357–784) and pcDNA–E4F–Sma (aa 1–161). E4F glutathione S-transferase (GST) fusions in pGEX4T (Pharmacia): pGEX–p120E4F (full-length E4F), pGEX–p50E4F (aa 1–357), pGEX–E4F–Cter (aa 357–784). GST–RB constructs have been described previously (27). The reporter E4F—TK–LUC was constructed by cloning a single copy of the double-stranded oligonucleotide [(+) GATCCTGACGTAACA/(−)GATCTGTTACGTCAG], corresponding to the E4F binding site of the adenoviral E4 promoter, upstream the HSV-TK (herpes simplex virus thymidine kinase) minimal promoter and the firefly luciferase gene in pGL2 (Promega).
Cell Culture, Protein Extracts, Transfections, and Luciferase Assays.
U2OS and NIH 3T3 were grown in DMEM, 10% FCS (Biomedia, Foster City, CA). Rb+/+, Rb−/−, and p107/p130−/− 3T3 fibroblasts were established from mouse embryo fibroblasts (MEFs) by M. Classon (Massachusetts General Hospital, Boston) and maintained as described previously (28). Cells were growth arrested at confluence and by serum starvation for 48 h. Nuclear and whole cell extracts were prepared as previously described (28). All transfections were performed by using the Lipofectamine Plus reagent following the manufacturer's instructions (GIBCO/Life Technologies). For the E4F—RB-mediated repression assays, NIH 3T3 cells or C33A cells were cotransfected with 0.2 μg of pCH110 (β-galactosidase expression vector) and 2 μg of E4Fs–TK–LUC reporter vector alone or with various combinations of 100 ng of pcDNA–p120E4F and pECE–RBp34 (29). β-Galactosidase and luciferase activity were measured 36 h later as previously described (28). Luciferase activity values were normalized to the β-galactosidase activity to account for variations in transfection efficiencies.
Two-Hybrid Screening.
Two-hybrid screening in yeast was performed essentially as previously described (23, 24). The yeast strain is EGY048/lacZ (23) carrying two reporters whose expression is regulated by LexA-responsive promoters; i.e., a chomosomally integrated LEU2 reporter gene (LexA:LEU2) and the 2 μ LacZ reporter plasmid pSH18–34 (23). pEG202–RB was used as a bait in EGY048/lacZ to screen the WI38 pjG4-5 prey library. A total of 2 × 106 yeast cotransformants were selected for galactose-induced reporter-dependent leucine prototrophy. Prey plasmids were rescued from positive clones and retransformed into EGY048/lacZ strains expressing pEG202–RB, pEG202–RB668W, pEG202–p107, pEG202–p130, or pEG202–bicoid. Cotransformants were re-selected for reporter-dependent leucine prototrophy and β-galactosidase production; positive clones were sequenced. β-Galactosidase activities of galactose-induced exponentially growing cultures were measured as previously described (23).
In Vitro and in Vivo Pull-Down Assays and Coimmunoprecipitation.
In vitro translation reactions of E4F were performed by using [35S]methionine (Amersham) in a TNT-coupled reticulocyte lysate (Promega) according to the manufacturer's instructions. The plasmids pcDNA–p120E4F, pcDNA–p50E4F, and pcDNA–E4F–Cter were used as templates to generate the fragment FL (aa 1–784), N (aa 1–357), and C (358–784), respectively. Template to generate fragment S of E4F (aa 358–486) was a PCR product amplified by using pcDNA–E4F–Cter as a matrix. Fragments F1 (aa 184–254), F2 (aa 428–521), and F3 (aa 515–584) of E4F were produced by using PCR templates containing the sequence of the T7 promoter, generated by using the following pairs of primers: F1, (+)TAATACGACTCACTATAGGGACATGAACAAGGATGGCCGC/(−)CTACTTTCCACACTTGGAGCACTT; F2, (+)TAATACGACTCACTATAGGGACATGTCAGCGGTGCCCAGG/(−)CTAGGGACAAGGGTAGGGCCGCAC; F3, (+)TAATACGACTCACTATAGGGACATGGAGCGGCCCTACCCT/(−)CTAGGCGAAGCCACGGCCGCACTT.
For in vitro pull-down assays, the indicated GST–RB fusion proteins were purified from Escherichia coli (XL1Blue) by using glutathione-Sepharose beads (Pharmacia) as described previously (30). GST–RB bound beads were incubated for 1 h at room temperature (RT) with the indicated 35S-labeled in vitro-translated E4F constructs in pull-down buffer [50 mM Hepes (pH 7.8), 150 mM NaCl, 0.1% Nonidet P-40, 1 mM DTT, and 1 mM PMSF]. Proteins retained on the GST–RB beads were washed five times with 1 ml of pull-down buffer, solubilized in SDS-loading buffer, and analyzed on 8% acrylamide SDS/PAGE gels. For the in vivo pull-down assay, 2 μg of GST–p120E4F coupled to glutathione-Sepharose beads were incubated with 100 μg of nuclear extracts prepared from NIH 3T3 cells (28) in pull-down buffer for 2 h at RT. After three washes, bound proteins were separated on 8% acrylamide SDS/PAGE gels, blotted, and probed with either anti-pRB, G3–245 (PharMingen), anti-p107, C18 (Santa Cruz Biotechnology), or anti-p130 (Transduction Laboratories, Lexington, KY) Abs.
