Abstract
Background
The gene regulatory information is hardwired in the promoter regions formed by cis-regulatory elements that bind specific transcription factors (TFs). Hence, establishing the architecture of plant promoters is fundamental to understanding gene expression. The determination of the regulatory circuits controlled by each TF and the identification of the cis-regulatory sequences for all genes have been identified as two of the goals of the Multinational Coordinated Arabidopsis thaliana Functional Genomics Project by the Multinational Arabidopsis Steering Committee (June 2002).
Results
AGRIS is an information resource of Arabidopsis promoter sequences, transcription factors and their target genes. AGRIS currently contains two databases, AtTFDB (Arabidopsis thaliana transcription factor database) and AtcisDB (Arabidopsis thaliana cis-regulatory database). AtTFDB contains information on approximately 1,400 transcription factors identified through motif searches and grouped into 34 families. AtTFDB links the sequence of the transcription factors with available mutants and, when known, with the possible genes they may regulate. AtcisDB consists of the 5' regulatory sequences of all 29,388 annotated genes with a description of the corresponding cis-regulatory elements. Users can search the databases for (i) promoter sequences, (ii) a transcription factor, (iii) a direct target genes for a specific transcription factor, or (vi) a regulatory network that consists of transcription factors and their target genes.
Conclusion
AGRIS provides the necessary software tools on Arabidopsis transcription factors and their putative binding sites on all genes to initiate the identification of transcriptional regulatory networks in the model dicotyledoneous plant Arabidopsis thaliana. AGRIS can be accessed from http://arabidopsis.med.ohio-state.edu.
Background
Control of gene expression is central to all cellular processes. Transcription factors (TFs) often function in networks, in which a regulatory protein controls the expression of another, which in turn may modulate the expression of other regulatory loci, or control genes encoding structural proteins or enzymes. These hierarchical arrangements allow specific signals to be amplified, and provide the information necessary for given sets of genes to be deployed with particular space and temporal patterns. Thus, an emerging theme in regulation of gene expression is the identification of regulatory networks in which TFs participate to specify the temporal and spatial expression of all genes in an organism. A first step in making this possible is the identification of the entire complement of TFs and the corresponding cis-regulatory elements in the promoters of all genes.
The completion of the sequencing and annotation of the entire Arabidopsis genome [1] has provided a unique opportunity to start exploring the regulatory networks present in plants. TFs are usually classified according to the presence of conserved DNA-binding domains [2]. The DNA-binding domain is responsible for recruiting TFs to specific cis-regulatory DNA sequences in the promoters of the genes they regulate. In Arabidopsis, previous studies identified about 1,500 TF genes grouped into more than 34 families [3]. Some of these families are specific to the plant kingdom or have dramatically expanded in the plants, compared to the animals (e.g. MYB and WRKY), suggesting that they may participate in regulatory networks unique to the plants. Other families are much larger in animals, as for example the homeo-domain family. Only a small fraction (~200) of the 1,500+ Arabidopsis TFs have been genetically characterized, and direct target genes for TFs (i.e. genes to which TFs bind and activate) have been identified in just a handful of those cases. Despite the general interest in investigating the function of all Arabidopsis genes by year 2010 http://www.nsf.gov/pubs/2001/nsf01162/nsf01162.html, and the knowledge on the number and classification of Arabidopsis TFs [3], no resource is currently available to obtain all the Arabidopsis TF sequences or information on their possible binding sites. While several resources are available to search for putative cis-regulatory motifs in a DNA sequence, no initiative has been taken in carrying out this process at a genome-wide level. For example, PlantCARE http://oberon.rug.ac.be:8080/PlantCARE/index.html, a cis-regulatory element database for plants, contains information about 435 different plant TF binding sites in 159 promoters. From these 435 binding sites, 281 correspond to dicots. Clearly, this information is just a fraction of the total cis-regulatory regions present in Arabidopsis.
