Abstract
Background
Reading disability (RD) is a common syndrome with a large genetic component. Chromosome 6 has been identified in several linkage studies as playing a significant role. A more recent study identified a peak of transmission disequilibrium to marker JA04 (G72384) on chromosome 6p22.3, suggesting that a gene is located near this marker.
Results
In silico cloning was used to identify possible candidate genes located near the JA04 marker. The 2 million base pairs of sequence surrounding JA04 was downloaded and searched against the dbEST database to identify ESTs. In total, 623 ESTs from 80 different tissues were identified and assembled into 153 putative coding regions from 19 genes and 2 pseudogenes encoded near JA04. The identified genes were tested for their tissue specific expression by RT-PCR.
Conclusions
In total, five possible candidate genes for RD and other diseases mapping to this region were identified.
Keywords: reading disability, dyslexia, 6p22.3, In silico, ESTs
Background
Reading disability (RD), or dyslexia, is a common syndrome with a significant genetic component. Genetic linkage studies have identified five potential RD loci located on chromosomes 1 [1,2], 2 [3,4], 6 [5-9], 15 [7,10] and 18 [11]. The linkage to 6p has been the most often reproduced in independent samples with five studies showing peaks of linkage to different regional marker sets [5-9]. We recently constructed a BAC/PAC contig of the 6p RD locus [12] and identified the precise location and order of 29 short tandem repeat (STR) markers spanning this region [13]. A subsequent study with this new marker panel identified a peak of transmission disequilibrium at marker JA04 (G72384) [14].
We searched the expressed sequence tag (EST) database, dbEST http://www.ncbi.nlm.nih.gov/dbEST/index.html, with the genomic sequence corresponding to the peak of transmission disequilibrium at marker JA04. ESTs are partial and usually incomplete cDNA sequences prepared from various tissues. Presently, nearly four million ESTs (dbEST release 04/19/02) are catalogued in dbEST representing about 80% or more of all human genes with at least one representative entry [15]. While not every gene is accounted for in dbEST, computer database searching, also known as in silico cloning, can identify new genes without actual physically manipulating DNA. These types of analyses can also characterize intron-exon boundaries, splice variants, tissue specific expression levels, and gene homologies [16]. Furthermore, clustering ESTs together to form a contiguous sequence can predict putative open reading frame (ORFs). The first map of the human genome, which contained over 30,000 genes, was generated by mapping EST clusters to human-hamster radiation hybrid cell lines [17]. Despite their wide-ranging utility, ESTs have two inherent drawbacks: (1) they are based on single sequence reads making them vulnerable to sequencing errors, and (2) they are generated from cDNA libraries that may contain unexpressed or incompletely spliced sequences derived from heteronuclear RNA or other artifacts. The clustering of ESTs to form ORFs could contain both expressed and unexpressed sequences and should be treated with caution. Fortunately, the high redundancy of entries in dbEST permits the alignment of multiple ESTs for most genes thus diminishing the effects of these drawbacks.
To identify candidate genes for RD and other disorders mapping to this region we downloaded and searched the two million base pairs of genomic sequence surrounding the peak of transmission disequilibrium. In addition to RD, risk loci for Behçet's disease [18], inflammatory bowel disease (IBD3) [19,20], hypotrichosis simplex (HSS) [21], insulin dependent type 1 diabetes mellitus [22], attention deficit hyperactivity disorder (ADHD) [23] and schizophrenia [24] have all been assigned to this general chromosomal location by genetic linkage analysis. Using in silico cloning we identified a total of 19 genes and 2 pseudogenes and mapped their precise physical location and direction of transcription. The expression pattern of each gene was characterized by examining the number of ESTs identified from various tissues as well as by qualitative RT-PCR with RNA from 20 different human tissues. This study also allowed us to test the usefulness of in silico cloning to identify and map new genes in a focussed region of the genome.
Results
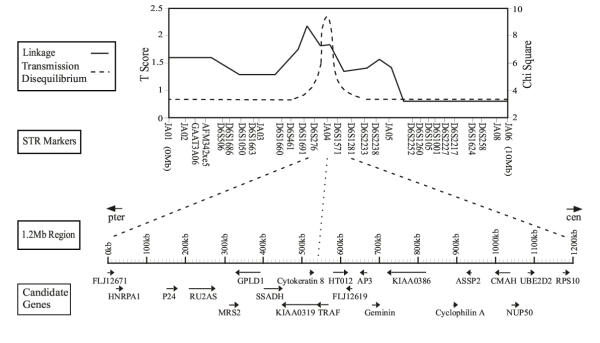
Using the blastc13 server at NCBI, we performed in silico cloning studies of the 6p RD locus to identify coding regions. In total, 623 ESTs from 80 different tissues were identified and aligned to 2 Mb of genomic sequence. These searches captured 157 putative coding regions from 19 genes and 2 pseudogenes concentrated in the central 1200 Kb shown in detail in Figure 1 with base pair 1 starting at the 5-prime end of FLJ12671 and ending 1 Kb centromeric to the 3-prime end of RPS10. Short tandem repeat marker JA04, which identified the peak of transmission disequilibrium for RD phenotypes in previous studies [14], is at 540 Kb. The most telomeric 200 Kb and the most centromeric 600 Kb of genomic sequence are void of coding regions. Intergenic distances range from less than 1 Kb (KIAA0319 and TRAF) to 110 Kb (HNRPA1 and P24). Cytokeratin 8, transcribed telomere to centromere, is located in the intron between exons 1 and 2 of KIAA0319 (transcribed centromere to telomere). Table 1 lists the genes identified in Figure 1, their size in Kb, the NCBI accession number for the corresponding mRNA or cDNA, number of exons, genomic mapping source, and putative function.