For coimmunoprecipitation of cellular pRB by E4F, 200 μg U2OS whole cell extracts were incubated with protein A beads coupled to either anti-E4F antibodies or preimmune serum in IP buffer [150 mM NaCl, 0.5% Nonidet P-40, and 50 mM Tris (pH 8)] for 1 h at RT. Bound proteins were washed three times in 1 ml IP buffer and separated on 8% acrylamide SDS/PAGE gels, blotted, and finally probed with anti-pRB (G3-245, PharMingen). For coimmunoprecipitation of cellular E4F by pRB, 500 μg U2OS nuclear cell extracts were incubated with agarose beads (AminoLink Coupling gel, Pierce) covalently coupled to either anti-pRB mAbs (21C9) or control mAbs [control 1: anti-HA tag, 12CA5; control 2: anti E2F5, 274 (46)] in IP buffer [150 mM NaCl, 0.5% Nonidet P-40, and 50 mM Tris (pH 8)] overnight at 8°C. Bound proteins were washed three times in 1 ml IP buffer and separated on 8% acrylamide SDS/PAGE gels, blotted, and finally probed with anti-E4F (88.2).
Electrophoretic Mobility Shift Assays (EMSA).
Suboptimal amounts of purified GST–p120E4F fusions (30) were incubated either alone or in combination with increasing amounts of recombinant human pRB purified from baculovirus-infected SF9 cells (31) for 15 min at RT in a total volume of 20 μl binding buffer [10 mM Tris-HCl (pH 7.9), 40 mM KCl, 10% glycerol, 0,05% Nonidet P-40 and 1 mM DTT, and 1 μg poly(dI⋅dC)] in presence of 1 ng of a T4–PNK end-labeled double-stranded oligonucleotide probe bearing the E4F binding site of the E4 promoter [(+)GATCCCGGATGTGGCAAAAGTGC]. DNA–protein complexes were separated by electrophoresis on a 4% polyacrylamide gel in 0.25× TBE buffer at RT and 10 V/cm.
Immunofluorescence.
Cells were grown on cover-slips; 24 h after transfection with the indicated plasmids they were incubated for 12 h with BrdUrd. After fixation and permeabilization with methanol, cells were treated with 1.5 M HCl for 10 min at RT and incubated with a purified anti-E4F rabbit polyclonal Ab (E4F 88.2) together with an anti-BrdUrd mouse mAb (Dako). Preparations were then incubated with a combination of Texas Red-conjugated anti-rabbit IgG and FITC-conjugated anti-mouse IgG. E4F 88.2 Ab was raised against the E4F peptide EEDEDDVHRCGRCQA (aa 50–64) and was affinity purified on an agarose-peptide column before use.
Results
Identification of p120E4F as a pRB Interacting Protein.
Our initial purpose was to use the yeast two-hybrid system to identify pRB interactors expressed in quiescent human fibroblasts. A LexA-pRB construct was used as a bait to screen a cDNA library of preys generated from confluent and serum-starved WI38 human fibroblasts (23–25). Two classes of clones were selected. One encoded E2Fs (E2F1 and E2F4). The second encoded the E1A-regulated transcription factor p120E4F, which was represented by three independent positive clones containing full-length p120E4F cDNAs. We then compared the binding of E2Fs and p120E4F to various pocket proteins in a two hybrid interaction assay by using a LacZ reporter strain (Table 1). E2F1 and E2F4 interacted, although with different intensities, with all three pocket proteins but not with a pocket mutant of pRB. By contrast to E2Fs, p120E4F bound strongly to both wild-type and mutant pRB, but interacted very weakly with p130 and not at all with p107. Thus, at least in this yeast assay, p120E4F displays a pocket protein binding specificity distinct from that of E2Fs.
Table 1.
Differential interactions of p120E4F, E2F1, and E2F4 with pocket protein family members in a two-hybrid assay
| Preys | Baits
|
||||
|---|---|---|---|---|---|
| pRB | RB61W | p107 | p130 | Bicoid | |
| p120E4F | +++ | +++ | − | + | − |
| E2F-1 | ++ | − | + | + | − |
| E2F-4 | ++ | − | ++ | ++ | − |
A yeast strain containing a LacZ reporter plasmid driven by a lexA responsive promoter was cotransfected with indicated combinations of baits and preys (23–25). LexA-fusion baits: human pRB, the pRB mutant RB61W that fails to bind E1A and E2Fs (38), the pRB-related proteins p107 and p130, and bicoid as a control (23). B42-fusion preys: human p120E4F, E2F1, and E2F4. β-Galactosidase activities that reflect the strength of the bait/prey interaction were measured on cultures of three isolates of each strain exponentially growing in 2% galactose. β-Galactosidase activities measured in Miller units were ordered as: +++, Miller units >1000; ++, >500; +, >50; and −, <50. Comparable levels of expression of the various baits were observed by using Abs directed against lexA (data not shown).