In the quest to determine Arabidopsis TF function and establish the regulatory networks that control the expression of all Arabidopsis genes, the development of a web-based resource that divides TFs into families and allows the rapid download of any Arabidopsis TF sequence and the fast identification of mutants, linked to the possible sequences to which these TFs may bind in the Arabidopsis genome is of pressing urgency. Providing these resources and starting to link TFs to target genes is the basic motivation behind the development of AGRIS. Here we describe AGRIS, constituted by AtTFDB and AtcisDB. AtTFDB is a downloadable database of 1,375 Arabidopsis TF sequences with information on available mutations in the corresponding genes. As information on the expression patterns of TFs is generated, the information will be added to AtTFDB. The first release of AtcisDB (AtcisDB 1.0) contains an annotation of the promoter sequences for all 27,975 annotated Arabidopsis genes. AtcisDB is linked to AtTFDB when binding sites for specific transcription factors have been identified. Together, AGRIS provides a first step in establishing the regulatory networks that regulate the expression of all Arabidopsis genes.
Results and Discussion
Arabidopsis Gene Regulatory Information Server (AGRIS)
AGRIS integrates data from a variety of different sources (see Methods section). AGRIS currently consists of two integrated databases, AtTFDB and AtcisDB.
AtTFDB Database
AtTFDB consists of a collection of Arabidopsis TF sequences (protein and DNA) grouped by family. Information on the TFs can be viewed through two portals: (i) by browsing the various families to get a complete list of TF genes in that family, or (ii) by searching for a specific gene by locus ID (in AtXgXXXX format), or using any word found in the description or family name. For each gene found through the search, a description is given and links are provided to display additional search results from TIGR, MATDB, SALK, and TAIR. Nucleotide or amino acid sequences for individual TF, for entire families or for the entire collection can be directly downloaded in FASTA format. There are also links to complete alignments, Hidden Markov Models and Motifs files of each family for direct download.
A previous analysis of the complete Arabidopsis thaliana genome sequence suggested the presence of about 1500 TFs with recognizable motifs [3]. A search of the TAIR website http://www.arabidopsis.org indicated that very few TF sequences can be retrieved using a search with the key words "transcription factor" or "regulatory protein". In addition, only two TF families (MYB & WRKY) were identified and listed within the "gene family page" http://www.arabidopsis.org/info/genefamily/genefamily.html. Thus, to identify TFs, a combination of BLAST and motif searches was used based on the available literature on known TFs, or on motifs conserved among TFs from a family. A few smaller families were identified directly from published literature (i.e.CCAAT-Dr1 & E2F-DP). Many families were found through a domain search and blast technique. Publications were found through PubMed and the conserved domain motif that characterizes each TF family was identified. Using the motif, a Blast was conducted in the TAIR website, the resultant sequences were then aligned and mismatches were discarded. Another approach, especially for large families where very few TFs had been identified, was an iterative Blast approach. A few representative proteins were used to perform a Blast on the TAIR website. The resultant sequences with an E-value smaller than 10-5 were considered a good match, then the last sequence was used in a subsequent Blast, and so forth until all similar sequences were identified, then they were aligned, and any mismatched sequences were discarded. There are some discrepancies between the previously estimated number of transcription factors per families from Riechmann et al. and those listed on AGRIS. These may be explained due to further genome refinement after the December 2000 publication or because of our more stringent analysis, although we continue to add more transcription factors on a daily basis. After identifying the sequences, a script performed an automatic gathering of information from public online repositories using the list of gene names to populate the database. This database was curated semi-automatically by discarding duplicated entries, unknown genes and outliers, the last ones being identified by hand looking at the alignments of the sequences of each individual family. In addition, transcription factors that were not readily aligned with a family, but have been documented and studied were placed together in the appropriately named "Orphans Family". The results included 1375 transcription factors, which were subsequently placed into one of 34 TF families. The breakdown of transcription factors into families and the queries used for their identification is shown in Table 1.
Table 1.
