Abstract
Vertebrate visual pigment proteins contain a conserved carboxylic acid residue in the third transmembrane helix. In rhodopsin, Glu113 serves as a counterion to the positively charged protonated Schiff base formed by 11-cis retinal attached to Lys296. Activation involves breaking of this ion pair. In UV cone pigments, the retinyl Schiff base is unprotonated, and hence such a salt bridge is not present; yet the pigment is inactive in the dark. Mutation of Glu108, which corresponds to rhodopsin’s Glu113, to Gln yields a pigment that remains inactive in the dark. The apoproteins of both the wild-type and mutant, however, are constitutively active with the mutant being of significantly higher activity. Thus one important role for preserving the negatively charged glutamate in the third helix of UV pigments is to maintain a less active opsin in a manner similar to rhodopsin. Ligand binding itself in the absence of a salt bridge is sufficient for deactivation.
Keywords: cone pigment, constitive activation, rhodopsin
List of abbreviations: CHAPS, 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate; SWS1, short-wavelength sensitive class II visual pigment; Meta II, Metarhodopsin II intermediate
1. Introduction
Visual pigment proteins fall into the superfamily of cell surface receptors known as G protein-coupled receptors. Rhodopsin, the photopigment in rods, is one of the best-studied members. The cone pigments are highly homologous to rhodopsin. Unlike other G protein-coupled receptors, the rod and cone pigments have their ligand, 11-cis retinal, bound covalently to the protein via a Schiff base linkage to the ɛ–amino group of the strictly conserved lysine in the seventh transmembrane helix. In rhodopsin and most cone pigments studied, the Schiff base is protonated resulting in a positive charge in the middle of the transmembrane domain (e.g., see review [1]). Vertebrate visual pigments contain a carboxylate (Glu in all cases except one, an Asp [2]) in the third helix. In rhodopsin, and presumably the cone pigments too, Glu113 serves as the counterion to the positively charged protonated Schiff base [3,4]. This ion pair has been implicated in two important properties of visual pigments: spectral tuning, and inactivation. The distance of the counterion from the Schiff base affects delocalization/localization of the positive charge along the polyene chain of retinal, and hence absorption maximum [5]. The ion pair has also been implicated as being crucial to the maintenance of the relatively inactive conformation of both the apoprotein and pigment of rhodopsin [6,7].
Recent studies on UV cone pigments, members of the short wavelength sensitive class I (SWS1) of visual pigments [8,9], have come to the conclusion that 11-cis retinal is bound to its opsin, not via a protonated Schiff base linkage, but via an unprotonated Schiff base linkage [10–12]. One advantage for having the ligand bound as an unprotonated Schiff base is that the absorption maximum of an unprotonated retinyl Schiff base is already in the near UV region at about 360 nm. Thus the protein would not have to modify the absorption properties of the chromophore much to tune the pigment to its desired wavelength.
Perhaps surprisingly, the carboxylate in the third helix counterion position is also present in the UV cone pigment. What role would it have if there is no salt bridge between the carboxylate and the Schiff base? Kusnetzow et al. [11] concluded that one role would be to serve as the counterion to the photoproducts leading up to the active Meta II state of a UV pigment for the proper timing of activation and subsequent deactivation. In this study, I have removed the counterion in the salamander UV pigment by introducing an E108Q (equivalent to E113Q in bovine rhodopsin) mutation in the salamander UV cone pigment. The apoprotein is constitutively active, while the pigment is inactive in the dark. Thus another role for conserving the counterion carboxylate residue is to maintain a relatively inactive opsin in the absence of its inverse agonist 11-cis retinal, similar to its role in rhodopsin.
2. Materials and Methods
2.1. Expression and purification of visual pigments
Cloning of the tiger salamander UV pigment cDNA, modification to include the anti-rhodopsin 1D4 epitope [13], and insertion into the pMT3 expression vector [14] have been described [15].