In Vivo Detection of p120E4F–pRB Complexes.
We next tested whether recombinant p120E4F was able to bind cellular pocket proteins in vitro. Pull-down assays were performed with GST–p120E4F or an unrelated protein, GST–ELK, and nuclear extracts prepared from confluent 3T3 fibroblasts established from RB+/+ and RB−/− MEF (a gift from M. Classon; refs. 28 and 32). Bound proteins were then examined for the presence of pRB by immunoblotting. pRB was clearly detected on GST–p120E4F beads and not on control beads (Fig. 1A). Furthermore, the pRB band was not observed using nuclear extracts from RB-deficient cells (Fig.1A). Interestingly, we failed to detect GST–p120E4F–pRB complexes in nuclear extracts prepared from exponentionally growing NIH 3T3 fibroblasts (Fig.1B) or from S phase-synchronized cells (data not shown), suggesting that the p120E4F–pRB interaction might be cell-cycle dependent. Consistent with this, p120E4F-bound pRB appeared to migrate as the hypophosphorylated pRB (Fig. 1B) which is the predominant and active form of the protein in growth-arrested cells (1–3).
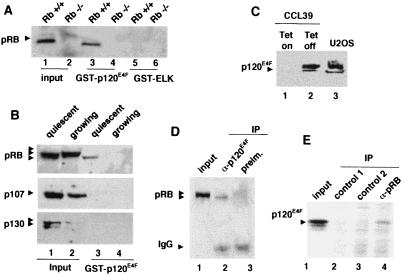
Figure 1.
p120E4F associates with pRB. (A) Recombinant p120E4F associates with cellular pRB. GST pull-down assays were performed with nuclear extracts prepared from confluent cultures of either RB+/+ (lanes 1, 3, and 5) or RB−/− NIH 3T3 mouse fibroblasts (lanes 2, 4, and 6). Extracts were incubated with either Sepharose-bound GST–p120E4F (lanes 3 and 4) or GST–ELK (negative control) (lanes 5 and 6). Bound proteins were released and probed for the presence of pRB by immunoblotting. One-third of the input extract was loaded in lane 1 and 2. (B) Recombinant p120E4F associates with the hypophosphorylated form of pRB present in arrested cells and does not associate at all with cellular p107 and p130. GST pull-down assays were performed as above by using nuclear extracts from either quiescent (lanes 1 and 3) or exponentionally growing (lanes 2 and 4) NIH 3T3 fibroblasts. Bound proteins (lanes 3 and 4) were probed for the presence of pRB (Top), p107 (Middle), or p130 (Bottom) by immunoblotting. One-third of the input extract was loaded in lane 1 and 2. (C) p120E4F is easily detectable in U2OS cells. Nuclear extract from U2OS cells is probed by immunoblotting by using an affinity-purified rabbit polyclonal antibody (E4F-88) (lane 3). Specificity of this Ab is tested on nuclear extracts prepared from a Chinese hamster cell line (CCL39-tet/E4F) expressing a tetracycline inducible p120E4F gene (lanes: 1, + tetracycline; 2, − tetracycline). (D and E) Coimmunoprecipitation of cellular p120E4F and pRB proteins from U2OS cells extracts. (D) p120E4F is immunoprecipitated from confluent U2OS nuclear extracts by using the E4F 88.2 Ab, and precipitate is probed for the presence of pRB by immunoblotting (lane 2). The same experiment is performed with the corresponding preimmune serum (lane 3). One-tenth of the input extract was loaded in lane 1. (E) pRB is immunoprecipitated from confluent U2OS nuclear extracts by using the anti-pRB mAb coupled to agarose beads and precipitate is probed for the presence of p120E4F by immunoblotting (lane 4). The same experiment is performed with control beads coupled to unrelated mAbs (lanes 2 and 3). One-thirtieth of the input extract was loaded in lane 1.
Interestingly and in agreement with the result of our two-hybrid experiment (Table 1), the other members of the pRB family, p107 and p130, were not found to associate with the GST–p120E4F beads (Fig. 1B). Thus, both sets of results suggest that pRB is the unique member of the pocket-protein family to associate with E4F in rodent fibroblasts.