Figure 1.
Transmission disequilibrium and genetic linkage analyses of the 6p21.3 reading disability locus, regional STR markers and transcription map. At the top of the figure is the result of the DeFries-Fulker linkage (T Score, solid line), and QTDT linkage disequilibrium (chi-square, dashed line) [14]. The location and order of the 29 STRs are shown below which identify a peak of transmission disequilibrium at marker JA04. Below the markers is a detailed representation of 1.2 Mb surrounding marker JA04. The 19 genes and 2 pseudogenes encoded in this region are shown with the telomere on the left and centromere on the right and their position and direction of transcription indicated by the arrows.
Table 1.
Genes encoded within 2 Mb of JA04 in order from telomere (top) to centromere.
| Gene | Gene Size (kb) | MRNA Accession # | # of Exons | Mapping Source | Function |
| FLJ12671 | 15.885 | NM_030980 | 3 | This paper | Function unknown |
| HNRPA1 (pseudogene) | 1.813 | XM_031157 | 1 | This paper | Functions as carriers for RNA during export to the cytoplasm |
| P24 | 21.142 | AF418980 | 5 | This paper | Transmembrane protein; may have a role in vesicular trafficking |
| RU2AS | 79.995 | AF181722 | 7 | [46] | Cellular defense response |
| MRS2L | 20.629 | XM_041317 | 11 | NT_017021a | Transporter protein |
| GPLD1 | 61.339 | XM_041310 | 25 | [47] | Hydrolyzes inositol-PO4 linkage in proteins anchored by phosphatidylinositol glycans |
| SSADH | 39.639 | NM_001080 | 10 | [48] | Required for 4-aminobutyric acid degradation |
| KIAA0319 | 102.051 | XM_011367 | 21 | NT_017021a | Function unknown |
| Cytokeratin 8 | 2.206 | XM_069402 | 2 | This paper | Regulation of exocrine pancreas homeostasis |
| TRAF | 16.902 | AF223469 | 7 | [49] | Inhibits NF-KappaB activation |
| HT012 | 34.662 | AF220186 | 3 | NT_017021a | Uncharacterized hypothalamus protein |
| FLJ12619 | 14.192 | XM_041290 | 5 | NT_017021a | Function unknown |
| AP3 | 1.667 | BC000804 | 3 | This paper | May be involved in intracellular protein transport |
| Geminin | 11.086 | NM_015895 | 9 | NT_017021a | Inhibits DNA replication during cell cycle S, G2, and M phase |
| KIAA0386 | 106.682 | AB002384 | 23 | NT_017021a | Stimulates the formation of a non-specific multinucleate syncytium from proliferative cytotrophoblasts during trophoblast differentiation |
| Cyclophilin A | 0.81 | XM_098483 | 2 | NT_017021a | May to be involved in protein folding and/or intracellular protein transport |
| ASSP2 (pseudogene) | 1.954 | XM_000845 | 1 | This paper | Catalyzes the ATP-dependent ligation of citrulline to aspartate, forms argininosuccinate in the urea cycle |
| CMAH | 56.489 | XM_041278 | 15 | [50] | Converts the nucleotide sugar donor N-acetylneuraminic acid to N-glycolylneuraminic acid |
| NUP50 | 1.737 | XM_018531 | 1 | NT_017021a | Allows bi-directional transport of macromolecules through the nuclear envelope |
| UBE2D2 | 1.732 | XM_030770 | 1 | This paper | Modifies proteins with ubiquitin for degradation |
| RPS10 | 1.428 | XM_069039 | 2 | This paper | Ribosomal protein |
a The gene has been mapped on NCBI accession NT_017021 but no citation is present
Thirteen genes were previously mapped to this region (RU2AS, MRS2L, GPLD1, SSADH, KIAA0319, TRAF, HT012, FLJ12619, Geminin, KIAA0386, Cyclophilin A, CMAH, and NUP50) and are present on the NCBI accession view (NT_017021) of 6p21.3-22. Our in silico studies identified an additional six genes (RPS10, FLJ12671, UBE2D2, AP3, Cytokeratin 8, and P24) and two pseudogenes (HNRPA1 and ASSP2) not represented on NT_017021. In addition we identified a putative fifth exon for P24 from hypothalamus (BG715502) and pineal gland (AA363698) cDNA libraries, located between exons 2 and 3 in the 5-prime untranslated region and flanked by non-traditional splice donor and acceptor bases [25]. The novel exon was not present in the full-length mRNA sequence (AF418980).
