Abstract
Background
micF RNA, a small regulatory RNA found in bacteria, post-transcriptionally regulates expression of outer membrane protein F (OmpF) by interaction with the ompF mRNA 5'UTR. Phylogenetic data can be useful for RNA/RNA duplex structure analyses and aid in elucidation of mechanism of regulation. However micF and associated genes, ompF and ompC are difficult to annotate because of either similarities or divergences in nucleotide sequence. We report by using sequences that represent "gene signatures" as probes, e.g., mRNA 5'UTR sequences, closely related genes can be accurately located in genomic sequences.
Results
Alignment and search methods using NCBI BLAST programs have been used to identify micF, ompF and ompC in Yersinia pestis and Yersinia enterocolitica. By alignment with DNA sequences from other bacterial species, 5' start sites of genes and upstream transcriptional regulatory sites in promoter regions were predicted. Annotated genes from Yersinia species provide phylogenetic information on the micF regulatory system. High sequence conservation in binding sites of transcriptional regulatory factors are found in the promoter region upstream of micF and conservation in blocks of sequences as well as marked sequence variation is seen in segments of the micF RNA gene. Unexpected large differences in rates of evolution were found between the interacting RNA transcripts, micF RNA and the 5' UTR of the ompF mRNA. micF RNA/ompF mRNA 5' UTR duplex structures were modeled by the mfold program. Functional domains such as RNA/RNA interacting sites appear to display a minimum of evolutionary drift in sequence with the exception of a significant change in Y. enterocolitica micF RNA.
Conclusions
Newly annotated Yersinia micF and ompF genes and the resultant RNA/RNA duplex structures add strong phylogenetic support for a generalized duplex model. The alignment and search approach using 5' UTR signatures may be a model to help define other genes and their start sites when annotated genes are available in well-defined reference organisms.
Keywords: gene annotation, phylogeny, RNA/RNA interactions, regulatory RNA
Background
The rapid determination of microbial genomic sequences poses a challenge in gene annotation and assignment of transcriptional start sites. Without experimental data, incorrect annotations can be made as well as erroneous determination of gene start sites. This is especially true for genes that are evolutionarily and structurally related such as the bacterial porin genes, ompF and ompC. For example, a BLAST search using the Salmonella typhimurium ompF gene sequence identifies Enterobacter cloacae ompC as well as Salmonella minnesota ompC (unpublished). However, when gene promoter sites, transcript 5'UTR sequences, or signatures within genes from reference organisms are used, an accurate assignment of a gene as well as a prediction of its start site can be made by a comparative approach. These regions serve specific functions in molecular processes, e.g., several 5' UTRs of mRNA transcripts are mRNA stability determinants [1,2]. Thus they can display sequence and/or secondary structure signatures and these defined segments can be more useful than using entire gene sequences for annotations. The use of domains for annotation of genomic sequences originated with analyses of protein coding regions [3-5]. In this paper, alignment and comparative methods have been used to annotate the small regulatory RNA gene micF and its associated genes, ompF and ompC in Yersinia species. Transcriptional start sites have also been assigned based on alignment data.
The micF transcript is a small non-protein coding RNA found in E. coli and related bacteria [6,7]. micF regulates outer membrane protein F (OmpF) synthesis in response to stress and other environmental signals [8] and these signals induce the transcription of micF RNA. micF RNA functions by interacting with the target ompF mRNA 5' UTR to form an RNA/RNA duplex. The micF transcript inhibits ompF expression post-transcriptionally by blocking translation and inducing degradation of the ompF message [9].
With the use of newly determined Yersinia genomic sequences [10] (see also Accession no. gnl|SANGER_34054|, Yersinia enterocolitica 8081), micF RNA/ompF mRNA 5' UTR duplex models have been deduced. These structures show a strong evolutionary conservation of the RNA/RNA duplex structure, but also reveal additional interacting sites. The new Yersinia sequences further support a phylogenetic conservation of the micF regulatory system in γ-proteobacteria.
Results and Discussion
Gene Annotation
BLAST searches were performed to find micF and its target ompF, as well as ompC in genomic sequences available on the GenBank site of the National Center for Biotechnology Information (NCBI). Due to similarities in sequence in protein coding regions of ompF and ompC, a BLAST search using known gene sequences can provide erroneous results. However the mRNA 5' UTR transcript sequences of ompF and ompC differ significantly and the ompF 5' UTR sequence provides a basis for detection in a BLAST search since several ompF 5' UTR sequence signatures are conserved and are ompF specific. Additional markers used were genomic positions relative to highly conserved genes such as asnS, the asparaginyl-tRNA synthetase, which is preceded by ompF in the E. coli and S. typhimurium chromosomes by about 600 bp. The ompC and micF genes are upstream of each other in Gram-negative genomic sequences and in E. coli, they are separated by a 253 bp regulatory promoter region [8]. Since these genes share a transcriptional regulatory region, rearrangements, where these genes may be repositioned and separated on the chromosome, are probably unlikely.
Y. pestis and Y. enterocolitica ompF mRNA 5' UTR sequences were found by a BLAST search on GenBank using the Serratia marcescens ompF 5' UTR sequence as a probe. Position 1600321 of the Y. pestis genomic sequence and position 1759190 of the Y. enterocolitica sequence were pinpointed as the 5' starts.
The S. marcescens ompC 5' UTR/regulatory promoter region/micF segment provided a suitable sequence for determination of genomic positions of Yersinia ompC and micF, however these have been partially annotated in Y. pestis (Accession # NC_003143.1). Assignment of 5' start sites of Yersinia species micF, ompF and ompC was based on sequence alignment using 5' start sites of other organisms (see below). Annotation of ompF, ompC and micF genes in Yersinia species is shown in Table 1.
Table 1.