We next explored whether crude nuclear extracts contained an endogenous p120E4F–pRB complex. To perform this analysis, we developed and affinity purified an antibody specific for human p120E4F (Fig. 1C). Coimmunoprecipitation experiments were not performed with mouse fibroblasts because endogenous p120E4F was hardly detectable in these cells (data not shown). We used whole cell or nuclear extracts prepared from confluent human U2OS cells where endogenous p120E4F and pRB were both easily visualized (Fig. 1C). On immunoprecipitation with the anti-p120E4F Ab (88.2), pRB was clearly and specifically detected by immunoblotting (Fig. 1D). The other way around, p120E4F was detected by immunoblotting among proteins bound to agarose beads covalently coupled to anti-pRB Abs (21C9) (Fig. 1C). Similar coimmunoprecipitation were also observed by using nuclear extracts from MCF7 cells blocked in G1 by leucine and serum depletion (data not shown). Together, these results strongly suggest that p120E4F–pRB complexes do exist in living cells.
Mapping the Binding Domains of pRB and p120E4F.
Several distinct protein-binding domains have been identified in pRB: The “large pocket,” composed of subdomains A and B, that binds E2Fs and proteins containing an LXCXE-motif, and the C-terminal domain that binds the nuclear c-Abl tyrosine kinase, UBF, and Jun proteins (Fig. 2B) (1–3). Pull-down assays were performed by using in vitro-translated radiolabeled p120E4F and GST fusion proteins encoding various domains of pRB (Fig. 2A). No binding was detected with pocket domain A alone, whereas GST–RB proteins containing either the three domains, A, B, and C, or only domains B and C, exhibited strong binding to p120E4F.
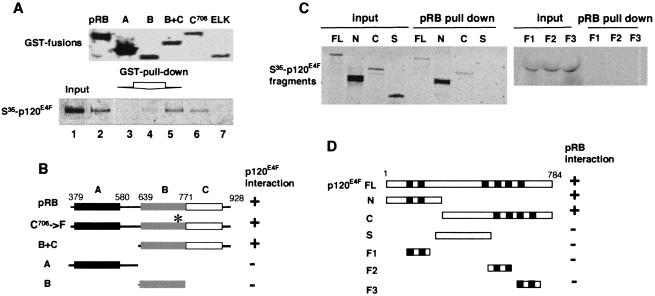
Figure 2.
Mapping of the binding domains of pRB and p120E4F. (A) GST pull-down assays using the indicated GST-pRB constructs (Upper) and 35S-labeled in vitro translated p120E4F (full-length) (lanes 2–6, Lower). Input of radiolabeled p120E4F is loaded lane 1 (Lower). As a control, GST pull-down assay is done with GST–ELK protein (lane 7). (B) Schematic representation of the various pRB protein constructs and summary of their capacity to bind p120E4F. Pocket domains A (aa 379–580), B (aa 639–771), C (aa 771–928) are indicated. The asterisk indicates the C→F mutation of pRBC706-F. (C) GST pull-down assays using the indicated 35S-labeled in vitro-translated p120E4F fragments and the GST–pRB construct containing domains A, B, and C of pRB. (D) Schematic representation of the various p120E4F fragments used in the assay and summary of their capacity to bind pRB. The zinc fingers domains are shown as black boxes. FL, full length (aa 1–784); N/p50E4F, aa 1–357; C, aa 358–784; S, aa 358–486; F1, aa 184–254; F2, aa 428–521; F3, aa 515–584.
A weak binding was also observed with domain B alone. Nevertheless, p120E4F was still able to bind to pRBC706F which bears a naturally occurring mutation in the pocket B of pRB that disrupts the binding to the transcription factor E2F (33). This finding suggested that intact pocket B was not required for interaction with p120E4F. No binding was observed with GST–ELK (Fig. 2A) or GST alone (data not shown), demonstrating the specificity of these p120E4F–pRB interactions. Together, these results suggest that the p120E4F–pRB interaction is mainly pocket A/B-independent and requires the pocket C-terminal region (Fig. 2B).
p120E4F is a GLI-Kruppel-related transcription factor containing six C2H2 zinc finger motives (Fig. 2D) which exists as a predominant 120-kDa protein p120E4F, and a less abundant, proteolytically derived 50-kDa N-terminal fragment (p50E4F) (15, 16). In vitro-translated radiolabeled p50E4F (N terminus) and p120E4F exhibited strong GST–pRB binding (Fig. 2C). Interestingly, the C-terminal domain of p120E4F, which is not present in p50E4F, also interacted with pRB. In contrast, no binding was detected with the various zinc finger motives and with the central domains of p120E4F (Fig. 2 C and D). Thus p120E4F can interact with pRB through at least two domains located in the C- and N-terminal parts of the protein, respectively.
pRB Up-Regulates the Transcriptional Activity of p120E4F.
