The molecular analysis would be fragmented without the rich information on the ecology of UV vision.
From the detection of a changing traffic light to the relaxing sight of Vincent van Gogh's ”Sunflower,” color vision plays an important role in our lives. But, to the eyes of a hummingbird or goldfish, we humans are partially color-blind, as they can see what is known as UV, to which we are simply blind. How did we lose this sensitivity, or, alternatively, how did they acquire it? In this issue of PNAS, Shi and Yokoyama (1) use techniques of paleomolecular biology to unravel the mystery of the evolution of UV vision in vertebrates.
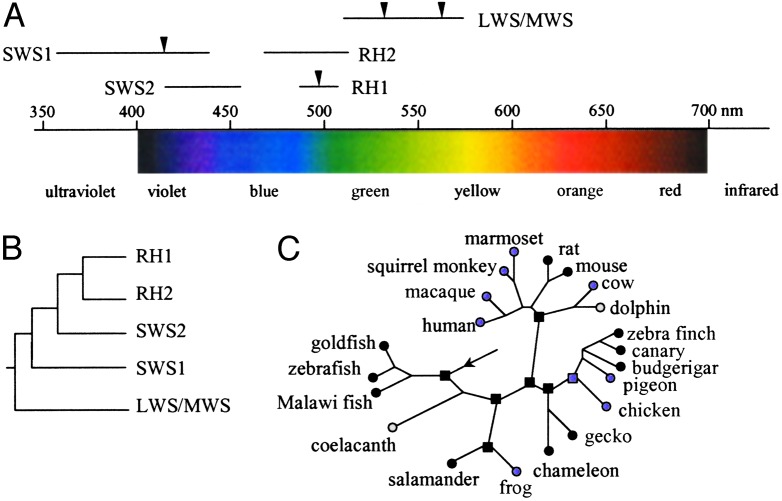
Vision starts when visual pigments are activated by the absorption of photons. A visual pigment is composed of a protein called opsin and a chromophore. The light sensitivity of a visual pigment is determined by the interaction between opsin and chromophore and is tuned to a particular wavelength of maximum absorption called λmax. Humans have four different visual pigments with the respective λmax being 414, 497, 530, and 560 nm. Thus, lights in the spectrum of ≈400–700 nm are visible, whereas those <400 nm (UV) or >700 nm (IR) are invisible to us (Fig. 1A).
Fig. 1.
Evolution of vertebrate visual pigments. (A) Variation in λmax of the five groups of visual pigments across vertebrates (2, 33). The arrows show the λmax for the human visual pigments. (B) Phylogenetic relationships of the five groups of visual pigment genes (2, 33). (C) Evolution of the vertebrate UV vision. Circles indicate extant organisms, and squares indicate ancestral species. Black filled symbols indicate UV vision, purple filled symbols indicate violet vision, and open symbols indicate nonfunctional SWS1 genes. The arrow shows the root of the tree and the common ancestor of vertebrates. The phylogeny follows (1, 11, 34). The UV/violet vision of the ancestral species was inferred by Shi and Yokoyama (1).
Because most vertebrates use only one type of chromophore (11-cis-retinal), spectral tuning is largely determined by the sequence and function of opsins. Vertebrate opsins may be classified into five groups according to their phylogenetic relationships: SWS1, SWS2, RH1, RH2, and LWS/MWS (Fig. 1B). Some species (e.g., chicken and goldfish) have all five types of opsins, but many have only three or four types. Evolutionary studies suggest the presence of five opsins belonging to each of the five groups in the common ancestor of vertebrates, but gene duplication and deletion led to gains and losses of certain types of opsin genes in various species (2). For example, the four opsins of humans belong to SWS1, RH1, and LWS/MWS groups; our ancestors lost SWS2 and RH2 genes but gained an additional LWS/MWS gene (Fig. 1 A). Variation across species extends beyond the presence or absence of a particular type of opsin, because, even for a given type, λmax can vary substantially among species. For instance, the λmax of LWS/ MWS opsins ranges from 508 nm in rat to 571 nm in chicken. Fig. 1 A illustrates large overlaps of the ranges of λmax among the five types of opsins. An interesting observation is that only SWS1 opsins may sense UV light. Thus, the evolution of UV vision can be studied by examining SWS1 opsins.