Annotation of ompF, ompC, and micF in Yersinia species
| Organism and Accession Number | Gene | Positions |
| Y. pestis strain CO92 NC_003143.1| | ompF 5'UTR | 1600321-1600226 |
| ompC 5'UTR | 1382092-1382165 | |
| micF | 1381842-1381759 | |
| Y. enterocolitica 8081 gnl|SANGER_34054 | ompF 5'UTR | 1759190-1759087 |
| ompC 5'UTR | 1571915-1571842 | |
| micF | 1571589-1571499 |
Sequence Alignments and comparisons
ompF and ompC 5' UTR
Figure 1 shows the alignment of DNA sequences representing six ompF 5' UTRs. opnP is the ompF homolog in Xenorhabdus nematophilus, an organism which resides in a specific ecological nitch but is phylogenetically related to the γ-proteobacteria [11-13]. Assignment of 5' start sites of Yersinia species mRNA transcripts was based on alignment and similarity with the other ompF 5' UTR sequences.
Figure 1.
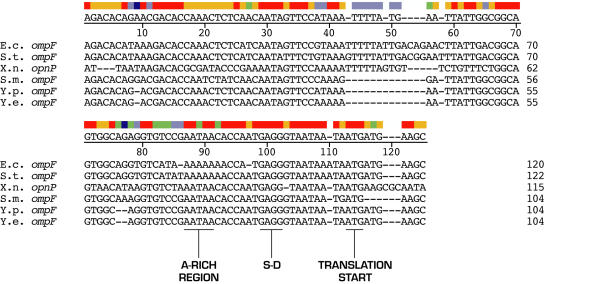
Alignment of ompF mRNA 5' UTRs from 6 bacterial species. Putative 5' end nucleotides (position one) of Yersinia species ompF transcripts were determined by sequence alignment. Sequences were aligned by the ClustalV method using the DNASTAR Inc MegAlign program. OpnP is the ompF homolog in X. nematophilus. Color bar indicates degree of similarity at each position with red signifying 100% identity, orange signifying that 5 out of 6 nucleotides are the same, green shows partial similarity whereas purples depict poor identity. The consensus sequence is shown under the color bar.
Y. pestis and Y. enterocolitica ompF 5' UTRs differ only by a T to A base substitution at position 39 (Figure 1). In addition, the nucleotide sequence identity between Yersinia species and other γ-proteobacteria is high (Figure 2). Consistent with bacterial evolutionary relatedness, the percent identity of Yersinia species ompF 5' UTRs appear to be closest to Serratia marcescens (Figure 2).
Figure 2.
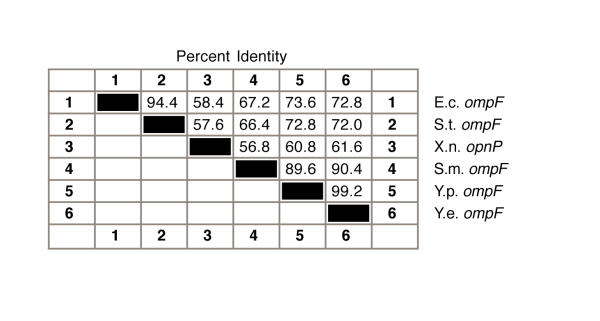
The % identity between bacterial ompF mRNA 5' UTR sequences shown in Figure 1.
High sequence conservation implies a functional role for conserved elements. The 5' UTRs of bacterial mRNAs are important determinants of mRNA stability and/or translational regulation [1,2,14-16]. In keeping with a functional role, the segment of ompF 5' UTR that forms the major RNA/RNA duplex pairing with micF (i.e., positions 96–125), is highly conserved amongst the species analyzed, including Yersinia species (Figure 1). In addition, this region contains the ribosome binding site [Shine-Dalgarno (S-D) sequence] and initiation codon ATG start site and these also contribute to evolutionary sequence stability. An additional signature shared by the Yersinia species is an A-rich region that precedes the S-D domain. The 5' end region of ompF mRNA 5' UTR contains a long stem-loop and evolutionary changes in this segment consist primarily of compensatory base-pair changes that maintain a stem-loop structure [17]. However the 5'-end region of the ompF UTR also has a high sequence conservation, with the exception of a 12 nt deletion that the Yersinia species share with S. marcescens (Figure 1).
Putative 5' start sites of Yersinia species ompC transcripts were also determined by sequence alignment (Figure 3). High similarity in the promoter regions also support start site predictions of both micF and ompC transcripts (see below).ompC mRNA 5' UTR sequences show more sequence divergence than ompF 5' UTR sequences, e.g., the percent identity between Y. pestis and Y. enterocolitica sequences is 81% and divergence between the two Yersinia ompC 5' UTR segments is greater than that of the closely related organisms, E. coli and S. typhimurium (Figure 4). As with ompF mRNA 5' UTR, the S-D sequence GAGG and AT rich region between the S-D site and ATG start codon are highly conserved (Figure 3). With exception of the ribosome binding site, a functional role for the ompC mRNA 5' UTR is not known.
Figure 3.
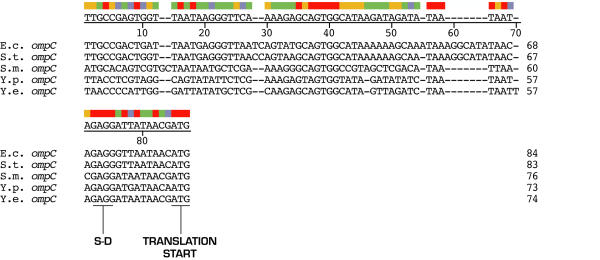
Alignment of ompC 5' UTR sequences (by J. Hein method). Putative 5' start sites of Yersinia species ompC were also determined by sequence alignment. The consensus sequence is shown below the color bar.
Figure 4.