The functions of the newly identified genes were inferred by their similarity to other known genes identified in the BLAST searches. Ribosomal protein L21 (RPS10) is 95% identical to the genomic and mRNA sequences of a known ribosomal gene. RPS10 is one of the many proteins that make up the ribosome macromolecule. Other highly homologous genes of RPS10 are RPS5, RPS9, RPS29, RPL5, RPl27a, and RPL28 [26]. FLJ12671s a hypothetical gene with an unknown function identified by the NEDO human cDNA-sequencing project [27]. Blasting the FLJ12671 sequence against the nr database also identified hits on chromosomes 1 and 11, suggesting that this gene has been duplicated on several chromosomes. Ubiquitin conjugating enzyme E2D2 (UBE2D2) is a protein that targets abnormal or short-lived proteins for degradation by the 26S proteasome [28]. The EST sequences identified for the UBE2D2 gene on chromosome 6 are 93% identical to the human UBC4/5 gene located on chromosome 5 suggesting that this gene could be a duplicate as well. Adapter-related protein complex 3 (AP3) may be involved in intracellular protein transport [29]. Cytokeratin 8 is 95% identical to both the genomic and mRNA sequences of keratin. Vesicular membrane protein (P24), a previously characterized but unmapped gene [30] has been localized in intracellular organelles of highly differentiated neural cells and may have a role in the neural organelle transport system. ASSP2 is one of 12 pseudogenes of argininosuccinate synthetase encoded on 10 chromosomes with the only functional sequence residing on chromosome 9 [31]. Heterogeneous nuclear ribonucleoprotein A1 (HNRPA1) is also a pseudogene with three other copies on chromosomes 3, 13, and 20 [32]. The copy on chromosome 12q13.1 is thought to encode the gene responsible for the functional HNRPA1, which serves as a carrier for RNA during export to the cytoplasm [33].
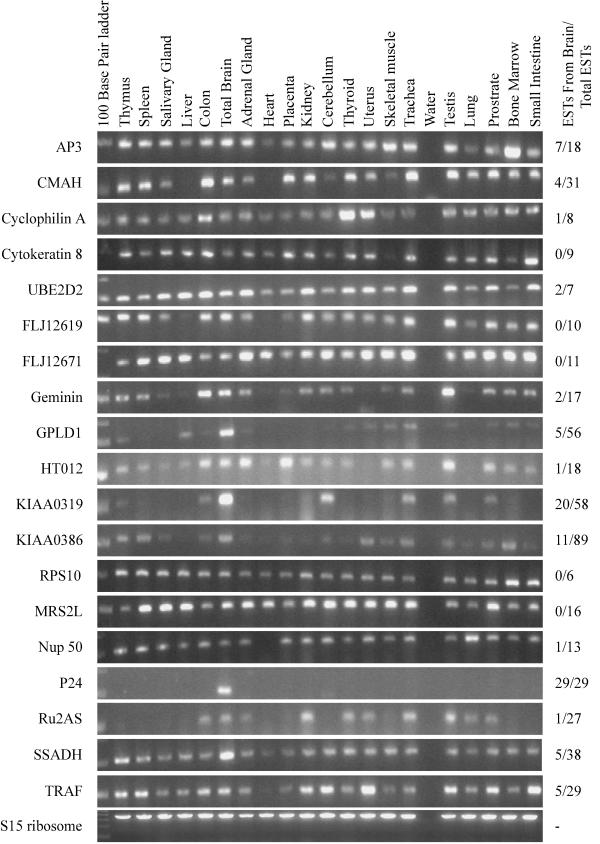
The results of the qualitative RT-PCR, though not quantitative, were useful for characterizing the pattern of tissue expression (Figure 2). While most genes were about equally represented in the mRNAs of the twenty tissues in the panel, three genes, P24, NAD (+)-dependent succinic semialdehyde dehydrogenase (SSADH), and KIAA0319 had exceptional patterns. P24 was almost exclusively expressed in brain by RT-PCR, correlating with the origin of all 29 publicly accessible ESTs from brain cDNA libraries (Figure 2). RT-PCR suggested that the expression of SSADH was greatest in brain, though it was ubiquitously expressed in all tissues tested. Correspondingly, only 5 of 38 SSADH ESTs accessible on public domain servers were from brain cDNA libraries, with the remainder of mixed origin. KIAA0319 had strong signal from brain and cerebellum mRNA, reflected by the 10 of 29 publicly accessible ESTs originating from brain cDNA libraries.
Figure 2.
RT-PCR analysis of genes within 2 Mb of JA04. The results of the qualitative PCR for the 19 genes identified in this region. RNA from 20 tissues was used along with a blank control (lane 16) for the RT-PCR step. The genes are listed on the left along side their corresponding gel. The number of ESTs identified in brain libraries is listed on the right along with the total number of ESTs for that gene.
Discussion
The primary goal of this study was to identify candidate genes surrounding the peak of transmission disequilibrium for RD on chromosome 6p and to characterize their patterns of expression. A secondary goal was to investigate the usefulness of the in silico approaches and specifically the dbEST database to identify and map new genes.