Because we humans cannot see UV, it is difficult for us to appreciate its importance. But UV plays critical roles in mate selection, foraging, and communication in species that see UV. Extensive phylogenetic surveys showed that UV vision is present in some, but not all, species of fishes, amphibians, reptiles, birds, and mammals (ref. 3; see Fig. 1C), which raised three interrelated questions. First, did the common ancestor of all vertebrates have UV vision? Second, how was the UV vision gained and lost in various organisms? Third, does the presence or absence of UV vision have adaptive values to organisms? Shi and Yokoyama (1) address all three questions.
The first question is not unfamiliar to evolutionary biologists, as they frequently infer morphological, physiological, and behavioral character states of ancestral organisms from those of extant ones, usually by the principle of parsimony. But the accuracy of such inferences is often unknown or low, even with improved methodologies (4). This uncertainty is in part caused by the complexity of the genetic basis of most morphological, physiological, and behavioral traits and a lack of knowledge of their evolutionary mechanisms. At the protein or DNA sequence level, the evolutionary mechanism is much simpler, and a substantial knowledge has been gained in the last two decades (5, 6). Indeed, computer simulation showed that inference of ancestral protein or DNA sequences from extant sequences can be quite reliable under certain circumstances (7). Thus, instead of directly inferring the λmax of the SWS1 opsin of the common ancestor of vertebrates from the λmax values of extant opsins, Shi and Yokoyama (1) first computationally inferred the protein sequence of the ancestral SWS1 opsin. They then used the recombinant DNA techniques and site-directed mutagenesis to make this ancestral opsin in the laboratory, incubated the opsin with chromophore, and measured the λmax of the regenerated visual pigment. The result showed that the visual pigment has a λmax of 361 nm, suggesting that the vertebrate ancestor could sense UV (Fig. 1C).
This conclusion is not without caveats. It is known that statistical inference of ancestral protein sequences can contain errors. Shi and Yokoyama used the Bayesian method (8) in their inference, which computes the posterior probability of every possible amino acid at each position of the ancestral protein and chooses the one with the highest probability. The computation is based on a number of assumptions about the amino acid substitution rate and pattern. When the assumptions are satisfied, the calculated posterior probabilities are unbiased, reflecting the reliability of the inference (7, 8). However, because the true substitution rate and pattern at any given amino acid position is poorly known, it is better to try more than one substitution model. In their work, Shi and Yokoyama used two empirically derived models (JTT and Dayhoff) and identified 11 amino acid positions where the two models gave different inferences of the ancestral amino acids. In addition, the inferences at several other positions have posterior probabilities <0.9, most of which, nevertheless, are located in functionally less important N- and C-terminal regions of the protein. To evaluate the robustness of the result on the ancestral λmax, the authors made ancestral proteins with the alternative inferences. Through meticulous examinations, they conclude that the ancestral opsin was UV-sensitive. The presence of a UV-sensitive opsin, however, is not equivalent to a color capacity of the organism in that part of the spectrum, because the opsin may detect the light, but its output may be wired into the brain in a manner that would not allow color vision (3). Statement of color vision requires direct behavioral corroboration, which is lacking for the ancestral vertebrates. Nonetheless, because UV vision is mediated by SWS1 opsins and is not fundamentally different from blue or violet vision, it is unlikely that a novel neural processing channel is required.
After the first question is answered, the second is relatively easy. Using the same approach, the authors reconstructed the ancestral proteins at six other interior nodes of the vertebrate tree, representing the respective ancestors of tetrapods, amphibians, amniotes, saurians, birds, and mammals (Fig. 1C). For all of these nodes except that of the common ancestor of birds, the reconstructed opsins have a λmax of ≈360 nm, suggesting that they were UV-sensitive as well. For the avian ancestor, however, λmax was 393 nm, which is at the boundary of UV and violet and is referred to as violet by Shi and Yokoyama. From these results and the phylogenetic distribution of UV and violet vision across vertebrates, one can see that violet vision originated at least four times, in frogs, birds, primates, and cetartiodactyls (Fig. 1C). In most extant vertebrates, UV vision was directly inherited from the vertebrate ancestor, but in budgerigar, zebra finch, and canary it was restored from violet vision secondarily. Based on this and several other studies (9–11), Shi and Yokoyama were also able to identify amino acid substitutions that affect the λmax of SWS1 opsins. If such information is accurate and complete, it would be possible to predict the λmax of a SWS1 opsin simply from its sequence. Using this strategy, Odeen and Hastad (12) recently obtained the SWS1 opsin sequences of 45 birds and predicted that UV vision was regained at least four times in birds. This finding suggests that it is relatively easy to change back and forth between UV and violet vision. Indeed, although the entire opsin sequence has ≈300 aa, only six positions are involved in such changes in birds. Substitutions at a few positions repeatedly occurred in several vertebrate lineages independently. This pattern of parallel substitution provides strong evidence for the action of positive Darwinian selection on the spectral tuning of SWS1 (13).