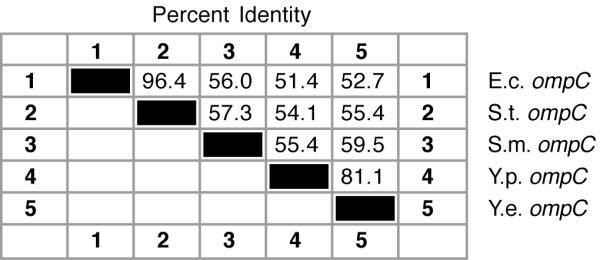
Percent identity between ompC 5' UTR sequences from five bacterial species.
micF sequence comparisons
5' ends of Yersinia micF genes were deduced by alignment with micF sequences from related species (Figure 5). The ρ-independent termination motif of the micF transcript delineates the 3' end. A comparison of micF sequences from 6 bacterial species reveals that the initial 13 nt from the 5' end are invariant; in addition, the first 32 nt are highly conserved (Figure 5). This 32 nt region represents the segment of the micF transcript that forms the major duplex interaction with the target ompF mRNA in E. coli [2]. Similar to target sequences in ompF mRNA 5' UTR (positions 96–125, see Figure 1), this binding role may account for the high evolutionary sequence conservation of the 32 nt segment at the 5' end of micF.
Figure 5.
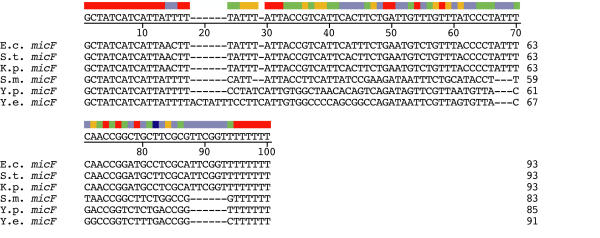
Alignment of micF gene sequences (by J. Hein method). The consensus sequence is shown below the color bar.
The variability of micF nucleotide sequence in the last two thirds of the gene may be due to several factors. For example, the ρ-independent termination stem-loop transcript structure may function on secondary and not primary structural features, and can tolerate compensatory base-pair changes in the stem as well as changes in loop sequences. In addition, variation in the type of RNA/RNA base pairing occurs with sequences in the highly variable middle section of micF RNA (positions 33–70, Figure 5). For example, nucleotide sequences within this region allow for formation of intra- and inter- molecular base-pairings in RNA/RNA interactions in Yersinia species. These base pairings are not seen in other species (see below).
Yersinia micF sequences have diverged more than the S. marcescens micF sequence, e.g., the percent identity between S. marcescens micF and E. coli micF is 65.5%; it is 52.9% between Y. pestis and E. coli micF genes (Figure 6). Evolutionary instability in sequence is consistent with the high genetic flux found in the Y. pestis genome [10]. Interestingly, the Y. enterocolitica micF sequence has diverged somewhat more (Figure 6). But of interest also is the large difference in sequence between the two Yersinia micF RNA genes, i.e., 15 base substitutions and a 6 base insertion at positions 18–23 in Y. enterocolitica (Figure 5). These sequence differences actually reinforce the Yersinia RNA/RNA duplex model (see below). The reason for the evolutionary drift in Y. enterocolitica micF sequence is not known.
Figure 6.
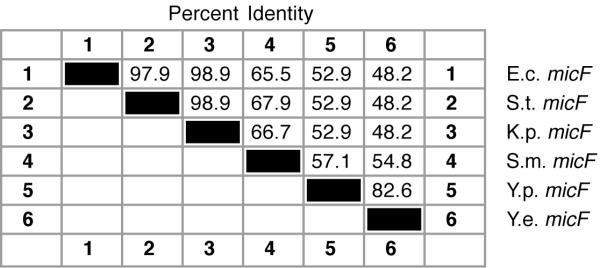
Percent sequence identity in micF bacterial genes.
The micF sequence identities differ markedly from those of ompF 5' UTR (compare Figure 2 and Figure 6). These differences are also evident in the phylogenetic trees (Figure 7 and Figure 8). Noteworthy are the differences between Yersinia species sequence s, i.e., there are 21 base changes between the two Yersinia species micF sequences (Figure 5) and only one base change (at position 39) between Yersinia ompF 5' UTR sequences (Figure 1). Thus this reveals a very unequal rate of nucleotide sequence change between Yersinia micF and ompF 5' UTR sequences. On the other hand there appears to be a uniform rate of nucleotide change between S. marcescens and E. coli sequences in ompF mRNA 5' UTR (67.2 % identity) and micF (65.5 % identity) (compare Figures 2 and 6).
Figure 7.
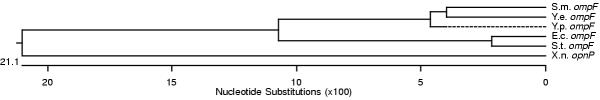
Phylogenetic tree of ompF mRNA 5' UTR sequences determined by DNASTAR program.
Figure 8.

Phylogenetic tree of micF sequences determined by DNASTAR program.
Upstream regulatory region
In E. coli, the 253 nt upstream sequence between micF and ompC contains promoters as well as binding sites for several transcription factors [8]. micF is part of a global regulatory network that responds to environmental stress conditions in E. coli [18,9,19]. It is thus not surprising that this region has a complex set of transcriptional regulatory sites. These sites comprise approximately one third of the upstream sequence between micF and ompC in E. coli.
A comparison of nucleotide sequences from related bacteria shows that these binding sites are highly conserved evolutionarily and that Yersinia species sequences have not appreciably diverged, either between themselves or in comparison to those in other bacterial sequences (Figure 9). OmpR, the transcription factor that activates transcription of both micF and ompC in response to osmolarity increase binds at three sites in E. coli, C1, C2, and C3 [18,20]. These sites are highly conserved between all bacterial species where 5–6 positions out of 10 are totally conserved (Figure 9). A high degree of sequence conservation is also found for integration host factor (IHF), a protein that participates in bending of DNA [21].
Figure 9.