We identified 19 genes within 2 Mb of the peak of transmission disequilibrium – but are some better candidates for RD than others? The patterns of tissue expression as profiled by RT-PCR and the frequency with which ESTs originated from brain cDNA libraries (Figure 2), serve to highlight five genes that are highly expressed in the brain: P24, SSADH, GPLD1, KIAA0386, and KIAA0319. Of these five, only one gene, SSADH, has been associated with a brain related phenotype. Two frameshift mutations, a G-to-T transversion in the intron 9 splice donor site, and a G-to-A transition in the intron 5 splice donor site, cause an exon to be skipped resulting in abnormal metabolism of GABA, an important neurotransmitter in the brain. The handful of described cases were originally diagnosed by anomalous GABA metabolites in the urine associated with developmental and speech delays, hyporeflexia, and behavioral problems including mild autism with clinical variation between affected family members [34]. There is no data, however, that links GABA or GABA metabolism to specific defects of reading independent of IQ. None of the other genes highly expressed in brain have associated diseases or clinical phenotypes. GPLD1 selectively hydrolyzes inositol phosphate linkages in vitro, releasing the protein bound to the plasma membrane via a glycosylphosphatidylinositol anchor into the cytosol [35]. P24 is a neuron specific membrane protein localized in intracellular organelles of highly differentiated neural cells and is involved in neural organelle transport. KIAA0386 encodes a protein that stimulates the formation of a non-mitotic multinucleated syncytium from proliferative cytotrophoblasts during trophoblast differentiation [36]. KIAA0319 encodes a protein of unknown function [37]. While HT012, an uncharacterized hypothalamus protein, could also be considered as a possible candidate gene, RT-PCR and EST searches (1 of 18 from brain cDNA libraries) do not suggest a high level or selective expression in the brain.
The in silico studies also identified candidate genes for other diseases that map to this region. The five brain candidate genes described above for RD are also reasonable candidates for the neurobehavioral disorders schizophrenia and ADHD. HSS results in the complete loss of scalp hair in childhood. Betz et al [21] described evidence for linkage to HSS with markers spanning D6S276 (400 Kb telomeric of JA04) through D6S1607 (5.6Mb centromeric of JA04). Neither the RT-PCR results nor the tissue origin for any single gene suggests any best candidates among the 19. Behçet's disease is an autoimmune disorder characterized by a systemic vasculitis that affects the joints, all sizes and types of blood vessels, the lungs, the central nervous system, and the gastrointestinal tract [38]. There is evidence for linkage with markers spanning nearly 28 Mb of 6p with JA04 in the middle [18]. Candidate genes for Behçet's would include those expressed in lymphocytes or perhaps bone marrow such as AP3 and FLJ12671 and other immune related genes such as TRAF and RU2AS. These genes may also serve as candidates for other autoimmune disorders that map to this region such as IBD3.
Overall the in silico method for identifying genes in a specific genomic region worked well here, yielding a reasonable gene density of one per 95 kb. This method is highly dependent upon the quality of the information in dbEST. Any contamination from non-coding DNA, bacterial DNA, cDNA from other species, vectors or mitochondria DNA could generate false gene assignments. Fortunately, the high redundancy of EST hits in dbEST increased our confidence that any identification was likely physiologic and that the searches were sensitive. It is possible however, that our in silico approach may have missed some genes, in particular those with small ORFs and/or large 5-prime and/or 3-prime UTRs [39]. As dbEST expands over the next few years, new genes may be identified with repeated in silico searches, or with biophysical approaches such as cDNA hybridization [40], exon trapping [41] and amplification [42], or by identification of evolutionary conserved sequences [43] and HTF islands [44].
Conclusion
In summary, we examined 2 Mb surrounding the transmission disequilibrium peak with RD at short tandem repeat marker JA04 on chromosome 6p. In silico searches of the dbEST database identified 19 possible candidate genes. While tissue expression patterns suggest five candidates that are highly expressed in brain – one with a known association with neurological disease – neither the RT-PCR data nor the EST information can absolutely rule out any of the 19 as culpable candidates. We conclude therefore that in silico cloning is a powerful and effective technique for quickly identifying existing and novel genes, which can then be used to develop cDNA single nucleotide polymorphism markers (cSNPs) for pinpointing a more precise location of the 6p RD gene, and other disease genes that map to this region.
Methods
In silico cloning
The two million base pairs of index genomic sequence surrounding marker JA04 was downloaded from the NCBI website (accession number NT_017021). A perl script was written to parse the sequence into 200 files each containing 10 Kb segments in FASTA format. Repeat sequences were masked with the RepeatMasker program (RepeatMasker at http://ftp.genome.washington.edu/RM/RepeatMasker.html). Each masked file was sent to the blastc13 server as a query for searching the dbEST database using the BLAST algorithm [45]. Only ESTs with an identity of 93% or greater were considered; all other hits were discarded. Surviving ESTs were then used to search the nr database to identify parental cDNA or mRNA matches, which assembled 623 non-overlapping ESTs into 21 gene or pseudogene antecedents. The BLAST search of nr also showed hits against chromosome 6 BAC or PAC sequence. ESTs not mapping to chromosome 6 were also discarded. The final assemblies of EST and cDNA or mRNA sequences were aligned to the index NT_017021 genomic sequence using the Martinez Needleman-Wunsch algorithm in MegAlign (DNA Star, Lasergene) permitting identification of exon-intron boundaries, new exons (P24), and the direction of transcription relative to the telomere.