Different transcription factors in AtTFDB Different transcription factor families in AtTFDB. The queries used to identify and classify TFs into different families are mentioned in column 3.
| Transcription Factor Family | Number of Members in AtTFDB | Members Identified Using |
| AB13VP1 | 18 | Identify domain and blast |
| Alfin-like | 7 | Iterative blast |
| AP2-EREBP | 118 | Identify domain and blast |
| ARF | 21 | Identify domain and blast |
| ARR-B | 12 | Identify domain and blast |
| bHLH | 147 | Iterative blast |
| bZIP | 70 | TAIR website |
| C2C2 (Zn) CO-like | 28 | Identify domain and blast |
| C2C2 (Zn) Dof | 33 | Identify domain and blast |
| C2C2 (Zn) GATA | 24 | Identify domain and blast |
| C2C2 (Zn) YABBY | 4 | Identify domain and blast |
| C2H2 | 91 | Iterative blast |
| C3H | 125 | Iterative blast |
| CCAAT-Dr1 | 2 | Identified in literature |
| CCAAT-HAP2 | 10 | Identify representative protein and blast |
| CCAAT-HAP3 | 10 | Identify representative protein and blast |
| CCAAT-HAP5 | 13 | Identify representative protein and blast |
| CPP (Zn) | 7 | Iterative blast |
| E2F-DP | 8 | Identified in literature |
| EIL | 6 | Iterative blast |
| G2-like | 41 | Identify domain and blast |
| GRAS | 24 | Identify domain and blast |
| HB | 60 | Identify domain and blast |
| HSF | 19 | Identify domain and blast |
| MADS | 98 | Iterative blast |
| MYB | 132 | TAIR website |
| MYB-related | 8 | Iterative blast |
| NAC | 89 | Identify domain and blast |
| SBP | 17 | Identify domain and blast |
| TCP | 25 | Iterative blast |
| Trihelix | 29 | Iterative blast |
| TUB | 10 | Identify representative protein and blast |
| WRKY | 69 | TAIR website |
| Orphans | 1 | Documentation |
AtcisDB Database
AtcisDB is an integrated database of promoter sequences with annotations of cis-regulatory elements. The current version of AGRIS contains only 5' promoter regions, binding sites have also been identified in 3' regions and introns, but they are limited in number and for the time being not represented. Users can search AtcisDB for a specific gene promoter or the direct target genes of a specific transcription factor. The current database version, AtcisDB 1.0, consists of promoter sequences of 29,388 annotated genes. The length (L) distribution of these promoter sequences are: 5,759 sequences have L = 500 nt, 5,322 have 500<L = 1000 nt, 6,761 have 1000<L = 2000 nt and the rest have 2000<L = 3000 nt. Nearly 4% of the annotated genes are located in head-to-head direction sharing bi-directional promoters of length less than 500 nt. The transcriptional regulation of such genes being organized as bidirectional gene pairs and what distinguishing them from other unidirectional promoter genes is mostly unclear.
One of the main features of AtcisDB is mapping of experimentally known and computationally predicted (see Methods for details) TF binding sites to their respective gene promoters, and display in graphical form. To date the website contains mainly predicted binding sites, but by August 1, 2003 the goal is to integrate experimentally proven binding sites highlighted to show their significance. In addition, a web page is being developed that will provide a complete list of transcription factor binding sites and a reference for each. While the experimentally known cis-regulatory information is rather limited, the use of high throughput micro-array expression studies in recent times have generated transcript profiles of Arabidopsis genes so as to infer the functions of known and putative TFs. Chen et al. [4] have showed that the promoter sequences of genes responsive to different treatments are enriched with known TF binding sites for those responses. They also identified a novel promoter motif in genes responding to a broad set of pathogen infection treatments. Klok et al. [5] identified genes involved in low-oxygen response to Arabidopsis root cultures by using cDNA microarray experiments, with similar sequence motifs in the promoters of genes with similar expression profiles. Similar studies [6-8] found common binding sites in the promoter regions of genes responsive to various conditions. AGRIS will be complemented by extensive searches of publicly available microarray expression data to include all such information about TFs and their regulatory motifs present in their target gene promoters.
We also provide a web page at our database, where the users of AtcisDB can report back any of the validated annotations of AtcisDB from their experiments. This facilitates the generation of a draft-annotated version of AtcisDB.