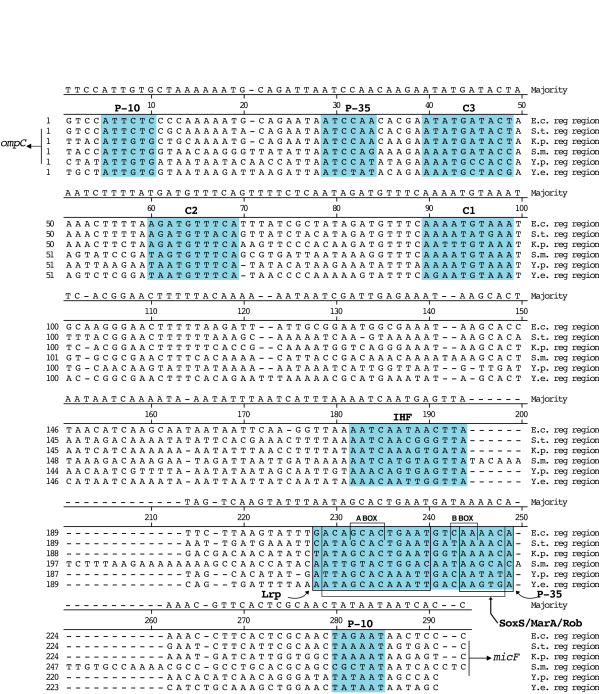
Alignment of sequences upstream of ompC and micF (promoter and transcription regulatory region). J. Hein alignment method was used, however positions 197–213 and 246–261 were aligned by eye to reflect known homologies. The consensus (Majority) sequence is shown above alignments. Promoters (P-10, P-35) and transcription factor binding sites are shown in color. ompC and micF are transcribed in opposite directions (shown with arrows).
SoxS, MarA, and Rob are part of the family of AraC/XylS transcription regulators and share the same DNA binding sites in E. coli [22]. These factors regulate E. coli micF transcription in response to different environmental factors. The SoxS/MarA/Rob binding site upstream of micF, termed the SoxS/MarA/Rob box, is not well conserved phylogenetically, however the major polymerase interaction site, the A box, is almost totally conserved in bacterial sequences analyzed, including those of Yersinia (Figure 9). MarA and Rob transcription regulators in E. coli have two helix-turn-helix (HTH) motifs that interact with A and B box sequences [23,24], and one HTH element of Rob has been shown to insert itself in the micF promoter DNA at the major groove of the A box region [24].
An alignment of six different gene promoters in E. coli that contain a SoxS/MarA/Rob box shows that they conform to a conserved 18–19 bp distance between the SoxS/MarA/Rob binding sequences and the -10 promoter [25,26]. This also holds true for micF promoters in related bacteria, including promoters of the Yersinia species, but the exception is the S. marcescens sequence, which has a 12 bp insertion between the -10 and -35 micF promoter sites and adjacent to the B box of (Figure 9). Thus this partially conserved 18–19 bp distance may not be as relevant as previously thought. Although sites of interaction of RNA polymerase (RNAP) with these transcription regulators have not been determined [27,28], the 12 bp pair insertion does pose the question of how putative S. marcescens SoxS/MarA/Rob factors interact with RNAP to activate transcription in S. marcescens.
The leucine response protein (Lrp), a global regulator of transcription [29] represses micF [30]. In E. coli the primary Lrp binding site [30] overlaps much of the SoxS/MarA/Rob site upstream of micF (Figure 9). The highest sequence conservation is primarily in the SoxS/MarA/Rob-related A box sequence GCAC. Since the Lrp site overlaps the SoxS/marA/Rob site, a high overall sequence conservation may be expected, but the degree of conservation is much less than that for example, of the OmpR sites. The putative Lrp binding site sequences between Yersinia species are nearly identical but differ almost uniformly from those of the other bacteria.
Some regions not pinpointed as sites for factor binding have diverged appreciably, e.g., the region between integration host factor (IHF) and Lrp binding sites (positions 195 to 228), but the region between C1 and IHF binding sites (100 to 182) has some partially conserved sequences (Figure 9). These may serve as sites for other transcription factors where binding sequences have not been defined. For example in E. coli, H-NS binds in an as yet unspecified site in the ompC/micF regulatory region [31]. In addition, nfxB encodes a transcriptional repressor and may act on micF as an nfxB mutant was found to reduce OmpF levels post-transcriptionally in E. coli [32]. nfxB has also been found in Pseudomonas aeruginosa [33]; thus a putative micF regulatory site for nfxB may also be evolutionarily conserved. Evolutionary conservation of transcription factor binding sites as seen in Figure 9 implies that these transcriptional regulators function in bacterial species being analyzed here. Some factors, such as OmpR have been found in related organisms [34].
Although transcription factor binding sites display a high uniformity in sequence, a sequence comparison of the entire upstream regulatory region shows that the Yersinia species have diverged significantly between each other and more than the E. coli/S. typhimurium/K. pneumoniae grouping has diverged within itself (data not shown). It remains to be seen if entire genome comparisons of Y. pestis and Y. enterocolitica will show this evolutionary trend, which has also been seen in micF, or if this divergence pattern is an anomaly.
OmpF mRNA 5'UTR/micF RNA duplex structures
Secondary structure probing by enzymatic and chemical techniques helped defined a micF RNA/ompF mRNA 5' UTR interaction [2,8]. In E. coli, micF RNA binds the ompF mRNA 5' UTR to form an RNA/RNA duplex that contains imperfect base-pairing [2,35]. In the present work, Yersinia ompF mRNA 5' UTR/ micF RNA duplex structure modeling was performed with the mfold program [36,37] (Figure 10 and Figure 11). Sequences between Yersinia micF RNA species differ in 21 out of 91 positions (shown in blue in Figure 11). Interestingly, these changes result in only minor variations in Yersinia RNA/RNA duplex structural models. For example, base substitutions at positions 39–42, and 77 in Y. enterocolitica are in looped regions (compare Figures 10 and 11). The 6 base insertion at positions 18–23 in Y. enterocolitica micF RNA, as well as the base substitution at position 61 expand duplex pairings in stems 1 and 3. Base changes at positions 38, 44, 46, 53, and 69 constitute compensatory changes that maintain base pairs in stems. Thus the large divergence in micF RNA sequence between Y. pestis and Y. enterocolitica does not significantly alter the RNA/RNA duplex structure. This strengthens the rationale for the duplex model.