Next we assessed what were the functional consequences of the p120E4F–pRB interaction on the binding and transrepression capabilities of p120E4F. Transient transfection assays, performed in NIH 3T3 cells, show that increasing amounts of p120E4F progressively decreased the activity of an E4F-responsive element-driven luciferase reporter construct, E4-TK-Luc (Fig. 3A). We then transfected a phosphorylation-defective constitutively active pRB (29) together with the maximum amount of p120E4F plasmid that was unable to inhibit the reporter activity (Fig. 3B, striped bar). This restored a strong reporter repression (Fig. 3B, dotted bar). This potentiating effect was specific for the presence of p120E4F because pRB alone had no effect on reporter activity (Fig. 3B, open bar). This effect was also specific for the E4F DNA-binding site because the same p120E4F–pRB combination did not affect a control reporter construct (Fig. 3B). To test whether this might be caused by an indirect effect of pRB on the cell cycle distribution of the transfected cells, we also performed similar reporter assays were in cervical carcinoma cell line C33A, which was previously shown to be resistant to pRB growth suppression (47). Results were identical to those obtained by using NIH 3T3 fibroblasts (data not shown).
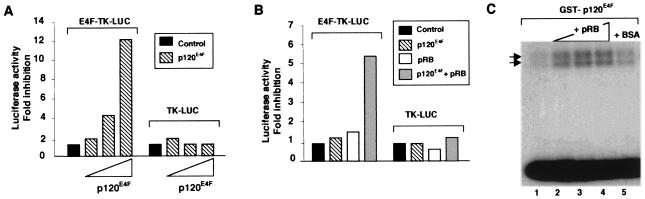
Figure 3.
pRB up-regulates the DNA binding and transrepression capabilities of p120E4F. (A) p120E4F is a site-dependent transcriptional repressor. NIH 3T3 cells are transiently cotransfected with an E4F-responsive reporter gene E4F-TK-Luc and increasing amounts of the pcDNA–p120E4F plasmid. TK-Luc (without E4F site) was used as a control. Luciferase activity is shown as fold inhibition of the activity obtained with E4F-TK-Luc or TK-Luc constructs transfected alone. (B) pRB up-regulates the transrepression capabilities of p120E4F. The maximum amount of p120E4F plasmid that was unable to inhibit the reporter activity as determined in A is cotransfected with either E4F-TK-Luc or TK-Luc and with (dotted bars) or without (stripped bars) a phosphorylation-defective constitutively active pRB (p34RB). As a control, E4s-TK-Luc and TK-Luc reporters were also transfected alone (filled bars) or with p34RB (open bars). (C) pRB up-regulates the DNA binding activity of p120E4F in vitro. EMSAs are performed with a probe containing an E4F DNA binding site. Purified GST–p120E4F is added alone (lane 1) or together with increasing amounts of baculovirus-expressed purified pRB protein (lanes 2–4) or BSA (lane 5).
One possibility was that pRB potentiated E4F-mediated repression through stimulation of DNA binding by p120E4F. To test this, EMSAs were performed by using a probe containing the p120E4F responsive element of the adenoviral E4 promoter and recombinant p120E4F and pRB proteins expressed in bacteria and baculovirus-infected cells, respectively (Fig. 3C). We used suboptimal concentrations of p120E4F (Fig. 3C, lane 1) with increasing amounts of pRB or of control proteins (BSA, GST–ELK) (Fig. 3C). Addition of pRB increased the binding of p120E4F to the probe, unlike BSA (Fig. 3C) or GST–ELK (data not shown). Recombinant pRB alone did not bind to the E4-probe and binding of p120E4F was competed by an excess of wild-type, but not of mutated, competitor (data not shown). However, several different pRB-specific antibodies did not supershift the p120E4F-specific complexes. Similar effects of pRB have been described previously with c-Jun, C/EBPs and NF-IL6 transcription factors (5, 6, 8). Thus, pRB somehow strongly facilitates and/or stabilizes binding of p120E4F to an E4F-site, as with the factors described above.
p120E4F-Mediated Growth Arrest Is pRB-Dependent.
Elevated levels of p120E4F inhibit the growth of mouse fibroblasts by blocking the cells in the G1 phase of their cell cycle (21). We explored whether the interaction with pRB plays a role in this p120E4F-mediated G1 arrest. To this end, we transfected a p120E4F expression vector into mouse NIH 3T3 fibroblasts derived from Rb−/− MEFs, from p107/p130−/− MEFs, or from the corresponding normal cells (Rb+/+), and measured their proliferation by BrdUrd incorporation (Fig. 4). As expected, Rb+/+ cells expressing p120E4F failed to incorporate BrdUrd (Fig.4A, Upper), quantitation of several experiments clearly showing that p120E4F blocked almost all transfected cells (Fig. 4B). Similarly, p107/p130−/− cells were also blocked efficiently by p120E4F (Fig. 4B). In contrast, a significant proportion of p120E4F-positive Rb−/− cells were also positive for BrdUrd labeling (Fig. 4A, Lower), an effect reproducibly observed in four independent experiments (Fig. 4B). Notably, this p120E4F-mediated arrest did not correspond to increased apoptosis, as shown by the lack of correlation between annexin-FITC staining and p120E4F overexpression (unpublished data). These results show that p120E4F overexpression can partially suppress cell growth in an Rb−/− background but that this effect is strongly enhanced in the presence of functional pRB. This observation is consistent with the results presented in Fig. 3 showing that pRB potentiates the p120E4F-mediated transcriptional repression.