The wild-type and mutant plasmids were transiently tranfected into COS cells using DEAE-dextran essentially as described previously [16] except the dimethyl sulfoxide treatment and final rinse were omitted. Typically, 20 10-cm plates were transfected for each reconstitution. Cells were harvested on the third day after transfection and pigments were immunopurified as described earlier [15] except the buffer used throughout the purification process was 10 mM sodium phosphate, 150 mM sodium chloride, pH 7.0.
COS cell membrane preparations containing wild-type and mutant salamander UV opsins were performed essentially as described by Robinson [17] except a Beckman SW 32 rotor was used throughout and the final pellet was resuspended in 200 μL of the buffer and stored in 25 μL aliquots. Concentrations were determined in triplicate by densitometric analysis from slot blots of the wild-type and mutant samples compared to known amounts of rhodopsin from bovine rod outer segments using the BioRad BioDot set-up. Samples were solubilized in 1% CHAPS in a Tris buffered saline buffer containing 25 mM N-ethyl maleimide for 1 h on ice. The samples and rod outer segment concentration standards were then centrifuged and blotted onto a nitrocellulose membrane in the slot blot apparatus; wells were washed twice with 0.5 mL Tris buffered saline. The blot was probed with the anti-rhodopsin 1D4 antibody, and the enhanced chemiluminescence was detected and analyzed using a BioRad Chemidoc XRS imaging system and its associated software, Quantity One.
2.2. UV-visible spectrophotometry
Absorption spectra were recorded on a Varian Cary300 spectrophotometer modified by the manufacturer for darkroom use. All spectra were recorded using a 50 μL ultramicrocuvette with a 1 cm path length containing 100 μL sample.
2.3. Transducin activation assay
The ability of the opsin and pigments to activate bovine rod transducin using a radioactive filter binding assay was measured essentially as described previously [18] except the pH of the reaction was 6.5 using 10 mM 2-[N-morpholino]-ethanesulfonic acid (MES) buffer instead of Tris. Light activation was initiated with a 6 s pulse of white light from a slide projector with a 300 watt lamp. Activity was converted to specific activity by normalizing to the pigment content in the assay for the purified pigment or the calculated opsin concentration for the membrane preparations.
3. Results
We have previously reported the expression and purification of the salamander UV cone pigment [15]. The E108Q mutant remains a UV pigment (Figure 1). This result is consistent with previous reports of the same mutation in the mouse UV cone pigment [10–12,19]. The absorption maximum of the salamander UV pigment E108Q mutant is blue shifted by about 8 nm to 348 nm. As with the wild type pigment, acid denaturation results in a red-shifted spectrum consistent with the spectrum for a protonated retinyl Schiff base in the absence of the protein environment. Normalizing the wild-type and E108Q spectra to the acid denatured spectra, it is evident that the extinction coefficient for the mutant pigment is slightly lower than for the wild-type (Figure 1). In agreement with data for mouse UV wild-type and E108Q mutants [12], attempts to protonate the Schiff base in a folded protein by lowering the pH was not possible as the protein appears to denature first (not shown). Babu et al. [19] reported a partially titrateable Schiff base with counterion mutants in the Xenopus violet cone pigment, which is in the same SWS1 class of visual pigments as the UV pigments. Fasick et al. [10] were able to titrate a mouse UV E108Q mutant pigment but only in a F81Y mutant background. The Schiff base of the F81Y mutant pigment is protonated at pH 6, and the additional E108Q mutation lowered the pK to about 6 [10].
Figure 1.