Qualitative PCR
A panel containing human total RNA from 20 different tissues was purchased from Clontech (BD Biosciences Clontech). RT-PCR was performed using the RETROscript first strand synthesis kit for RT-PCR (Ambion). 2μg of RNA was denatured with 0.5μM of random decamer primers in a total volume of 12μl, and heated to 77°C for three minutes and placed on ice. First strand cDNA was then synthesized with the addition of 2μl 10 × RT buffer, 4μl dNTP mix, 1 Unit RNase inhibitor, and 100 Units MMLV-RT in a total volume of 20μl. The reaction was incubated for one hour at 44°C, and then 92°C for ten minutes and then stored at -20°C.
To check the quality of the first strand cDNA, 1μl of cDNA was used in a PCR reaction to amplify the rig/S15 ribosomal gene. 1.5μl of 10 × PCR buffer (Qiagen), 250μM of each dNTP, 0.5 Units of HotstarTaq polymerase (Qiagen) and 0.5μM of primer (Ambion) were used in a 15μl reaction. The reaction was denatured for 15 minutes at 95°C, then ten cycles of 94°C for 30 seconds, 65°C for 30 seconds (-1°C/cycle), and 72°C for 30 seconds, 20 additional cycles of 94°C for 30 seconds, 55°C for 30 seconds and 72°C for 30 seconds, and a final extension of 72°C for ten minutes. PCR reactions were performed in a MJ Research thermocycler. PCR products were electrophoresed on 2% agarose gels.
Primers (primer sequences are listed in Table 2) were designed to amplify the mRNA sequences of each of the 20 genes identified in the transcript map (Figure 1). Each amplicon was designed to be between 90 and 150 base pairs in length. For each amplicon, 1μl of cDNA, 1.5μl of 10 × PCR buffer (Qiagen), 250μM of each dNTP, 0.5 units of HotstarTaq polymerase (Qiagen) and 0.5μM of primer (Life Technologies) were used in a 15μl reaction. PCR reactions were performed as above. One lane (Figure 2, lane 16) contained water during the RT-PCR step to check for RNA contamination. Products were electrophoresed on 2% agarose gels and stained with ethidium bromide.
Table 2.
Primer sequences for RT-PCR amplification of regional genes.
| Gene | Forward primer | Reverse primer | Product size |
| AP3 | GTTTGTTGTCAGATCCTCCA | ATCATCAGGGAGACTTTCCA | 97 bp |
| CMAH | TGGAAAACAAAGCATAGCTG | CGAAAACAGACCATTTCCAG | 105 bp |
| Cyclophilin A | TTCGAGTTGTCCACAGTCAG | CAAAGTGAAAGAAGGCATGA | 106 bp |
| Cytokeratin 8 | CCTCATCAAGAAGGATGTGGA | GCCTGAGGAAGTTGATCTCG | 95 bp |
| UBE2D2 | ATGGCAGCATTTGTCTTGAT | TTGGGATCACACAACAGAGA | 100 bp |
| FLJ12619 | CTTTACAAGGGTGTTTTGGAG | TGCACAAAATGAGTTCTGCT | 108 bp |
| FLJ12671 | CTCCACCAGCTCCACATCTT | GTTGGGTTCTGACGTGAAGG | 12 bp |
| Geminin | CGTGTGCTGAAGGAACTGTA | GGAGGTAAACTTCGGCAGTA | 103 bp |
| GPLD1 | CATGGTGAGCATTTTGATGG | GTGGCTGTTGGTGTGTGTCT | 103 bp |
| HT012 | TTTCCCAGGTGTTTTGTGTG | GCACATGTTCTGAAGCAAGG | 110 bp |
| KIAA0319 | GGATTCATACCCATGGCATT | TCACTGGCTTGTTGAGTGGT | 105 bp |
| KIAA0386 | TCAGGTCCCATTATTGCTTC | CCCCACCTTGTACACTCTTT | 117 bp |
| RPS10 | CATCAAGGGAATGGGTACTG | TTTACAACAATGCCAACAGC | 108 bp |
| MRS2 | ATGAGGCAACACAAGCACAG | GGCTTGTCAAGTTCCTACGC | 100 bp |
| NUP50 | GAAGATGGAGGTGGCATCTG | TAATGAGCCCAAAACAGAGC | 109 bp |
| P24 | CATGCATACACAGGGTGCTT | GAATGCGCCATTAAAAGGAA | 119 bp |
| RU2AS | TAGCAGGTGAACCTGGTGTG | GCTTGCAGGACTCGGAGTAG | 97 bp |
| SSADH | GGAGCACCTCTCCCTATTCC | TGAAGTGGCAAACAGGACAG | 107 bp |
| TRAF | TTCAGGATGTGAGGCCATTT | CAATGGGGTACCTGTTTGGA | 100 bp |
| S15 | TTCCGCAAGTTCACCTACC | CGGGCCGGCCATGCTTTACG | 361 bp |
Authors' Contributions
ERL designed and performed the experiments and wrote the manuscript. HM aided in analyzing the data. JRG designed the experiments, analyzed the data, and edited the manuscript. All of the authors have reviewed the manuscript.
