Abstract
Site-specific recombination is responsible for a broad range of biological phenomena, including DNA inversion, resolution of transposition intermediates, and the integration and excision of bacteriophage genomes. Integration of mycobacteriophage L5 is catalyzed by a phage-encoded integrase with recombination occurring between specific attachment sites on the phage and mycobacterial chromosomes (attP and attB, respectively). Although some site-specific recombination systems simply involve binding of the recombinase to the sites of strand exchange, synapsis, and recombination, phage systems typically require the assembly of higher-order structures within which the recombinational potential of integrase is activated. The requirement for these structures derives from the necessity to regulate the directionality of recombination—either integration or excision—which must be closely coordinated with other aspects of the phage growth cycles. We show herein that there are multiple pathways available for the assembly of L5 recombination complexes, including the early synapsis of the attP and attB DNAs. This process is in contrast to the model for lambda integration and illustrates the different usage of molecular machineries to accomplish the same biological outcome.
The temperate mycobacteriophage L5 forms stable lysogens in its mycobacterial hosts, including Mycobacterium tuberculosis and Mycobacterium smegmatis (1–3). Establishment of lysogeny involves the integration of a single copy of the L5 genome into the host chromosome and the repression of lytic gene expression (1, 3, 4). Comparison of the sequences of the phage and bacterial attachment sites (attP and attB, respectively) and the attachment junctions present in a lysogen (attL and attR) shows that these sites contain a common 43-bp region within which strand exchange occurs (1). Integration occurs by site-specific recombination between the attP and attB common core sites, is catalyzed by the phage-encoded integrase (Int-L5), and requires the mycobacterial integration host factor (mIHF; refs. 2 and 5).
As demonstrated by DNase I protection assays, Int-L5 binds to two types of sites in the L5 attP region (6, 7). These include a site overlapping the common core and seven arm-type sites that flank the core (P1–P7) and have a readily identifiable 10-bp consensus sequence. Only four of the arm-type sites are required for integrative recombination: P1/P2 arranged as a pair to the left of the core and P4/P5 as a pair to the right of the core (Fig. 1A; ref. 6). Int-L5 is a member of the family of tyrosine recombinases and contains the four catalytic residues and other regions that correspond to less-well conserved motifs in this family of proteins (1, 8–10). The presence of these features in Int-L5 implies that it shares similar secondary structural motifs to those of the well studied λ-integrase, being composed of two major domains, each possessing a distinct DNA binding activity (11, 12). The small N-terminal domain of λ-integrase binds to the arm-type sites within the λ-attP site, whereas a larger C-terminal domain contains the core-binding specificity and catalytic functions (11). The C-terminal domain of the tyrosine recombinases can be separated further into several smaller domains, including a region involved in core recognition (13), the catalytic region, and a small C-terminal tail involved in Int–Int interactions (as seen in the crystal structures of the HP1 integrase and the P1 Cre recombinase; refs. 14 and 15). Chymotrypsin cleavage assays reveal a proteolytically sensitive domain border in Int-L5 (J. Smith and G.F.H., unpublished results), suggesting that Int-L5 shares a similar domain structure.
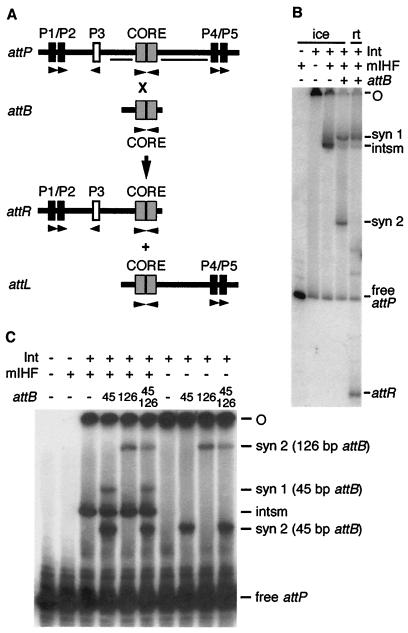
Figure 1.
Substrates, products, and complexes in L5 integrative recombination. (A) Scheme for recombination in vitro with linear substrates. Linear attP DNA radiolabeled on either one or both ends is incubated with a 45-bp attB DNA, Int-L5, and mIHF. Recombination of the substrates yields two products of intermediate sizes, attR and attL. Arm-type sites required for integration are in black, and the dispensable P3 site is in white (the dispensable P6/P7 pair of sites to the right of P4/P5 is not shown), with relative directionalities indicated by arrows (6). The gray boxes indicate loose inverted repeats at the core that are protected from DNase I by Int-L5 (6). The horizontal bars within attP are protected by mIHF in in situ footprinting of the intasome and SC1 (7). (B) The products of integrative recombination. Reactions containing attP DNA radiolabeled on the P1 end were performed either on ice or at room temperature (rt; as indicated) as described in Materials and Methods and loaded onto a native polyacrylamide gel. The positions of complexes, attR product, free attP, and intasome (intsm), and the origin of electrophoresis (O) are indicated. (C) Synaptic complex 2 (SC2) is mIHF-independent and contains one attB molecule. Complexes were formed on ice as described for B by using linear attB DNAs of 45 bp, 126 bp, or both (as indicated). The positions of SC1 as formed with the 45-bp attB and of SC2 as formed with either the 45-bp or 126-bp attB are indicated. SC1 formed with the 126-bp fragment migrates slowly and is not separated from the loading wells. Because only two SC2 bands are seen with two differently sized attBs, SC2 contains attP and only one molecule of attB. Note that the mobilities of the complexes also vary with the size of attP DNA used (compare with B).