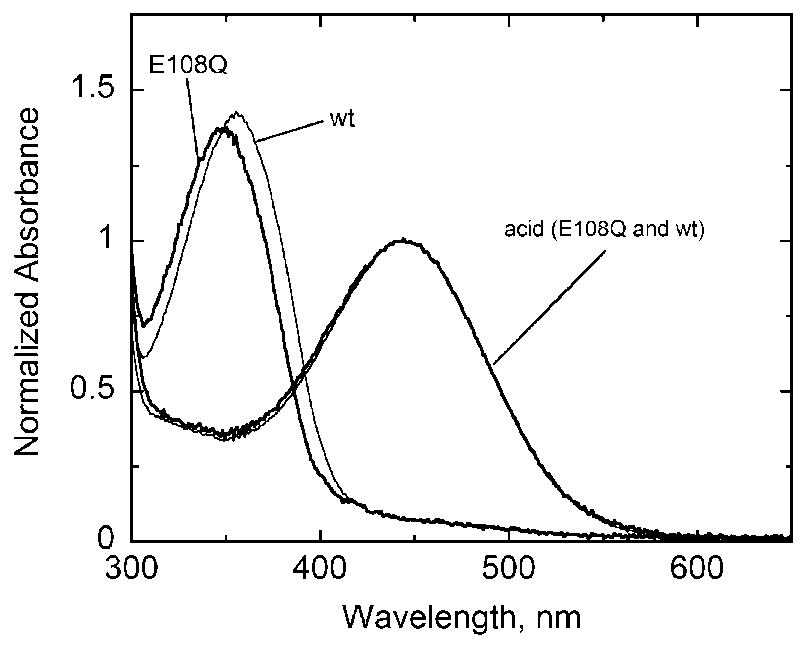
Absorption spectra of wild-type (thin black lines) and E108Q mutant (thick black lines) salamander UV cone pigment in the dark and acid-denatured forms. Spectra were normalized to the acid-denatured species.
Both the purified E108Q and wild-type UV cone pigments are not active in the dark but are able to activate transducin in a light-dependent manner (Figure 2) in agreement with previous transducin activation assays with UV cone pigments [10,11,15,19]. Activation by the wild-type pigment reaches a plateau quickly due to rapid decay of the active Meta II species in SWS1 pigments [11,15,20,21], and consequently there is no further accumulation of activated transducin. The initial rate of light-dependent activation by the wild-type pigment is about 100 min−1 per mol pigment and 45 min−1 per mol pigment for the E108Q mutant as determined using methods described previously [21]. Transducin activation by the mutant rises in a monophasic manner reflecting the decay of the Meta II intermediate. Time constants for the decay of Meta II calculated as described previously [21] are 0.4 and 1.6 min for the wild-type and E108Q mutant, respectively. This result is consistent with the longer-lived active Meta II intermediate of the counterion mutant shown in previous studies with bovine rhodopsin [4] and mouse UV cone pigment [11]. However, the relative activities here are opposite of those reported for the wild-type and E108Q mutant of the mouse UV pigment where the mutant displayed a much higher ability to activate transducin than the wild-type [11]. This difference is possibly due to differences in assay and analysis conditions. Measurements by Kusnetzow et al. [11] were conducted at a higher pH than my assays, and the kinetic analysis of the light-driven activation began after a 1 min illumination period during which a significant amount of transducin would have been activated and wild-type Meta II undergone at least two half-times. Here all of the kinetic analyses for light-driven activation begin with a much shorter 6 s illumination.
Figure 2.
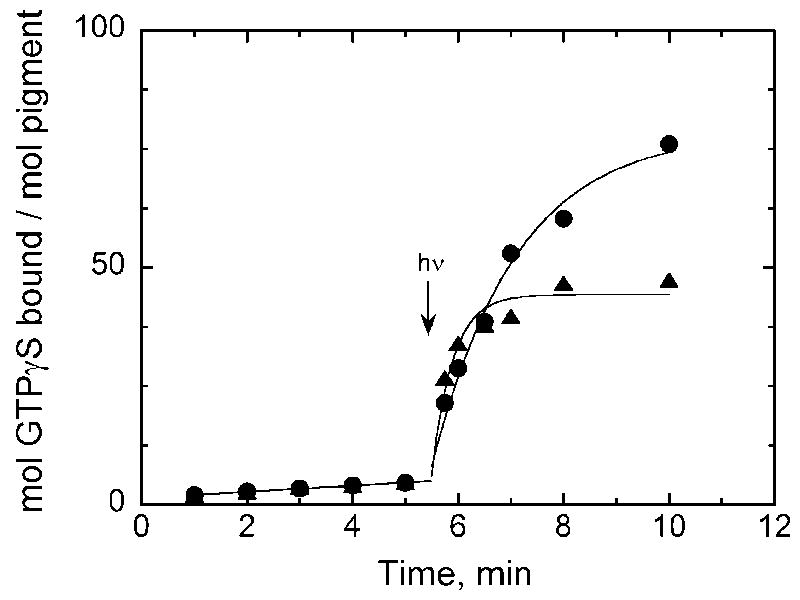
Transducin activation by purified wild-type (triangles) and E108Q mutant (circles) salamander UV cone pigment. Both pigments are able to activate transducin in a light-dependent manner. Note the identical lack of activation by the pigments in the dark. The reaction mixtures both contained 10 nM pigment as determined from the acid-denatured spectrum of the samples from Figure 1.