Note Added In Proof
Since submission of the manuscript for review, contig NT_017021 has been incorporated into NCBI contig NT_007592.13.
Acknowledgments
Acknowledgments
Support for JRG is through The Charles H. Hood Foundation, and National Institutes of Health (NIH) grants R01 DA12849, R01 DA12690, R01 NS43530, and U01 HD98002. Support for HM is through The Charles H. Hood Foundation. Support for EL is through The Robert Leet and Clara Guthrie Patterson Trust. The authors also wish to thank Thomas O'Reilly-Pol for help with characterizing NT_007592.13.
Contributor Information
Eric R Londin, Email: Eric.Londin@stonybrook.edu.
Haiying Meng, Email: Haiying.Meng@yale.edu.
Jeffrey R Gruen, Email: Jeffrey.Gruen@yale.edu.
References
- Rabin M, Wen XL, Hepburn M, Lubs HA, Feldman E, Duara R. Suggestive linkage of developmental dyslexia to chromosome 1p34-p36. Lancet. 1993;342:178. doi: 10.1016/0140-6736(93)91384-X. [DOI] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Pauls JE, Hart LA, Pauls DL. Linkage studies suggest a possible locus for developmental dyslexia on chromosome 1p. Am J Med Genet. 2001;105:120–129. doi: 10.1002/1096-8628(20010108)105:1<120::AID-AJMG1075>3.3.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Fagerheim T, Raeymaekers P, Tonnessen FE, Pedersen M, Tranebjaerg L, Lubs HA. A new gene (DYX3) for dyslexia is located on chromosome 2. J Med Genet. 1999;36:664–669. [PMC free article] [PubMed] [Google Scholar]
- Petryshen TL, Kaplan BJ, Hughes ML, Tzenova J, Field LL. Supportive evidence for the DYX3 dyslexia susceptibility gene in Canadian families. J Med Genet. 2002;39:125–126. doi: 10.1136/jmg.39.2.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC. Quantitative trait locus for reading disability on chromosome 6. Science. 1994;266:276–279. doi: 10.1126/science.7939663. [DOI] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP. A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet. 1999;64:146–156. doi: 10.1086/302190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Hart LA, Speed WC, Shuster A, Pauls DL. Susceptibility loci for distinct components of developmental dyslexia on chromosomes 6 and 15. Am J Hum Genet. 1997;60:27–39. [PMC free article] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Pauls DL. Chromosome 6p influences on different dyslexia-related cognitive processes: further confirmation. Am J Hum Genet. 2000;66:715–723. doi: 10.1086/302755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SD, Kimberling WJ, Pennington BF, Lubs HA. Specific reading disability: identification of an inherited form through linkage analysis. Science. 1983;219:1345–1347. doi: 10.1126/science.6828864. [DOI] [PubMed] [Google Scholar]
- Nopola-Hemmi J, Taipale M, Haltia T, Lehesjoki AE, Voutilainen A, Kere J. Two translocations of chromosome 15q associated with dyslexia. J Med Genet. 2000;37:771–775. doi: 10.1136/jmg.37.10.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Francks C, Marlow AJ, MacPhie IL, Newbury DF, Cardon LR, Ishikawa-Brush Y, Richardson AJ, Talcott JB, Gayan J, Olson RK, Pennington BF, Smith SD, DeFries JC, Stein JF, Monaco AP. Independent genome-wide scans identify a chromosome 18 quantitative- trait locus influencing dyslexia. Nat Genet. 2002;30:86–91. doi: 10.1038/ng792. [DOI] [PubMed] [Google Scholar]
- Ahn J, Won TW, Zia A, Reutter H, Kaplan DE, Sparks R, Gruen JR. Peaks of linkage are localized by a BAC/PAC contig of the 6p reading disability locus. Genomics. 2001;78:19–29. doi: 10.1006/geno.2001.6645. [DOI] [PubMed] [Google Scholar]
- Ahn J, Won TW, Kaplan DE, Londin ER, Kuzmic P, Gelernter J, Gruen JR. A detailed physical map of the 6p reading disability locus, including new markers and confirmation of recombination suppression. Hum Genet. 2002;111:339–349. doi: 10.1007/s00439-002-0771-x. [DOI] [PubMed] [Google Scholar]
- Kaplan DE, Gayan J, Ahn J, Won TW, Pauls D, Olson RK, DeFries JC, Wood F, Pennington BF, Page GP, Smith SD, Gruen JR. Evidence for linkage and association with reading disability on 6p21.3- 22. Am J Hum Genet. 2002;70:1287–1298. doi: 10.1086/340449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL. Est! Est!! Est!!! Bioessays. 1996;18:1021–1023. [Google Scholar]
- Jongeneel CV. Searching the expressed sequence tag (EST) databases: panning for genes. Brief Bioinform. 2000;1:76–92. doi: 10.1093/bib/1.1.76. [DOI] [PubMed] [Google Scholar]
- Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tomé P, Hui L, Matise TC, McKusick KB, Beckmann JS, Bentolila S, Bihoreau M-T, Birren BB, Browne J, Butler A, Castle AB, Chiannilkulchai N, Clee C, Day PJR, Dehejia A, Dibling T, Drouot N, Duprat S, Fizames C, Fox S, Gelling S, Green L, Harrison P, Hocking R, Holloway E, Hunt S, Keil S, Lijnzaad P, Louis-Dit-Sully C, Ma J, Mendis A, Miller J, Morissette J, Muselet D, Nusbaum HC, Peck A, Rozen S, Simon D, Slonim DK, Staples R, Stein LD, Stewart EA, Suchard MA, Thangarajah T, Vega-Czarny N, Webber C, Wu X, Hudson J, Auffray C, Nomura N, Sikela JM, Polymeropoulos MH, James MR, Lander ES, Hudson TJ, Myers RM, Cox DR, Weissenbach J, Boguski MS, Bentley DR. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- Gul A, Hajeer AH, Worthington J, Ollier WE, Silman AJ. Linkage mapping of a novel susceptibility locus for Behcet's disease to chromosome 6p22-23. Arthritis Rheum. 2001;44:2693–2696. doi: 10.1002/1529-0131(200111)44:11<2693::AID-ART449>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Cho J. Linkage of inflammatory bowel disease to human chromosome 6p. Inflamm Bowel Dis. 2000;6:259–261. doi: 10.1097/00054725-200008000-00016. [DOI] [PubMed] [Google Scholar]
- Dechairo B, Dimon C, van Heel D, Mackay I, Edwards M, Scambler P, Jewell D, Cardon L, Lench N, Carey A. Replication and extension studies of inflammatory bowel disease susceptibility regions confirm linkage to chromosome 6p (IBD3) Eur J Hum Genet. 2001;9:627–633. doi: 10.1038/sj.ejhg.5200687. [DOI] [PubMed] [Google Scholar]
- Betz RC, Lee YA, Bygum A, Brandrup F, Bernal AI, Toribio J, Alvarez JI, Kukuk GM, Ibsen HH, Rasmussen HB, Wienker TF, Reis A, Propping P, Kruse R, Cichon S, Nothen MM. A gene for hypotrichosis simplex of the scalp maps to chromosome 6p21.3. Am J Hum Genet. 2000;66:1979–1983. doi: 10.1086/302934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie BA, Todd JA, Pociot F, Nerup J, Akselsen HE, Joner G, Dahl-Jorgensen K, Ronningen KS, Thorsby E, Undlien DE. The predisposition to type 1 diabetes linked to the human leukocyte antigen complex includes at least one non-class II gene. Am J Hum Genet. 1999;64:793–800. doi: 10.1086/302283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr CL, Shulman R, Wigg K, Schachar R, Tannock R, Roberts W, Malone M, Kennedy JL. Linkage study of polymorphisms in the gene for myelin oligodendrocyte glycoprotein located on chromosome 6p and attention deficit hyperactivity disorder. Am J Med Genet. 2001;105:250–254. doi: 10.1002/ajmg.1302. [DOI] [PubMed] [Google Scholar]
- Maziade M, Roy MA, Rouillard E, Bissonnette L, Fournier JP, Roy A, Garneau Y, Montgrain N, Potvin A, Cliche D, Dion C, Wallot H, Fournier A, Nicole L, Lavallee JC, Merette C. A search for specific and common susceptibility loci for schizophrenia and bipolar disorder: a linkage study in 13 target chromosomes. Mol Psychiatry. 2001;6:684–693. doi: 10.1038/sj.mp.4000915. [DOI] [PubMed] [Google Scholar]
- Mount SM. A catalogue of splice junction sequences. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MD, Dubnick M, Kerlavage AR, Moreno R, Kelley JM, Utterback TR, Nagle JW, Fields C, Venter JC. Sequence identification of 2,375 human brain genes. Nature. 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- Yudate HT, Suwa M, Irie R, Matsui H, Nishikawa T, Nakamura Y, Yamaguchi D, Peng ZZ, Yamamoto T, Nagai K, Hayashi K, Otsuki T, Sugiyama T, Ota T, Suzuki Y, Sugano S, Isogai T, Masuho Y. HUNT: launch of a full-length cDNA database from the Helix Research Institute. Nucleic Acids Res. 2001;29:185–188. doi: 10.1093/nar/29.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JP, Bates PW, Yang M, Vierstra RD, Weissman AM. Identification of a family of closely related human ubiquitin conjugating enzymes. J Biol Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. AP-3: an adaptor-like protein complex with ubiquitous expression. Embo J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Niiya A, Masaki R, Yamamoto A, Araki M, Taketani S. A newly identified membrane protein localized exclusively in intracellular organelles of neurons. Brain Res Mol Brain Res. 1997;46:265–273. doi: 10.1016/S0169-328X(97)00023-5. [DOI] [PubMed] [Google Scholar]