Database Queries, Visualization and Analysis Tools
A Web interface for AGRIS has been developed using the J2EE technology (JSP and Servlet). Users can search the database and retrieve the promoter sequence, and annotation information of a specific gene in several ways. For example, users can use a gene name or gene symbol to search and retrieve the promoter of a specific gene and a set of genes regulated by the same transcription factor (Web Figure 1). A typical search result for a specific promoter, using Promoter Id (same as corresponding Gene Id of TAIR), is listed in Table 2. The table has seven entries, which are Promoter Id (unique id for each promoter), Site Name (the transcription factor binding site name represented by transcription factor name), Gene Symbol and Gene Description (detailed information for gene symbol and gene description for that promoter). In order to further organize the database information, we provide two links for Promoter Id entry and Gene Symbol entry. The detailed visualization and annotation table can be obtained by clicking the link on Promoter Id entry. Users can also access the promoter gene annotation by clicking the links on Gene Symbol entry.
Figure 1.
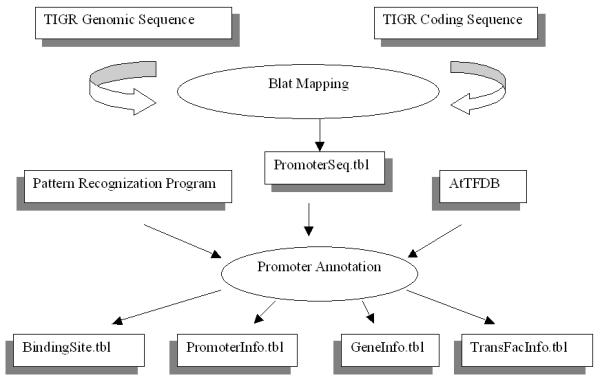
Data flow of AtcisDB The rectangular boxes with right shadow represents the data sources, the rectangular boxes with left shadow represent the final data tables, arrows represent the direction of data flow, eclipses represent computational operations implemented in data-mining pipeline.
Table 2.
A search result of AtcisDB using Promoter ID Promoter ID is specified in column1. Column 2 contains the annotated binding sites that are present in the corresponding promoter. Gene symbol and description are given in columns 3 & 4. Example search results for search engine
| Promoter Id | Site Name | Gene Symbol | Gene Description |
| At1g01010.1 | abre | NAC domain protein | similar to NAC domain protein NAM |
| At1g01010.1 | ebox | NAC domain protein | similar to NAC domain protein NAM |
| At1g01010.1 | gbox | NAC domain protein | similar to NAC domain protein NAM |
| At1g01010.1 | gccBox | NAC domain protein | similar to NAC domain protein NAM |
| At1g01010.1 | GT-elements | NAC domain protein | similar to NAC domain protein NAM |
| At1g01010.1 | MBS2 | NAC domain protein | similar to NAC domain protein NAM |
| At1g01010.1 | wbox | NAC domain protein | similar to NAC domain protein NAM |
Web Figure 2 shows a graphical organization of the regulatory regions for each promoter. We used in-house developed Genome Data Visualization Toolkit (GDVTK; Sun & Davuluri, submitted for publication; http://bioinformatics.med.ohio-state.edu/GDVTK for providing graphical view of the annotations. A line with small arrow indicates the position of the predicted or experimentally demonstrated transcription start site; (TSS). A set of small color squares represent the transcription factor binding sites, when the user move the mouse on these colored squares, a contextual menu will pop out and show the detailed information for that binding site. The user has the ability to zoom in or out of the promoter sequence and see the gene symbol for that promoter, and binding site information with relative position to TSS and their DNA sequence. The sequence of the promoter can be obtained by clicking the link on Get Promoter Sequence.
Figure 2.
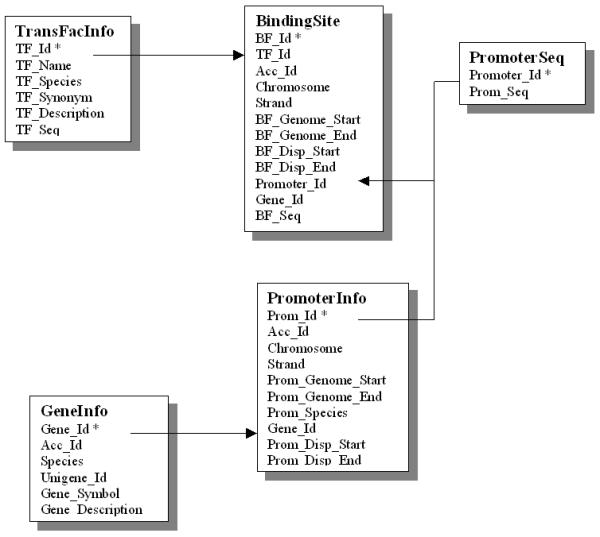
Database schema of AtcisDB Schematics diagram illustrating the interrelationship between the tables of the AtcisDB. The * represents the primary key and line with arrow link the foreign keys that associate each relational table.