Figure 3 shows that both the E108Q mutant and wild-type opsins are constitutively active. As with rhodopsin, 11-cis retinal acts as an inverse agonist by binding to and inactivating the protein. Unlike the assays shown in Figure 2, the opsins were not detergent solubilized and immunopurified. As summarized by Robinson [17], opsins are not stable in dodecyl maltoside micelles and need a membrane environment in order to remain viable. Therefore, preparations from a discontinuous sucrose gradient of COS cell membranes as described by Robinson [17] were used. Activation by the protein with and without ligand could be directly compared since aliquots from the same preparation were equally divided and then 11-cis retinal (240 μM final concentration) in ethanol or the same volume of ethanol without retinal was added. The presence of 11-cis retinal is sufficient to quench the ability of the opsins to activate transducin in the dark. Quenching is due to pigment formation because light-dependent activity can be elicited. Similar to rhodopsin [6], the counterion mutant opsin is more constitutively active than the wild-type opsin, about 2-fold for the UV pigment (Figure 3).
Figure 3.
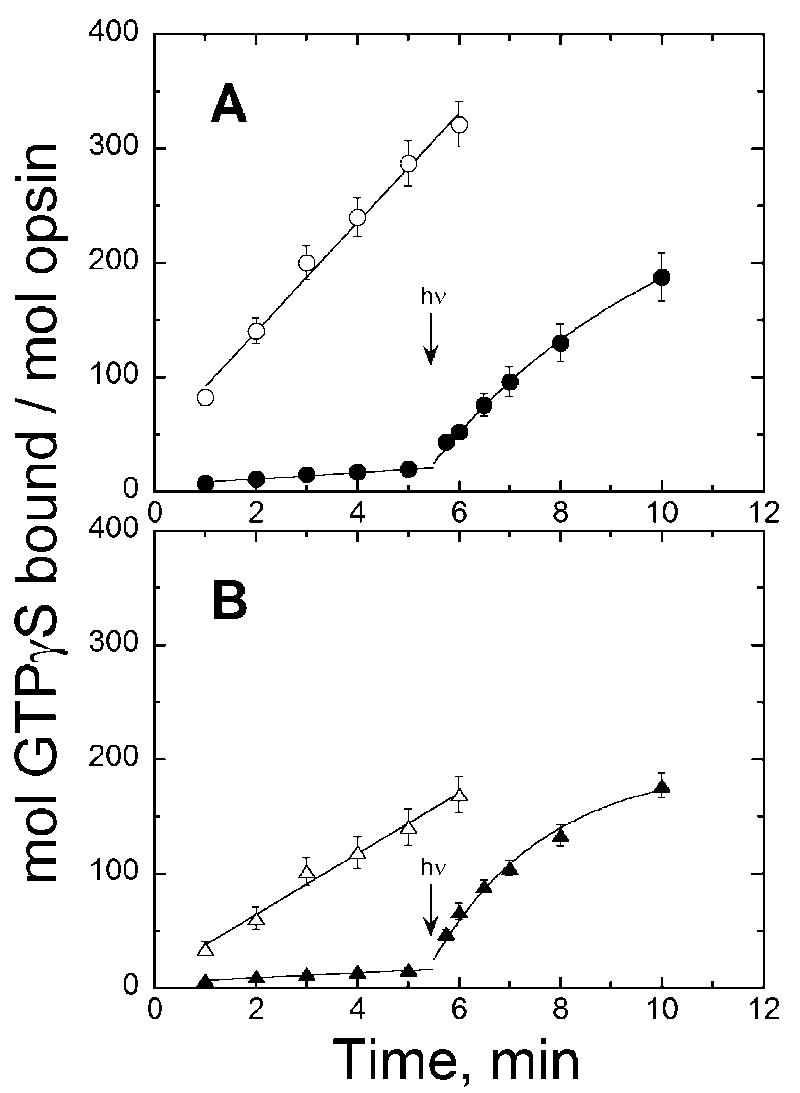
Transducin activation by (A) E108Q mutant and (B) wild-type salamander UV cone opsins (open symbols) and pigments (filled symbols). To the opsin sample, 1 μL ethanol was added; to the pigment sample, 1 μL 11-cis retinal in ethanol was added to a final concentration of 240 μM and incubated at least 1 h. Error bars reflect standard deviation of the means for both activity and opsin concentration.