Figure 10.
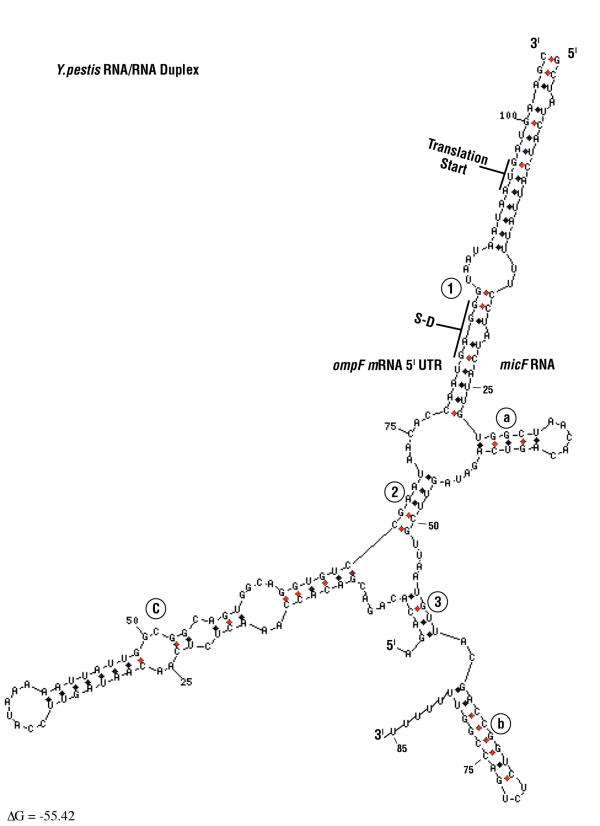
Y. pestis ompF mRNA 5' UTR/micF RNA duplex model. Secondary structures determined by mfold program of Zuker and co-workers. D. Stewart and M. Zuker graphics program was used (web site: http://www.bioinfo.rpi.edu/applications/mfold/). The Y. pestis duplex structure represents the first alternate structure by mfold modeling. Numbers 1–3 in the figure refer to inter-molecular stems formed by ompF mRNA 5' UTR and micF RNA sequences. Letters a-c refer to intra-molecular micF or ompF mRNA 5'UTR stem loops.
Figure 11.
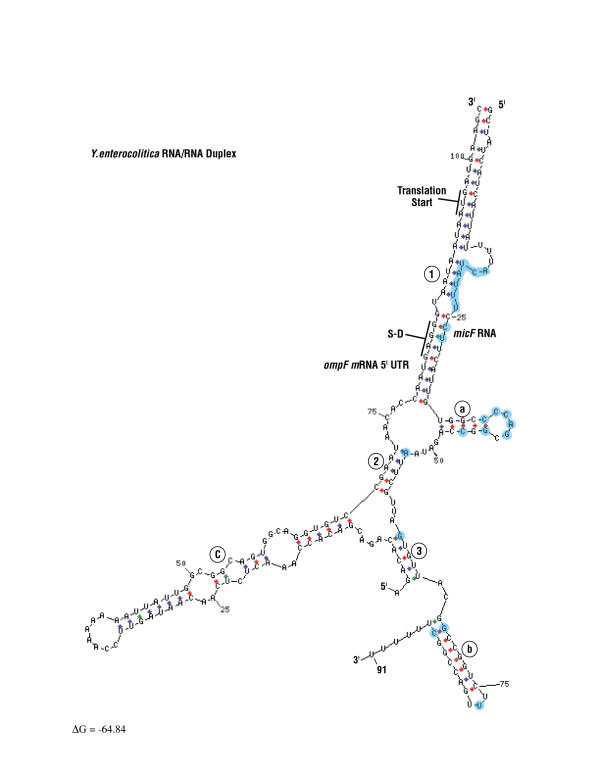
Y. enterocolitica ompF mRNA 5' UTR/micF RNA duplex model. The Y. enterocolitica duplex is structure one by mfold modeling. Numbers 1–3 in the figure refer to inter-molecular stems formed by ompF mRNA 5' UTR and micF RNA sequences. Letters a-c refer to intra-molecular micF or ompF mRNA 5'UTR stem loops. Positions shown in blue color are those that differ between Y. enterocolitica and Y. pestis micF RNAs.
Figure 12 shows a diagrammatic representation of the Yersinia RNA/RNA duplex models along with models of E. coli and S. marcescens duplexes. Both Yersinia structures conform to a generalized ompF mRNA 5'UTR /micF RNA structural model, but the Yersinia species form additional intra- and inter- molecular base pairings, i.e., stem loop a and stem 2 (Figure 12). Compensatory base pair changes in stem loop a add strong phylogenetic support for the presence of this structure in Yersinia species. The question is, does this stem loop have a function in regulation of ompF expression in Yersinia? Mfold modeling does not predict formation of thermodynamically stable stem loop a or stem 2 structures in E. coli or S. marcescens RNA/RNA duplexes. However structure probe data of the E. coli micF RNA/ompF mRNA 5' UTR duplex do not necessarily support or preclude Watson-Crick pairings that can form stem 2 in E. coli [2].
Figure 12.