Methods
Implementation of AGRIS
Transcriptional regulatory information from multiple databases are transformed and integrated into AGRIS. The main data sources of AGRIS are TAIR [[9]http://www.arabidopsis.org], PlantCARE [[10]http://oberon.rug.ac.be:8080/PlantCARE/index.html], PLACE [[11]http://www.dna.affrc.go.jp/htdocs/PLACE], TRANSFAC [[12]http://transfac.gbf.de/TRANSFAC/], and PubMed [[13]http://www.ncbi.nlm.nih.gov]. TAIR provides information about gene annotations of Arabidopsis genome sequence [1]. PlantCARE, PLACE, and TRANSFAC provide information about transcription factors and their binding sites. PubMed provides information about published literature. Initial population of the AGRIS database took place in March of 2003 using the available version of TAIR and the other support databases. Since the other public databases are not updated on a regular basis, automatic updates and synchronization with the other databases has not been enabled, instead manual updates will be used when the new versions are available.
We implemented the two databases AtTFDB and AtcisDB using MySQL http://www.mysql.com. The data flow of the AtcisDB is summarized in Figure 1, and its development is detailed in the following steps.
1. Locating Promoter Sequence on Arabidopsis Genome
We downloaded the annotated coding sequences and the chromosomal sequences from ftp://ftp.tigr.org (updated April 17, 2003). We mapped the coding sequences to the chromosomal sequences by BLAT [14]. Then, for each gene, if the upstream intergenic region is greater than 3 kb, we retrieved the sequence upstream of ATG of length 3 kb. Otherwise, we consider that intergenic region as promoter of the downstream gene, to exclude any coding region of upstream genes. We will repeat this process immediately after the release of every new annotation by TIGR so that the promoter sequences in AtcisDB are based on latest annotations.
2. Generation of a map of cis-regulatory sequences for all Arabidopsis promoters
We have collected consensus-binding sequences for known transcript factors from literature and maintain that information as a database in AtTFDB. We are providing a draft annotation of AtcisDB 1.1 by computational methods. We have developed computer programs to scan the promoter sequences in AtcisDB for transcription factor binding sites based on one of the following two approaches: (a) Scanning for consensus binding sites: AtTFDB contains the TFs and their consensus binding sites. We have developed Perl scripts to scan the promoters for consensus binding site occurrence. (b) Position Weight Matrix Methods (PWM) to scan known binding sites: We have prepared position-weight matrices [15] of those Arabidopsis transcription factors for which at least 5–10 experimentally known binding sites exist. While the number of Arabidopsis TFs with these properties is still small, it is rapidly growing. PWMs have been used to estimate the likelihood that a given sequence binds to a specific TF [16]. If a PWM exists for a TF, we follow approach (b). Otherwise approach (a) is carried out. We are refining these annotations by combining information from the AtTFDB database.
Database schema
The database schema AtcisDB and AtTFDB are shown in Figures 2 and 3. Six tables are used to store the promoter sequence and their cis-regulatory element annotation information.
Figure 3.
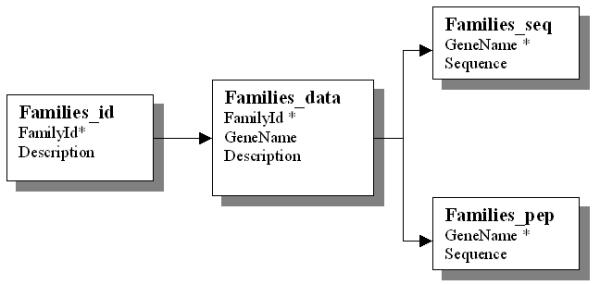
Database schema of AtTFDB Schematics diagram illustrating the interrelationship between the tables of the AtTFDB. This first step was the most time intensive and accounted for approximately 75% of the total development time.