Figure 3 also shows that the membrane environment prolongs the lifetime of Meta II in both the wild-type and mutant. The Meta II decay time constants were determined to be 2 and 5 min for the wild-type and mutant, respectively, using methods described previously [21]. In order to discount the possibility of continued activation by the membrane-embedded opsin after Meta II decay or photoexcitation of regenerated pigment, the transducin activation assay was conducted under dim red light conditions throughout (except for the 6 s pulse of white light) and in the presence of a large excess of 11-cis retinal which would immediately deactivate any opsin as it forms. Again, the initial activation rate by the wild-type (~ 70 min−1 per mol opsin) is higher than by the mutant (~50 min−1 per mol opsin) although less dramatic than by the detergent solubilized pigments. The slower Meta II decay of the wild-type in membranes could account for a better measure of initial activation. Nevertheless, activation by the apoprotein does occur and is greater by the mutant opsin than by the wild-type as reflected by both a comparison to each other and relative to its light-dependent activation rate where the mutant opsin is able to activate transducin with essentially the same efficiency as the light activated species; whereas, the wild-type opsin activates transducin about 40% that of the light-activated species.
Beta ionone, essentially a truncated retinoid analog, has been shown to increase the ability of rod opsin to activate transducin without covalently binding to the protein [22]. In other words, beta ionone acts as a weak agonist. Similarly, addition of beta ionone to membranes containing wild-type UV opsin stimulates its ability to activate transducin 1.3-fold over opsin alone; however, it has no effect on the mutant opsin’s activity (data not shown). This result is consistent with the wild-type opsin not being in a fully active conformation as opposed to the E108Q mutant opsin.
4. Discussion
Although there are several examples of rhodopsin mutations that are constitutively active (e.g., review by Rao and Oprian [7]), reports of constitutively active cone pigments are quite unusual. The only other example is our description of a triple mutant of the same pigment that is also dark active [18]. The E108Q opsin, however, can be deactivated by binding 11-cis retinal in the dark.
That the apoprotein of the E108Q mutant is constitutively active (active independent of ligand and light) suggests that a reason for conserving the glutamate is to maintain a less active opsin by forming a salt bridge with the Lys291 (equivalent to rhodopsin’s Lys296) much like with the rhodopsin apoprotein. This behavior is like that of the inactive E113Q rhodopsin mutant pigment containing an unprotonated Schiff base [4] although the wild-type UV opsin is quite a bit more active than wild-type rod opsin. The lack of an effect by beta ionone on the E108Q mutant further suggests that the absence of the counterion in opsin results in a fully active conformation of the protein not seen in the wild-type opsin.