It also leads to the third question raised earlier, which is about the utility and adaptive value of UV vision. Shi and Yokoyama make a convincing case that the presence of UV vision is associated strongly with the availability of UV light in the environment and with the UV-dependent behaviors of the animals (1). One striking example is the pseudogenization of the SWS1 gene in coelacanth, the so-called ”living fossil” that lives at depths ≈200 m in the ocean, where short-wavelength lights such as UV cannot reach (Fig. 1C). Presumably for a similar reason, the dolphin also lost the gene. Although UV vision is important to many organisms, UV light may also damage retinal tissues. In humans and many other species, screening pigments in lenses or corneas effectively obviate most UV light so our retinas are protected (3). It may be because of this and other reasons that humans, cow, chicken, and many terrestrial vertebrates do not possess UV vision (1).
The powerful strategy of reconstructing ancestral genes and proteins used by Shi and Yokoyama was envisioned 40 years ago by Pauling and Zuckerkandl (14) and was first realized in the mid-1990s when technological advances made such experiments feasible. To date, only a few people have used it in molecular evolutionary studies (15–20). Nevertheless, the limited examples have already yielded interesting and significant evolutionary insights that otherwise would be difficult to gain. For instance, this method helped to discover complementary advantageous substitutions in protein evolution (19), show how a digestive protein evolved along with the emergence of foregut fermentation in ruminants (16), and demonstrate what made an active transposon silent (18). Laboratory reconstruction of ancestral genes by site-directed mutagenesis is tedious work. This is particularly so when the ancestral protein has a large number of amino acid differences from extant ones. Under certain conditions, one may reconstruct pseudoancestral proteins by mutating only those sites that are known or highly suspected to be functionally relevant. Several studies of this kind have been conducted, and they yielded biologically significant results (21, 22). In molecular evolution literature, many papers end with untested speculations or hypotheses on the functional significance of particular nucleotide changes in evolution, and the approach of paleomolecular biology is highly useful in addressing such questions.
Last, but not least, Shi and Yokoyama's paper exemplifies the beauty of integrative biology. The molecular analysis they performed would be fragmented or irrelevant without the rich information they assembled on the ecology of UV vision, and the rich ecology of UV vision would still not be satisfactory if a clear understanding of the underlying genetic basis is not obtained. Recent years have seen the emergence of research on the molecular genetic basis of organismal evolution, diversity, and adaptation (23–32). Such studies often integrate molecular biology, evolution, paleontology, ecology, organismal biology, physiology, and other fields. I expect that a golden age of integrative biology will arrive with the enlistment of molecular evolutionary genetics, and our understanding of the living world will then become more vivid, accurate, and complete.
See companion article on page 8308.
References
- 1.Shi, Y. & Yokoyama, S. (2003) Proc. Natl. Acad. Sci. USA 100 8308-8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama, S. (2000) Prog. Retinol Eye Res. 19 385-419. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs, G. H. (1992) Am. Zool. 32 544-554. [Google Scholar]
- 4.Schluter, D., Price, T., Mooers, A. O. & Ludwig, D. (1997) Evolution 51 1699-1711. [DOI] [PubMed] [Google Scholar]
- 5.Li, W. H. (1997) Molecular Evolution (Sinauer, Sunderland, MA).
- 6.Nei, M. & Kumar, S. (2000) Molecular Evolution and Phylogenetics (Oxford Univ. Press, New York).