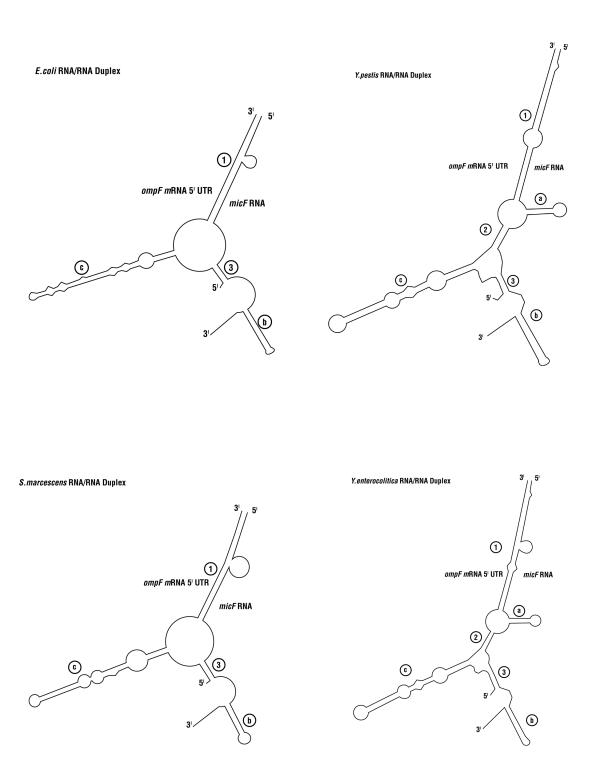
Diagrammatic representation of duplex models from four bacterial species. The E. coli RNA/RNA duplex model was determined by structure probing [2]. The S. marcescens RNA/RNA duplex model is according to the long range pairing algorithm of [38,8].
In E. coli, stem 1 of the RNA/RNA duplex is formed from regions that are largely single-stranded in free (uncomplexed) RNAs [2,39], but a minor stem loop in ompF 5' UTR and a thermodynamically weak stem loop in micF RNA unfold to form the RNA/RNA interaction [8]. In the E. coli RNA/RNA duplex, structure probe data show that intra-molecular stem loop c in ompF 5' UTR and stem loop b in micF RNA (the ρ-independent terminator), which are present in uncomplexed RNAs, are unaltered in the RNA/RNA duplex [2]. Mfold analyses of Yersinia species duplexes also predict that these stem loops are maintained in RNA/RNA duplex structures. Perhaps evolutionary pressure to maintain the stability of these intra-molecular stem loops is greater than a progression to unfolding that may create more sites for RNA/RNA interactions. However we cannot rule out a functional role for stem loops b and c in ompF mRNA inactivation, e.g., protein factor binding during micF RNA-induced ompF mRNA destabilization.
Conclusions and Future Prospects
With rapid sequencing of bacterial genomes, it is a challenge to annotate the thousands of genes and especially determine gene start sites (5' ends of RNA transcripts). In this work, BLAST searches using segments of genes, such as the ompF 5' UTR sequence resulted in the annotation of Yersinia genes. This was achieved in part because the ompF mRNA 5' UTR sequence has functional domains which are involved in translational regulation. Use of signatures to annotate genomic sequences has been widely reported before. This includes PRINTS/BLOCKS/Pfam data bases [3-5,40] and CDART [41]. In these examples, protein coding regions are employed to search for sequences that display similar domains.
In this work, sequence alignment programs have provided putative start sites as well as definition of transcription factor binding sites in the upstream regulatory region of micF. Additionally, with these sequences, new Yersinia ompF mRNA 5' UTR/micF RNA duplex structures have been proposed. These add strong phylogenetic support for previously determined duplex structures.
The approach used here to locate genes may also be useful in additional annotations. For example, a sequence has been located in Desufovibrio desulfuricans that displays 74% similarity to a putative mRNA 5' UTR sequence from K. pneumoniae (unpublished data). This is of particular of interest since Desufovibrio desulfuricans is phylogenetically in the δ-subdivision of the proteobacteria [42]; E. coli, K. pneumoniae, S. marcescens, and Yersinia species, are part of the γ-proteobacteria. micF genes in distantly related bacteria have not yet been found due to evolutionary divergence of micF, ompC mRNA 5' UTR, and the upstream sequences, in spite of the presence of conserved elements such as the invariant 13 nt at the 5' end of micF and conserved putative transcription factor binding sites in currently analyzed organisms. However a combination of sequence and secondary structure motifs [43,44] will be utilized in future studies for additional searches for micF.
Methods
BLAST searches were performed to find micF and target ompF genes as well as ompC in genomic sequences available on GenBank sites of the National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov/. Genomic data were scanned by alignment methods in GenBank. To search for particular genes, a highly conserved gene was first located and then alignment methods were used to locate the desire gene in adjacent sequences. mRNA signatures such as AUG start, the Shine-Dalgarno (S-D) ribosome binding site and other signatures were also used as a guide. Where there were questions of sequence errors in genes analyzed, these sequences were not used. BLAST searches for sequences homologous to micF and ompF also included sequences in Haemophilus influenzae, Klebsiella pneumoniae, Pasteurella multocida, Desulfovibrio desulfricans, and Schwanella oneidensis as well as those from Yersinia species.
BLAST sequence similarity search programs also provided in GenBank were used [45]. Microbial genome searches with BLASTN 2.2.3 and 2.2.4 using expect values of 10–1000 with the Advanced BLAST program was used to find micF and associated genes sequences in Yersinia species: ref #|NC_003143.1| Yersinia pestis strain CO92, complete genome, and gnl|SANGER_34054| Yersinia enterocolitica 8081. Yersinia species ompF and ompC genes were located by BLAST searches with ompF or ompC mRNA 5' UTR sequences.