In the absence of the attB recombinational partner DNA, Int-L5 and mIHF bind to attP DNA to form an intasome complex containing an intramolecular Int-mediated bridge between the attP core site and the P4/P5 arm-type sites. The mIHF protein, which does not bind specifically to attP DNA by itself, is required for the formation of this complex and seems to facilitate the formation of a DNA bend between the core site and the P4/P5 pair of sites (5). Curiously, the P1/P2 pair of sites is unoccupied in the intasome complex, even though Int-L5 occupies these sites (when present at the same concentration) in the absence of mIHF (5, 7). However, when attB is also present, a complex containing all four components is observed (complex 1) in which the P1/P2 arm-type sites within attP are partially protected in DNase I footprinting assays (7). From kinetic studies, it is not obvious that complex 1 is a simple obligatory intermediate in the reaction (7).
In this paper, we explore the recombinational potential of the Int–DNA complexes and the roles that they may play in assembly pathways. Although multiple pathways may be available, one of these seems to involve early synapsis of attP and attB DNA; this involvement is in contrast to lambda integration, where synapsis involves capture of naked attB DNA by the attP intasome (16, 17). We also show that the attP arm-type sites play highly specialized roles in complex formation, with the P1/P2 sites forming intermolecular bridges with attB DNA and the P4/P5 sites forming intramolecular bridges with the core-type sites of attP.
Materials and Methods
Plasmids.
The pCPΔL series, pCPΔR series, and plasmids pCP9 and pMH12.2 have been described (1, 6). Plasmid pCP9 contains the wild-type L5 attP site. pCPΔL1 and pCPΔL2 contain the L5 attP site with deletions of 72 bp and 83 bp, respectively, to the left of P1. pCPΔL3 contains a 128-bp deletion that removes the P1/P2 pair, and pCPΔL8 contains a 277-bp deletion removing P1, P2, P3, and the core-type site. Plasmids pCPΔR11 and pCPΔR13 both contain attP with the P6/P7 sites deleted, whereas pCPΔR56 contains a large deletion that removes all of attP except the P1/P2 pair. Plasmid pMH12.2 contains a 1.7-kilobase SalI attB fragment from M. smegmatis.
A set of mutants containing insertions in the intercore-P2 region of the L5 attP site was constructed. An insertion of 4 bp (to make plasmid pMK4) was made by digestion of plasmid pCP32 (containing attP with a substitution of the disposable P3 site; ref. 6) with XhoI, 3′ filling, and re-ligation of the resulting blunt ends. Insertions of 6 bp (pMK13), 9 bp (pMK12), 11 bp (pMK6), and 17 bp (pMK11) were constructed from plasmid pGL1 (containing wild-type attP; ref. 6) by site-directed mutagenesis with the Muta-Gene Phagemid In Vitro Mutagenesis system (Bio-Rad), introducing the unique restriction site NcoI. Insertions of 13 bp (pMK17), 15 bp (pMK18), and 21 bp (pMK16) were made by NcoI digestion of plasmids pMK12, pMK6, and pMK11, respectively, 3′ filling to generate blunt ends, and re-ligation.
DNA Fragments.
attP DNAs containing sites P1–P5 were generated by cutting plasmids pCPΔL1 or pCPΔL2 with BamHI and XcmI to give fragments of 379 bp and 368 bp, respectively, or by cutting plasmids pCPΔR11 or pCPΔR13 with BamHI and EcoRI to give fragments of 359 bp and 342 bp, respectively. A 353-bp DNA fragment containing sites P3–P5 was generated by cutting plasmid pCPΔL3 with HindIII and XcmI. DNA fragments containing only one pair of L5 attP arm-type sites and no core-type sites were generated as follows. A 497-bp fragment containing only the P1/P2 pair of sites was cut from plasmid pCPΔR56 by using BamHI and NaeI. A 318-bp fragment containing the P4/P5 pair only was cut from plasmid pCPΔL8 by using XcmI and XhoI. The P6/P7 pair of sites was cut from plasmid pCP9 by using ApaLI and XcmI to generate a fragment of 876 bp. attP DNA fragments containing insertions between the core and P2 (and including P1 through P5) were generated by digesting plasmids pMK4 (insertion of +4 bp), pMK13 (+6 bp), pMK6 (+11 bp), pMK17 (+13 bp), pMK18 (+15 bp), pMK11 (+17 bp), and pMK16 (+21 bp) with BamHI and XcmI to give fragments ranging from 451 to 470 bp, depending on the insertion size.
attB DNA fragments were generated by annealing pairs of oligonucleotides (to give 45-bp fragments; ref. 18) or were cut from plasmid pMH12.2 by using AvaII and MseI to give a 126-bp fragment.