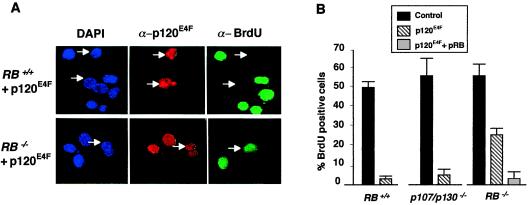
Figure 4.
p120E4F-mediated growth arrest is pRB-dependent. (A) Rb+/+ (Upper) or Rb−/− (Lower) mouse fibroblasts were transfected with the pcDNA–p120E4F plasmid. Twenty-four hours after transfection with the indicated plasmids, cells were assessed for BrdUrd uptake in their DNA (14 h in BrdUrd) as a marker of cell proliferation. Cells were fixed and labeled with 4′,6′-diamidino-2-phenylindole (DAPI) (blue). p120E4F-expressing cells were identified by immunofluoresence by using anti-human E4F (E4F 88.2) (red) and monitored for BrdUrd incorporation by using an anti-BrdUrd-specific Ab (green). (B) Diagram showing the average of four independent experiments (200 cells counted per experiment) performed as reported in A. Average of similar experiments performed in p107/p130−/− cells are also shown as indicated. As a control, Rb−/− cells are also transfected with p34(RB).
Discussion
pRB interacts with a variety of transcription factors and thereby regulates both cell growth and differentiation through its ability to coordinate multiple transcriptional events, acting as a competence factor to allow or impede various cellular transitions. Here we show that pRB also binds the E1A-regulated p120E4F transcription factor both in vitro and in vivo. Recombinant p120E4F and pRB proteins bind each other directly in vitro. In cells, the pRB–p120E4F complex was detected only in growth-arrested cells where it contained the hypophosphorylated form of pRB. Thus, as previously observed for most pRB-binding proteins, p120E4F interacts preferentially with the transcriptionally active form of pRB. An open question also remains about the role of p120E4F phosphorylation in this interaction because it has been reported that serum stimulation or adenovirus infection of arrested cells affects p120E4F phosphorylation (13, 15, 16, 18).
p120E4F displays pRB binding properties distinct from that of E2Fs or of LXCXE-containing proteins. Indeed, we found that mutations or deletions within the pocket region of pRB that abrogate pRB binding to E2F did not affect interactions with p120E4E in vitro. Moreover, we failed to detect any interaction between p120E4F and the other member of the pRB protein family, p107 or p130. Consistent with both observations, we found that p120E4F–pRB interaction involved the C-terminal part of pRB, downstream of the A/B pocket domains, which is not related in structure and sequence to that of p107 and p130. Beside p120E4F, this C-terminal region of pRB was reported to bind other proteins important for cell proliferation, including the high mobility group (HMG) box-containing transcription factor UBF (37), the c-Abl tyrosine kinase (34–36) and the transcription factor c-Jun (8). Consistent with the key role played by these factors in growth control, it was also shown that the C-terminal part of pRB contributes to RB-mediated growth suppression (1, 3, 34–37).
There are increasing evidence that the biological activity of pRB not only depends on the inhibition of its targets but also on its ability to properly assemble specific protein complexes on DNA. As is the case for C/EBPs, NF-IL6, ATF2, myoD, and c-Jun (1, 3, 5–8), we found that pRB stimulates the binding of p120E4F to its cognate DNA site. Surprisingly, it is worth to note here that five of the bZIP factors mentioned above are members of protein families, i.e., ATF/CREB, C/EBPs, and AP-1, that can also bind the core sequence of the E4F site with however, a lower affinity than p120E4F (13–15, 19). Although the biological significance of this observation remains to be explored, it is conceivable that pRB could control a specific class of regulatory elements through its association with multiple and nonrelated factors that recognize these elements. Nevertheless, we could not detect pRB in p120E4F–DNA complexes measured by gel shift assays, as has also been observed with C/EBPs, NF-IL6, and c-Jun in presence of pRB (4–9). The biochemical basis of this facilitation process is not understood, although it has been proposed in the case of C/EBPs, NF-IL6, and c-Jun, that pRB might promote conformational changes, dimerization and increased concentration of dimerized factors, resulting in a more efficient binding of the low abundance active form of the transcription factor (5, 6, 8). A similar mechanism occurs in our system as we found that pRB associates with the N-terminal region of p120E4F, which contains a domain required for its stable association to DNA (18).