• BindingSite table stores the transcription factor binding site's information.
• TransFacInfo table stores the data of the transcription factor information associated with binding site table.
• PromoterSeq stores the data for promoter sequence.
• GeneInfo stores the data of the gene annotation for each promoter.
• PromoterInfo stores the coordinate information of the promoter mapped on genome and relative information for promoter annotation.
Web interface
A web interface was developed to allow user interaction with the information in the databases. The web pages were developed using JavaServer Page (JSP) technology, because of its rapid development and easy maintainability qualities in development of dynamically-generated web pages and takes advantage of the java technology provided by the Apache Tomcat server http://jakarta.apache.org/tomcat. Figure 4 contains a simplified diagram revealing the interaction of the different components behind the AGRIS online resource. The result is a unified public domain online resource consisting of all the currently identified Arabidopsis TFs, with regular updates to the database as additional data is discovered.
Figure 4.
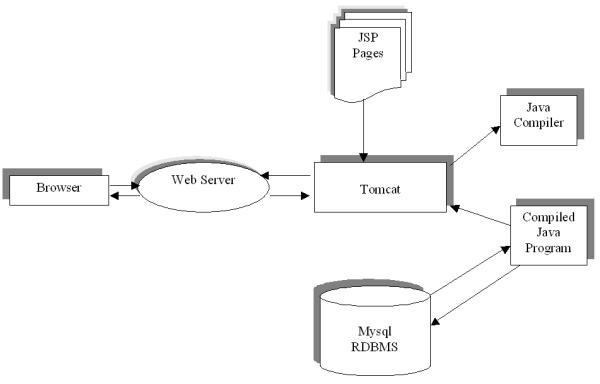
AGRIS system diagram A simplified diagram representing the interaction of different components of AGRIS.
Figure 5.
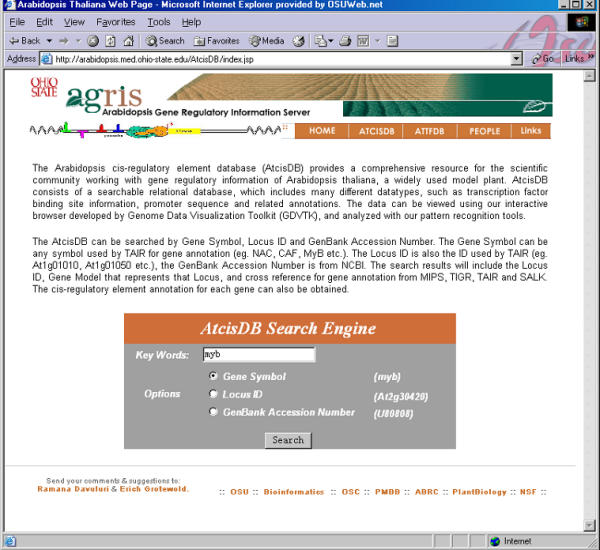
Web page for searching AtcisDB Users can use a gene name or gene symbol to search the promoter of a specific gene and a set of genes regulated by a specific transcription factor.
Figure 6.