The 5' start sites of micF and, ompF and ompC 5'UTRs, were predicted by nucleotide sequence alignment and similarity. MegAlign alignment programs were from DNASTAR, Inc. http://www.dnastar.com/. Parameters used were either that of J. Hein with gap penalty 11, gap length, 3; ClustalV, with gap penalty 10, gap length 10; or ClustalW with gap penalty 15, gap length, 6.66. Percent identities, consensus (majority) sequences, and phylogenetic trees were also based on programs from DNASTAR, Inc. Alignment methods chosen were for the most part based on results with well established identities in previously determined sequences. When an alignment showed obvious discrepancies, such a lack of alignment of stretches of near perfect identity, it was discarded.
Standard default parameters, with the exception of assignment of the number of alternate structures to 10 were use to fold RNAs into duplex structures with the Zuker/Turner mfold program, version 3.1. The program is available at internet address: http://www.bioinfo.rpi.edu/applications/mfold/
Abbreviations
Organisms
E.c., Escherichia coli; S.t., Salmonella typhimurium; S.m. Serratia marcescens; K.p., Klebsiella pneumoniae. Y.e., Yersinia enterocolitica, Y.p., Yersinia pestis;H.i.,Haemophilus influenzae; P.m.,Pasteurella multocida, D.d., Desulfovibrio desulfricans.
Proteins
OmpF, outer membrane protein F; OmpC, outer membrane protein C; Lrp, leucine-responsive protein; IHF, integration host factor; HTH, helix-turn-helix; RNAP, RNA polymerase.
Nucleic Acids
nt, nucleotides; bp, base pairs
Web Site References
GenBank sites of the National Center for Biotechnology Information: http://www.ncbi.nlm.nih.gov/subtils/Entrez/genom_table_cgi and http://www.ncbi.nlm.nih.gov/BLAST/
The Wellcome Trust Sanger Institute:
http://www.sanger.ac.uk/Projects/Microbes/
Washington University Genome Sequencing Center:
DNASTAR, Inc:
M. Zuker mfold program:
Acknowledgments
Acknowledgements
I thank Dr. Nicholas Thomson from the Wellcome Trust Sanger Institute for discussions concerning annotation of Yersinia genes and Dr. John Spieth, Washington University, GSC Center for discussions of ompF-like sequences in K. pneumoniae. Work supported by DRAC Award, School of Medicine, SUNY, Stony Brook.
References
- Chen LH, Emory SA, Bricker AL, Bouvet P, Belasco JG. Structure and function of a bacterial mRNA stabilizer: analysis of the 5' untranslated region of ompA mRNA. J Bacteriol. 1991;173:4578–4586. doi: 10.1128/jb.173.15.4578-4586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Zheng P, Delihas N. Secondary structures of Escherichia coli antisense micF RNA, the 5'-end of the target ompF mRNA, and the RNA/RNA duplex. Biochemistry. 1995;34:3621–3631. doi: 10.1021/bi00011a017. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Pietrokovski S. Blocks+: a non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics. 1999;15:471–479. doi: 10.1093/bioinformatics/15.6.471. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, Durbin R, Falquet L, Fleischmann W, Gouzy J, Hermjakob H, Hulo N, Jonassen I, Kahn D, Kanapin A, Karavidopoulou Y, Lopez R, Marx B, Mulder NJ, Oinn TM, Pagni M, Servant F, Sigrist CJ, Zdobnov EM, InterPro Consortium. InterPro – an integrated documentation resource for protein families, domains and functional sites. Bioinformatics. 2000;16:1145–1150. doi: 10.1093/bioinformatics/16.12.1145. [DOI] [PubMed] [Google Scholar]
- Attwood TK, Blythe MJ, Flower DR, Gaulton A, Mabey JE, Maudling N, McGregor L, Mitchell AL, Moulton G, Paine K, Scordis P. PRINTS and PRINTS-S shed light on protein ancestry. Nucleic Acids Res. 2002;30:239–241. doi: 10.1093/nar/30.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Chou M-Y, Inouye M. Regulation of gene expression by a small RNA transcript (mic RNA) in Escherichia coli. Proc Jpn Acad. 1983;59:335–339. [Google Scholar]
- Esterling L, Delihas N. The regulatory RNA gene micF is present in several species of gram-negative bacteria and is phylogenetically conserved. Mol Microbiol. 1994;12:639–646. doi: 10.1111/j.1365-2958.1994.tb01051.x. [DOI] [PubMed] [Google Scholar]
- Delihas N, Forst S. micF : an antisense RNA gene involved in response of Escherichia coli to global stress factors. J Mol Biol. 2001;313:1–12. doi: 10.1006/jmbi.2001.5029. [DOI] [PubMed] [Google Scholar]
- Andersen J, Forst SA, Zhao K, Inouye M, Delihas N. The function of micF RNA: micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J Biol Chem. 1989;264:17961–17970. [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Basham D, Bentley SD, Brooks K, Cerdeno-Tarraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougan G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston PC, Quail M, Rutherford K, Simmonds M., Skelton J, Stevens K, Whitehead S, Barrell BG. Genome sequence of Yersinia pestis, the causative agent of plague. Nature. 2001;413:523–527. doi: 10.1038/35097083. [DOI] [PubMed] [Google Scholar]
- Forst S, Waukau J, Leisman G, Exner M, Hancock R. Functional and regulatory analysis of the OmpF-like porin, OpnP, of the symbiotic bacterium Xenorhabdus nematophilus. Mol Microbiol. 1995;18:779–789. doi: 10.1111/j.1365-2958.1995.mmi_18040779.x. [DOI] [PubMed] [Google Scholar]
- Forst S, Nealson K. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol Rev. 1996;60:21–43. doi: 10.1128/mr.60.1.21-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz J, Meinert F, Wyss U, Ehlers RU, Stackebrandt E. Development and application of oligonucleotide probes for molecular identification of Xenorhabdus species. Appl Environ Microbiol. 1990;56:181–186. doi: 10.1128/aem.56.1.181-186.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambronne ED, Schneewind O. Yersinia enterocolitica type III secretion: yscM1 and yscM2 regulate yop gene expression by a posttranscriptional mechanism that targets the 5' untranslated region of yop mRNA. J Bacteriol. 2002;184:5880–5893. doi: 10.1128/JB.184.21.5880-5893.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Weiner A. Measuring the immeasurable. Mol Cell. 2002;10:437–439. doi: 10.1016/s1097-2765(02)00661-5. [DOI] [PubMed] [Google Scholar]
- Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Delihas N. Antisense micF RNA and 5'-UTR of the target ompF RNA: phylogenetic conservation of primary and secondary structures. Nucleic Acids Symp Ser. 1997;36:33–35. [PubMed] [Google Scholar]
- Forst S, Inouye M. Environmentally regulated gene expression for membrane proteins in Escherichia coli. Annu Rev Cell Biol. 1988;4:21–42. doi: 10.1146/annurev.cellbio.4.1.21. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Hsing W, Gibson KE, Silhavy TJ. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol. 1996;20:911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x. [DOI] [PubMed] [Google Scholar]
- Tsung K, Brissette RE, Inouye M. Identification of the DNA-binding domain of the OmpR protein required for transcriptional activation of the ompF and ompC genes of Escherichia coli by in vivo DNA footprinting. J Biol Chem. 1989;15:10104–10109. [PubMed] [Google Scholar]
- Rice PA. Making DNA do a U-turn: IHF and related proteins. Curr Opin Struct Biol. 1997;7:86–93. doi: 10.1016/S0959-440X(97)80011-5. [DOI] [PubMed] [Google Scholar]
- Gallegos MT, Schleif R, Bairoch A, Hofmann K, Ramos JL. Arac/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S, Martin RG, Rosner JL, Davies DR. Protein, nucleotide, structure A novel DNA-binding motif in MarA: the First structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HJ, Bennik MH, Demple B, Ellenberger T. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nature Struct Biol. 2000;5:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- Martin RG, Gillette WK, Rhee S, Rosner JL. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol Microbiol. 1999;34:431–441. doi: 10.1046/j.1365-2958.1999.01599.x. [DOI] [PubMed] [Google Scholar]
- Martin RG, Gillette WK, Rosner JL. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol Microbiol. 2000;35:623–634. doi: 10.1046/j.1365-2958.2000.01732.x. [DOI] [PubMed] [Google Scholar]
- Griffith KL, Shah IM, Myers TE, O'Neill MC, Wolf RE., Jr Evidence for "pre-recruitment" as a new mechanism of transcription activation in Escherichia coli: the large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem Biophys Res Commun. 2002;291:979–986. doi: 10.1006/bbrc.2002.6559. [DOI] [PubMed] [Google Scholar]
- Martin RG, Gillette WK, Martin NI, Rosner JL. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol Microbiol. 2002;43:355–370. doi: 10.1046/j.1365-2958.2002.02748.x. [DOI] [PubMed] [Google Scholar]
- Calvo JM, Matthews RG. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario M, Ernsting BR, Borst DW, Wiese DE, 2nd, Blumenthal RM, Matthews RG. The leucine-responsive regulatory protein of Escherichia coli negatively regulates transcription of ompC and micF and regulates translation of ompF. J Bacteriol. 1995;177:103–113. doi: 10.1128/jb.177.1.103-113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ueguchi C, Mizuno T. H-NS regulates OmpF expression through micF antisense RNA in Escherichia coli. J Bacteriol. 1996;178:3650–3653. doi: 10.1128/jb.178.12.3650-3653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper DC, Wolfson JS, Souza KS, Ng EY, McHugh GL, Swartz MN. Mechanisms of quinolone resistance in Escherichia coli: characterization of nfxB and cfxB, two mutant resistance loci decreasing norfloxacin accumulation. Antimicrob Agents Chemother. 1989;33:283–290. doi: 10.1128/aac.33.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T, Ishiguro K, Takemoto N, Koibuchi H, Sugimoto K. Purification and characterization of the Pseudomonas aeruginosa NfxB protein, the negative regulator of the nfxB gene. J Bacteriol. 1995;177:5872–5877. doi: 10.1128/jb.177.20.5872-5877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang IS, Audia JP, Park YK, Foster JW. Auto induction of the ompR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol Microbiol. 2002;44:1235–1250. doi: 10.1046/j.1365-2958.2002.02937.x. [DOI] [PubMed] [Google Scholar]
- Delihas N, Rokita SE, Zheng P. Natural antisense RNA/target RNA interactions: possible models for antisense oligonucleotide drug design. Nat Biotechnol. 1997;15:751–753. doi: 10.1038/nbt0897-751. [DOI] [PubMed] [Google Scholar]
- Zuker M, Mathews DH, Turner DH. Algorithms and Thermodynamics for RNA Secondary Structure Prediction: A Practical Guide. In: Barciszewski J, editor. In RNA Biochemistry and Biotechnology. Clark BFC: NATO ASI Series, Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Tabaska JE, Cary RB, Gabow HN, Stormo GD. An RNA folding method capable of identifying pseudoknots and base triples. Bioinformatics. 1998;14:691–699. doi: 10.1093/bioinformatics/14.8.691. [DOI] [PubMed] [Google Scholar]
- Lindell M, Romby P, Wagner EGH. Lead(II) as a probe for investigating RNA structure in vivo. RNA. 2002;8:534–541. doi: 10.1017/S1355838201020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Conserved Domain Architecture Retrieval Tool. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubinoux J, Valente FM, Pereira IA, Costa A, Grimont PA, Le Faou AE. Reclassification of the only species of the genus Desulfomonas, Desulfomonas pigra, as Desulfovibrio piger comb. nov. Int J Syst Evol Microbiol. 2002;52:1305–1308. doi: 10.1099/ijs.0.02175-0. [DOI] [PubMed] [Google Scholar]
- Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Boguski MS, Lipman DJ, Ostell J, Ouellette BF, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 1999;27:12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]