Another reason for this study is to address the role of a salt bridge (or lack of one) in the visual pigment because of the somewhat confusing literature about its necessity for maintaining an inactive pigment and reports that suggest otherwise. For example, breaking such a salt bridge in the holoprotein was postulated as the underlying basis for dark activity of photoreceptor cells [23,24]. This notion has been difficult to demonstrate. Jin et al. [25] using transgenic Xenopus rods containing E113Q rhodopsin showed that the rods were desensitized consistent with persistent activation of the signaling cascade. However, their data suggested that this was due to constitutive activity of the apoprotein, not dark activity of the pigment, since addition of 11-cis retinal resensitized the rod suggesting that ligand binding itself was sufficient to inactivate the protein independent of the salt-bridge. These results are consistent with earlier in vitro data on inactivation of the E113Q mutant after binding 11-cis retinal [4,6]. Recently, Kim et al. [26] concluded that breaking the salt bridge itself is not sufficient to form an active pigment, but further conformational changes induced by chromophore isomerization or additional mutations were needed. Similarly, attempts by Sampath and Baylor [27] to quench the dark activity of red cones by changing the pH further inferred that the dark noise does not correlate with deprotonation of the Schiff base although it should be noted that there was no direct demonstration of changes in Schiff base protonation states.
Thus the salt bridge between a carboxylate in helix 3 and protonated Schiff base is not necessary to hold a visual pigment in an inactive conformation. Binding of the inverse agonist 11-cis retinal is sufficient. However, the conserved carboxylate is important in keeping the apoprotein from being overly active and stimulating unwanted visual signaling in both rod opsin and UV cone opsin.
Acknowledgments
I would like to thank Margaret Lewis and Patrice Goletz for technical help; Rosalie Crouch and the National Eye Institute for the 11-cis retinal; and Yiannis Koutalos and Rosalie Crouch for helpful discussions. This work was funded by NIH grant R01-EY013748 (MK). This work was also partially supported from an unrestricted grant to the Storm Eye Institute (MUSC) from Research to Prevent Blindness and NIH grant R24-EY014793 (Dept. of Ophthalmology).
References
- 1.Kochendoerfer GG, Lin SW, Sakmar TP, Mathies RA. How color visual pigments are tuned. Trends Biochem Sci. 1999;24:300–305. doi: 10.1016/s0968-0004(99)01432-2. [DOI] [PubMed] [Google Scholar]
- 2.Starace DM, Knox BE. Cloning and expression of a Xenopus short wavelength cone pigment. Exp Eye Res. 1998;67:209–220. doi: 10.1006/exer.1998.0507. [DOI] [PubMed] [Google Scholar]
- 3.Zhukovsky EA, Oprian DD. Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science. 1989;246:928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 4.Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci, USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honig B, Ebrey TG. Protein-chromophore interactions as spectroscopic and photochemical determinants. Methods Enzymol. 1982;88:462–470. [Google Scholar]
- 6.Robinson PR, Cohen GB, Zhukovsky EA, Oprian DD. Constitutively active mutants of rhodopsin. Neuron. 1992;9:719–725. doi: 10.1016/0896-6273(92)90034-b. [DOI] [PubMed] [Google Scholar]
- 7.Rao VR, Oprian DD. Activating mutations of rhodopsin and other G protein-coupled receptors. Annu Rev Biophys Biomol Struct. 1996;25:287–314. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]