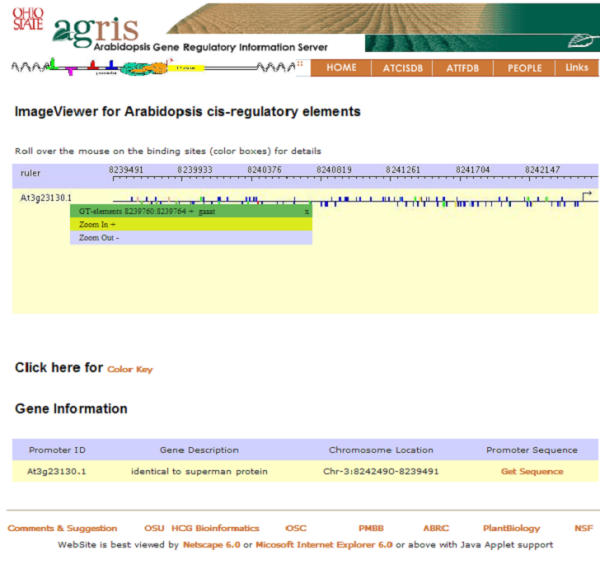
A screen shot of an Arabidopsis gene promoter with annotation of cis-regulatory elements Users can use a gene name or gene symbol to search the promoter of a specific gene and a set of genes regulated by a specific transcription factor. The graphical interface was developed using GDVTK. The position of translation start site (ATG) is indicated by a line with small arrow. The region upstream of ATG is considered as the promoter region of the corresponding gene. A set of small colored boxes represent the transcription factor binding sites. The boxes that are above the line represent the sites on positive strand and those that are below the line represent sites on negative strand. When the user moves the mouse on the colored squares, a contextual menu pops up to show the detailed information for that binding site. The sequence of the promoter can be obtained by clicking the link on Get Promoter Sequence.
Authors' contributions
HS participated in the design of AGRIS and AtcisDB database. SP performed the web implementation. NM, CM and MK collected the transcription factors data and implemented AtTFDB. RD and EG conceived of AGRIS, and participated in its design and coordination. All authors read and approved the final manuscript.
Contributor Information
Ramana V Davuluri, Email: Davuluri-1@medctr.osu.edu.
Hao Sun, Email: sun.143@osu.edu.
Saranyan K Palaniswamy, Email: Palaniswamy-1@medctr.osu.edu.
Nicole Matthews, Email: nmm1007@yahoo.com.
Carlos Molina, Email: molina.31@osu.edu.
Mike Kurtz, Email: kurtz.64@osu.edu.
Erich Grotewold, Email: grotewold.1@osu.edu.
References
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053. doi: 10.1146/annurev.biochem.61.1.1053. [DOI] [PubMed] [Google Scholar]
- Reichmann J, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineada O, Ratcliffe O, Samah R, Creelman R, Pilgrim M, Broun P, Zhang J, Ghandehari D, Sherman B, Yu G-L. Arabidopsis Transcription Factors: Genome-Wide Caomparative Analysis Among Eukaryotes. Science. 2000;209:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Chen W, Provart N, Glazebrook J, Katagiri F, Chang H, Eulgem T, Mauch F, Luan S, Zou G, Whitham S, Budworth P, Tao Y, Xie Z, Chen X, Lam S, Kreps J, Harper J, Si-Ammour A, Mauch-Mani B, Heinlein M, Kobayashi K, Hohn T, Dangl J, Wang X, Zhu T. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok E, Wilson I, Wilson D, Chapman S, Ewing R, Somerville S, Peacock W, Dolferus R, Dennis E. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell. 2002;14:2481–2494. doi: 10.1105/tpc.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong Y, Chang H, Gupta R, Wang X, Zhu T, Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner R, Hailing J, Cominelli E, Denekamp M, Fuertes A, Greco R, Kranz H, Penfield S, Petroni K, Urzainqui A, Martin C, Paz-Ares J, Smeekens S, Tonelli C, Weisshaar B, Baumann E, Klimyuk V, Jones J, Pereira A, Wisman E, Bevan M. Function search in a large transcription factor gene family in Arabidopsis: Assesing the potential of reverse genetics to identify insertional mutations in R2R3 MYB genes. Plant Cell. 1999;11:1827–1840. doi: 10.1105/tpc.11.10.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 2000;124:1525–1531. doi: 10.1104/pp.124.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S, Beavis W, Berardini T, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, Miller N, Mueller LA, Mundodi S, Reiser L, Tacklind J, Weems D, Wu Y, Xu I, Yoo D, Yoon J, Zhang P. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D, Church D, Federhen S, Lash A, Madden T, Pontius J, Schuler G, Schriml L, Sequeira E, Tatusova T, Wagner L. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. BLAT – the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. 10.1101/gr.229202. Article published online before March 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- Frech K, Herrmann G, Werner T. Computer-assisted prediction, classification, and delimitation of protein binding sites in nucleic acids. Nucleic Acids Res. 1993;21:1655–1564. doi: 10.1093/nar/21.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]