- 7.Zhang, J. & Nei, M. (1997) J. Mol. Evol. 44 Suppl. 1, S139-S146. [DOI] [PubMed] [Google Scholar]
- 8.Yang, Z., Kumar, S. & Nei, M. (1995) Genetics 141 1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkie, S. E., Robinson, P. R., Cronin, T. W., Poopalasundaram, S., Bowmaker, J. K. & Hunt, D. M. (2000) Biochemistry 39 7895-7901. [DOI] [PubMed] [Google Scholar]
- 10.Yokoyama, S., Radlwimmer, F. B. & Blow, N. S. (2000) Proc. Natl. Acad. Sci. USA 97 7366-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi, Y., Radlwimmer, F. B. & Yokoyama, S. (2001) Proc. Natl. Acad. Sci. USA 98 11731-11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odeen, A. & Hastad, O. (2003) Mol. Biol. Evol. 20 855-861. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, J. & Kumar, S. (1997) Mol. Biol. Evol. 14 527-536. [DOI] [PubMed] [Google Scholar]
- 14.Pauling, L. & Zuckerkandl, E. (1963) Acta Chem. Scand. 17 Suppl. 1, S9-S16. [Google Scholar]
- 15.Adey, N. B., Tollefsbol, T. O., Sparks, A. B., Edgell, M. H. & Hutchison, C. A., III (1994) Proc. Natl. Acad. Sci. USA 91 1569-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jermann, T. M., Opitz, J. G., Stackhouse, J. & Benner, S. A. (1995) Nature 374 57-59. [DOI] [PubMed] [Google Scholar]
- 17.Chandrasekharan, U. M., Sanker, S., Glynias, M. J., Karnik, S. S. & Husain, A. (1996) Science 271 502-505. [DOI] [PubMed] [Google Scholar]
- 18.Ivics, Z., Hackett, P. B., Plasterk, R. H. & Izsvak, Z. (1997) Cell 91 501-510. [DOI] [PubMed] [Google Scholar]
- 19.Zhang, J. & Rosenberg, H. F. (2002) Proc. Natl. Acad. Sci. USA 99 5486-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang, B. S., Jonsson, K., Kazmi, M. A., Donoghue, M. J. & Sakmar, T. P. (2002) Mol. Biol. Evol. 19 1483-1489. [DOI] [PubMed] [Google Scholar]
- 21.Dean, A. M. & Golding, G. B. (1997) Proc. Natl. Acad. Sci. USA 94 3104-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun, H., Merugu, S., Gu, X., Kang, Y. Y., Dickinson, D. P., Callaerts, P. & Li, W. H. (2002) Mol. Biol. Evol. 19 1490-1500. [DOI] [PubMed] [Google Scholar]
- 23.Carroll, S. B., Grenier, J. K. & Weatherbee, S. D. (2001) From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design (Blackwell, Malden, MA).
- 24.Ting, C. T., Tsaur, S. C., Wu, M. L. & Wu, C. I. (1998) Science 282 1501-1504. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, J., Zhang, Y. P. & Rosenberg, H. F. (2002) Nat. Genet. 30 411-415. [DOI] [PubMed] [Google Scholar]
- 26.Krieger, M. J. & Ross, K. G. (2002) Science 295 328-332. [DOI] [PubMed] [Google Scholar]
- 27.Terai, Y., Mayer, W. E., Klein, J., Tichy, H. & Okada, N. (2002) Proc. Natl. Acad. Sci. USA 99 15501-15506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roelofs, W. L., Liu, W., Hao, G., Jiao, H., Rooney, A. P. & Linn, C. E., Jr. (2002) Proc. Natl. Acad. Sci. USA 99 13621-13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volny, V. P. & Gordon, D. M. (2002) Proc. Natl. Acad. Sci. USA 99 6108-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, J., Webb, D. M. & Podlaha, O. (2002) Genetics 162 1825-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enard, W., Przeworski, M., Fisher, S. E., Lai, C. S., Wiebe, V., Kitano, T., Monaco, A. P. & Paabo, S. (2002) Nature 418 869-872. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, J. & Webb, D. M. (2003) Proc. Natl. Acad. Sci. USA 100 8337-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebrey, T. & Koutalos, Y. (2001) Prog. Retinol Eye Res. 20 49-94. [DOI] [PubMed] [Google Scholar]
- 34.Murphy, W. J., Eizirik, E., O'Brien, S. J. Madsen, O., Scally, M., Douady, C. J., Teeling, E., Ryder, O. A., Stanhope, M. J., de Jong, W. W. & Springer, M. S. (2001) Science 294 2348-2351. [DOI] [PubMed] [Google Scholar]