The presence of pRB but not of the other pocket proteins strongly enhances the negative effect of p120E4F on transcription and proliferation. Consistent with this, overexpression of p120E4F blocks the cell cycle more efficiently in RB+/+ or p107/p130−/− cells than in RB−/− cells and, importantly, this block is not associated with increased apoptosis. Similarly, pRB interacts with and triggers activation of the negative regulators of transcription, BRCA1 (39), HBP1 (40), and p202 (41), whose overexpression in cells also induces cell cycle arrest in G1. Thus, it is conceivable that one of the cellular function of pRB could be to act as a competence factor for multiple growth suppressor, including p120E4F. A question then arises about the physiological function of these factors. Interestingly, one possible shared downstream effector for all these growth suppressors, and therefore for pRB, might be the cyclin-cdk inhibitor p21(WAF1/CIP1/SDI1). Indeed, it has been reported that p120E4F-, BRCA1-, HBP1-, and p202-arrested cells contain elevated levels of p21 protein (21, 42–44). The mechanism by which p120E4F exerted a control on p21 remains unclear. It was first reported to result from a p53-independent posttranscriptional stabilization of the p21 protein (21). However, others have found recently that p120E4F-induced arrest requires p53 (48). Anyhow, this is likely not the only mechanism for p120E4F induced growth arrest because we have recently observed that p21−/− and p53−/− primary fibroblasts (45) are still blocked in G1 on ectopic expression of p120E4F (L.F. and C.S., unpublished observation). Further studies are currently underway in our laboratory aiming to identify bona fide target genes of p120E4F involved in cell cycle regulation.
In summary, we provide evidence for a novel function of pRB that might promote cell-cycle arrest, namely potentiating the repressive effect of the ubiquitous cellular factor p120E4F.
Acknowledgments
We thank M. Classon, E. Harlow, and T. Jacks for their generous gifts of RB-, p107/p130-, and p21-deficient fibroblasts. We thank R. Hipskind and A. Le Cam for critical reading of the manuscript. This work was funded by grants from the French Centre National de la Recherche Scientifique (ATIPE no. 3), from l'Association pour la Recherche contre le Cancer, from La Ligue contre le Cancer, and from the Human Frontier in Science Program. J.P. was supported by a Boehringer Ingelheim doctoral fellowship, L.L.C. by a predoctoral fellowship from La Ligue contre le Cancer, and L.F. by a EEC/TMR postdoctoral fellowship.
Abbreviations
- C/EBP
CCAAT/enhancer binding proteins
- AP-1
activator protein-1
- GST
glutathione S-transferase
- MEF
mouse embryo fibroblast
- RT
room temperature
- EMSA
electrophoretic mobility shift assays
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130198397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130198397
References
- 1.Sardet C, Le Cam L, Fabrizio E, Vidal M. In: Progress in Gene Expression: Oncogenes as Transcriptional Regulators. Yaniv M, Ghysdael J, editors. Vol. 2. Basel: Birkhäuser; 1997. pp. 1–63. [Google Scholar]
- 2.Dyson N. Genes Dev. 1998;12:2245–2261. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 3.Grana X, Garriga J, Mayol X. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 4.Gu X, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 5.Chen P L, Riley D J, Chen K S, Lee W H. Proc Natl Acad Sci USA. 1996;93:465–469. doi: 10.1073/pnas.93.1.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P L, Riley D J, Chen K S, Lee W H. Genes Dev. 1996;10:2794–2728. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 7.Kim S J, Wagner S, Liu F, O'Reilly M A, Robbins P D, Green M R. Nature (London) 1992;358:331–334. doi: 10.1038/358331a0. [DOI] [PubMed] [Google Scholar]
- 8.Nead M A, Baglia L A, Antinore M J, Ludlow J W, McCance D J. EMBO J. 1998;17:2342–2352. doi: 10.1093/emboj/17.8.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishitani J, Nishinaka T, Cheng C H, Rong W, Yokoyama K K, Chiu R. J Biol Chem. 1999;274:5454–5461. doi: 10.1074/jbc.274.9.5454. [DOI] [PubMed] [Google Scholar]
- 10.Tevosian S G, Shih H H, Mendelson K G, Sheppard K A, Paulson K E, Yee A S. Genes Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- 11.Jones N. Curr Top Microbiol Immunol. 1995;199:59–80. doi: 10.1007/978-3-642-79586-2_4. [DOI] [PubMed] [Google Scholar]
- 12.Nevins J R. Curr Top Microbiol Immunol. 1995;199:25–32. doi: 10.1007/978-3-642-79586-2_2. [DOI] [PubMed] [Google Scholar]