- Su TS, Nussbaum RL, Airhart S, Ledbetter DH, Mohandas T, O'Brien WE, Beaudet AL. Human chromosomal assignments for 14 argininosuccinate synthetase pseudogenes: cloned DNAs as reagents for cytogenetic analysis. Am J Hum Genet. 1984;36:954–964. [PMC free article] [PubMed] [Google Scholar]
- Saccone S, Biamonti G, Maugeri S, Bassi MT, Bunone G, Riva S, Della Valle G. Assignment of the human heterogeneous nuclear ribonucleoprotein A1 gene (HNRPA1) to chromosome 12q13.1 by cDNA competitive in situ hybridization. Genomics. 1992;12:171–174. doi: 10.1016/0888-7543(92)90424-q. [DOI] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature- dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Chambliss KL, Hinson DD, Trettel F, Malaspina P, Novelletto A, Jakobs C, Gibson KM. Two exon-skipping mutations as the molecular basis of succinic semialdehyde dehydrogenase deficiency (4-hydroxybutyric aciduria) Am J Hum Genet. 1998;63:399–408. doi: 10.1086/301964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallon BJ, Fung WJ, Tsang TC, Li S, Kado-Fong H, Huang KS, Kochan JP. Primary structure and functional activity of a phosphatidylinositol- glycan-specific phospholipase D. Science. 1991;252:446–448. doi: 10.1126/science.2017684. [DOI] [PubMed] [Google Scholar]
- Morrish DW, Dakour J, Li H. Functional regulation of human trophoblast differentiation. J Reprod Immunol. 1998;39:179–195. doi: 10.1016/S0165-0378(98)00021-7. [DOI] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Nakajima D, Ohira M, Seki N, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. VII. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1997;4:141–150. doi: 10.1093/dnares/4.2.141. [DOI] [PubMed] [Google Scholar]
- Sakane T, Takeno M, Suzuki N, Inaba G. Behcet's disease. N Engl J Med. 1999;341:1284–1291. doi: 10.1056/NEJM199910213411707. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, Szustakowki J, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Parimoo S, Patanjali SR, Shukla H, Chaplin DD, Weissman SM. cDNA selection: efficient PCR approach for the selection of cDNAs encoded in large chromosomal DNA fragments. Proc Natl Acad Sci U S A. 1991;88:9623–9627. doi: 10.1073/pnas.88.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyk GM, Kim SW, Myers RM, Cox DR. Exon trapping: a genetic screen to identify candidate transcribed sequences in cloned mammalian genomic DNA. Proc Natl Acad Sci U S A. 1990;87:8995–8999. doi: 10.1073/pnas.87.22.8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler AJ, Chang DD, Graw SL, Brook JD, Haber DA, Sharp PA, Housman DE. Exon amplification: a strategy to isolate mammalian genes based on RNA splicing. Proc Natl Acad Sci U S A. 1991;88:4005–4009. doi: 10.1073/pnas.88.9.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco AP, Neve RL, Colletti-Feener C, Bertelson CJ, Kurnit DM, Kunkel LM. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Van Den Eynde BJ, Gaugler B, Probst-Kepper M, Michaux L, Devuyst O, Lorge F, Weynants P, Boon T. A new antigen recognized by cytolytic T lymphocytes on a human kidney tumor results from reverse strand transcription. J Exp Med. 1999;190:1793–1800. doi: 10.1084/jem.190.12.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield JN, Rademacher TW. Structure and expression of the human glycosylphosphatidylinositol phospholipase D1 (GPLD1) gene. Biochim Biophys Acta. 2000;1494:189–194. doi: 10.1016/S0167-4781(00)00194-9. [DOI] [PubMed] [Google Scholar]
- Trettel F, Malaspina P, Jodice C, Novelletto A, Slaughter CA, Caudle DL, Hinson DD, Chambliss KL, Gibson KM. Human succinic semialdehyde dehydrogenase. Molecular cloning and chromosomal localization. Adv Exp Med Biol. 1997;414:253–260. [PubMed] [Google Scholar]
- Pype S, Declercq W, Ibrahimi A, Michiels C, Van Rietschoten JG, Dewulf N, de Boer M, Vandenabeele P, Huylebroeck D, Remacle JE. TTRAP, a novel protein that associates with CD40, tumor necrosis factor (TNF) receptor-75 and TNF receptor-associated factors (TRAFs), and that inhibits nuclear factor-kappa B activation. J Biol Chem. 2000;275:18586–18593. doi: 10.1074/jbc.M000531200. [DOI] [PubMed] [Google Scholar]
- Irie A, Suzuki A. CMP-N-Acetylneuraminic acid hydroxylase is exclusively inactive in humans. Biochem Biophys Res Commun. 1998;248:330–333. doi: 10.1006/bbrc.1998.8946. [DOI] [PubMed] [Google Scholar]