- 8.Ebrey T, Koutalos Y. Vertebrate photoreceptors. Prog Retinal Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- 9.Yokoyama S. Molecular evolution of vertebrate visual pigments. Prog Retinal Eye Res. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- 10.Fasick JI, Applebury ML, Oprian DD. Spectral tuning in the mammalian short-wavelength sensitive cone pigments. Biochemistry. 2002;41:6860–6865. doi: 10.1021/bi0200413. [DOI] [PubMed] [Google Scholar]
- 11.Kusnetzow AK, Dukkipati A, Babu KR, Ramos L, Knox BE, Birge RR. Vertebrate ultraviolet visual pigments: Protonation of the retinylidene Schiff base and a counterion switch during photoactivation. Proc Natl Acad Sci, USA. 2004;101:941–946. doi: 10.1073/pnas.0305206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Radlwimmer FB, Yokoyama S. Molecular genetics and the evolution of ultraviolet vision in vertebrates. Proc Natl Acad Sci, USA. 2001;98:11731–11736. doi: 10.1073/pnas.201257398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molday RS, MacKenzie D. Monoclonal antibodies to rhodopsin: Characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 14.Franke RR, Sakmar TP, Oprian DD, Khorana HG. A single amino acid substitution in rhodopsin (Lysine 248 --> Leucine) prevents activation of transducin. J Biol Chem. 1988;263:2119–2122. [PubMed] [Google Scholar]
- 15.Ma JX, Kono M, Xu L, Das J, Ryan JC, Hazard ES, III, Oprian DD, Crouch RK. Salamander UV cone pigment: Sequence, expression, and spectral properties. Vis Neurosci. 2001;18:393–399. doi: 10.1017/s0952523801183057. [DOI] [PubMed] [Google Scholar]
- 16.Oprian DD. Expression of opsin genes in COS cells. Methods Neurosci. 1993;15:301–306. [Google Scholar]
- 17.Robinson PR. Assays for the detection of constitutively active opsins. Methods Enzymol. 2000;315:207–218. doi: 10.1016/s0076-6879(00)15845-8. [DOI] [PubMed] [Google Scholar]
- 18.Kono M, Crouch RK, Oprian DD. A dark and constitutively active mutant of the tiger salamander UV pigment. Biochemistry. 2005;44:799–804. doi: 10.1021/bi047898f. [DOI] [PubMed] [Google Scholar]
- 19.Babu KR, Dukkipati A, Birge RR, Knox BE. Regulation of phototransduction in short-wavelength cone visual pigments via the retinylidene Schiff base counterion. Biochemistry. 2001;40:13760–13766. doi: 10.1021/bi015584b. [DOI] [PubMed] [Google Scholar]
- 20.Starace DM, Knox BE. Activation of transducin by a Xenopus short wavelength visual pigment. J Biol Chem. 1997;272:1095–1100. doi: 10.1074/jbc.272.2.1095. [DOI] [PubMed] [Google Scholar]
- 21.Das J, Crouch RK, Ma J-x, Oprian DD, Kono M. Role of the 9-methyl group of retinal in cone visual pigments. Biochemistry. 2004;43:5532–5538. doi: 10.1021/bi036097u. [DOI] [PubMed] [Google Scholar]
- 22.Palczewski K, Jäger S, Buczylko J, Crouch RK, Bredberg DL, Hofmann KP, Asson-Bartres MA, Saari JC. Rod outer segment retinol dehydrogenase: substrate specificity and role in phototransduction. Biochemistry. 1994;33:13741–13750. doi: 10.1021/bi00250a027. [DOI] [PubMed] [Google Scholar]
- 23.Barlow RB, Birge RR, Kaplan E, Tallent JR. On the molecular origin of photoreceptor noise. Nature. 1993;366:64–66. doi: 10.1038/366064a0. [DOI] [PubMed] [Google Scholar]
- 24.Birge RR, Barlow RB. On the molecular origins of thermal noise in vertebrate and invertebrate photoreceptors. Biophys Chem. 1995;55:115–126. doi: 10.1016/0301-4622(94)00145-a. [DOI] [PubMed] [Google Scholar]
- 25.Jin S, Cornwall MC, Oprian DD. Opsin activation as a cause of congenital night blindness. Nature Neurosci. 2003;6:731–735. doi: 10.1038/nn1070. [DOI] [PubMed] [Google Scholar]
- 26.Kim JM, Altenbach C, Kono M, Oprian DD, Hubbell WL, Khorana HG. Structural origins of constitutive activation in rhodopsin: Role of the K296/E113 salt bridge. Proc Natl Acad Sci, USA. 2004;101:12508–12513. doi: 10.1073/pnas.0404519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampath AP, Baylor DA. Molecular mechanism of spontaneous pigment activation in retinal cones. Biophys J. 2002;83:184–193. doi: 10.1016/S0006-3495(02)75160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