- 13.Raychaudhuri P, Rooney R, Nevins J R. EMBO J. 1987;6:4073–4081. doi: 10.1002/j.1460-2075.1987.tb02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rooney R J, Raychaudhuri P, Nevins J R. Mol Cell Biol. 1990;10:5138–5149. doi: 10.1128/mcb.10.10.5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandes E R, Rooney R J. Mol Cell Biol. 1997;17:1890–1903. doi: 10.1128/mcb.17.4.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fognani C, Della Valle G, Babiss L E. EMBO J. 1993;12:4985–4992. doi: 10.1002/j.1460-2075.1993.tb06192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raychaudhuri P, Bagchi S, Nevins J R. Genes Dev. 1989;3:620–627. doi: 10.1101/gad.3.5.620. [DOI] [PubMed] [Google Scholar]
- 18.Rooney R J, Rothammer K, Fernandes E R. Nucleic Acids Res. 1998;26:1681–1688. doi: 10.1093/nar/26.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rooney R J, Daniels R R, Jenkins N A, Gilbert D J, Rothammer K, Morris S W, Higgs D R, Copeland N G. Mamm Genome. 1998;9:320–323. doi: 10.1007/s003359900758. [DOI] [PubMed] [Google Scholar]
- 20.Herbst R S, Pelletier M, Boczko E M, Babiss L E. J Virol. 1990;64:161–172. doi: 10.1128/jvi.64.1.161-172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandes E R, Zhang J Y, Rooney R J. Mol Cell Biol. 1998;18:459–467. doi: 10.1128/mcb.18.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandes E R, Rooney R J. Mol Cell Biol. 1999;19:4739–4749. doi: 10.1128/mcb.19.7.4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 24.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. Nucleic Acids Res. 1995;23:1123–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sardet C, Vidal M, Cobinik D, Geng Y, Onufrik C, Chen A, Weinberg R A. Proc Natl Acad Sci USA. 1995;92:2403–2424. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Nature (London) 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 28.Le Cam L, Polanowska J, Geng Y, Fabbrizio E, Olivier M, Philips A, Ng-Eaton E, Classon M, Sardet C. EMBO J. 1999;18:1878–1890. doi: 10.1093/emboj/18.7.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamel P A, Gill R M, Phillips R A, Gallie B L. Mol Cell Biol. 1992;12:3431–3438. doi: 10.1128/mcb.12.8.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabbrizio E, Le Cam L, Polanowska J, Kackzorek M, Lamb N, Brent R, Sardet C. Oncogene. 1999;18:4357–4363. doi: 10.1038/sj.onc.1202825. [DOI] [PubMed] [Google Scholar]
- 31.Dowdy S F, Hinds P W, Louie K, Reed S I, Arnold A, Weinberg R A. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 32.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Nature (London) 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 33.Kratzke R A, Otterson G A, Lin A Y, Shimizu E, Alexandrova N, Zajac-Kaye M, Horowitz J M, Kaye F J. J Biol Chem. 1992;267:25998–6003. [PubMed] [Google Scholar]
- 34.Welch P J, Wang J Y. Genes Dev. 1995;9:31–46. doi: 10.1101/gad.9.1.31. [DOI] [PubMed] [Google Scholar]
- 35.Welch P J, Wang J Y. Cell. 1993;75:779–790. doi: 10.1016/0092-8674(93)90497-e. [DOI] [PubMed] [Google Scholar]
- 36.Whitaker L L, Su H, Baskaran R, Knudsen E S, Wang J Y. Mol Cell Biol. 1998;18:4032–4042. doi: 10.1128/mcb.18.7.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voit R, Schafer K, Grummt I. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kratzke R A, Otterson G A, Hogg A, Coxon A B, Geradts J, Cowell J K, Kaye F J. Oncogene. 1994;5:1321–1326. [PubMed] [Google Scholar]
- 39.Aprelikova O N, Fang B S, Meissner E G, Cotter S, Campbell M, Kuthiala A, Bessho M, Jensen R A, Liu E T. Proc Natl Acad Sci USA. 1999;96:11866–11871. doi: 10.1073/pnas.96.21.11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih H H, Tevosian S G, Yee A S. Mol Cell Biol. 1998;8:4732–4743. doi: 10.1128/mcb.18.8.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choubey D, Lengyel P. J Biol Chem. 1995;270:6134–6140. doi: 10.1074/jbc.270.11.6134. [DOI] [PubMed] [Google Scholar]
- 42.Gartel A L, Goufman E, Tevosian S G, Shih H, Yee A S, Tyner A L. Oncogene. 1998;17:3463–3469. doi: 10.1038/sj.onc.1202240. [DOI] [PubMed] [Google Scholar]
- 43.Somasundaram K, Zhang H, Zeng Y X, Houvras Y, Peng Y, Zhang H, Wu G S, Licht J D, Weber B L, El-Deiry W S. Nature (London) 1997;389:187–190. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 44.Gutterman J U, Choubey D. Cell Growth Differ. 1999;10:93–100. [PubMed] [Google Scholar]
- 45.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Nature (London) 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 46.Polanowska J, Le Cam L, Orsetti B, Fabbrizio E, Fajas L, Taviaux S, Theillet C, Sardet C. Genes Chomosomes Cancer. 2000;28:126–130. [PubMed] [Google Scholar]
- 47.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 48.Sandy P, Gostissa M, Fogal V, Cecco L D, Szalay K, Rooney R J, Schneider C, Sal G D. Oncogene. 2000;19:188–199. doi: 10.1038/sj.onc.1203250. [DOI] [PubMed] [Google Scholar]