DNA fragments were radiolabeled either by phosphorylation or by end fill with Klenow.
Complex Formation and in Vitro Integrative Recombination.
Recombination assays were similar to those described previously (2, 7) and were performed in 10-μl volumes. Unless otherwise noted, ≈0.024 pmol of linear, radiolabeled attP substrate (342–470 bp, encompassing P1–P5) was preincubated with 0.07–0.23 pmol purified Int-L5 and 3.6–12.0 pmol purified mIHF for 15–30 min on ice; 0.06 pmol of a 45-bp (unless otherwise noted) attB DNA (18) was added (where indicated), and the entire reaction was incubated either on ice for 15–30 min or at room temperature for 2 h. Reactions were electrophoresed through a 5% polyacrylamide gel in 1× TBE (100 mM Tris/84 mM borate/1 mM EDTA), and products were visualized by autoradiography.
In Gello Recombination.
Portions of lanes containing protein–DNA complexes were excised from a wet gel and soaked for 3 h at room temperature either in reaction buffer alone or with ≈120 nM 45-bp attB DNA, 14.4 nM Int-L5, and/or 720 nM mIHF (as indicated). Protein–DNA complexes were denatured by soaking gel slices in 0.5% SDS for 10 min. The gel slices were then laid horizontally across the top of and electrophoresed in a second dimension through a 5% polyacrylamide/0.05% SDS gel in 1× TBE.
Results
Identification of SCs.
We have noted previously that, although L5 integration is stimulated by DNA supercoiling, linear DNA substrates will undergo recombination (see Fig. 1B; refs. 7 and 19). However, the event is strongly temperature-dependent, and if the reactions are kept on ice, then no recombinant products are observed (Fig. 1B). Under these conditions, two of the observed complexes are the same as those seen at room temperature: the intasome, which contains Int-L5, mIHF, and attP DNA, and SC1, which has been described (7). Although SC1 seems to contain both proteins and both partner DNAs and is present under conditions in which strand exchange occurs, it is not clear whether it is a recombinational intermediate (7). Moreover, we cannot rule out the possibility that this band—or any other band on these gels—is composed of more than one distinct type of complex, each with different recombinogenic properties.
When the reactions are incubated on ice rather than at room temperature, a third complex, which we will refer to as SC2, is also present (Fig. 1B). The reason why this complex is not seen at room temperature is presumably because it either readily converts back into its constituents or acts as a precursor in the formation of additional complexes. Further analysis showed that this complex contains a ratio of 1:1 attB:attP molecules, because the addition of two differently sized attB DNAs does not generate complexes with hybrid mobilities (Fig. 1C). Moreover, unlike all of the other complexes, it does not contain mIHF and mIHF is not required for its formation (Fig. 1C). This complex can be dissociated by SDS (data not shown) and presumably contains attP and attB DNA held together via an integrase-mediated intermolecular bridge. Identification of such a complex is important, because it suggests the possibility that synapsis of the attP and attB sites can occur without obligatory and prior formation of the intasome.
Roles of the attP Arm-Type Sites in Synapsis.
The nature of SC2 was investigated by examining the substrate requirements for its formation (Fig. 2). It was found that removal of the P1/P2 pair of arm-type sites eliminates the formation of both SC1 and SC2 without affecting intasome formation (Fig. 2A). In contrast, mutational inactivation of P4 and P5 results in loss of intasome and SC1 formation, without affecting SC2 formation (Fig. 2A). Because the P1/P2 sites are required for SC2 formation, it is likely that they are involved specifically in Int-mediated intermolecular bridges with attB DNA, although it is unclear why the P4/P5 sites would not also do so, given their similarity to P1 and P2 (Fig. 2B). We note that if the concentration of either attB DNA or Int-L5 is increased above that used under standard recombination conditions, then an additional complex (SC3) is also formed, presumably by the addition of another molecule of attB DNA via a second intermolecular bridge with the P4/P5 sites (Fig. 2C). The P1/P2 sites may thus have an inherent preference over the P4/P5 sites for bridging with attB DNA.
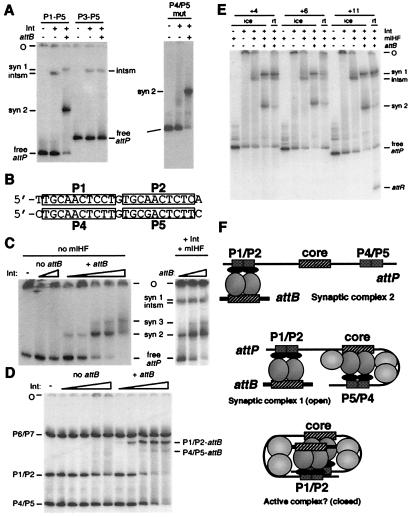
Figure 2.
The role of arm-type sites. (A) Formation of integration complexes with mutant attP substrates. Reactions were performed on ice as in Fig. 1B with an attP DNA containing P1–P5 or P3–P5 (Left) or mutant P4/P5 sites (Right), Int-L5 (as indicated), mIHF, and attB, as indicated. (B) Comparison of the sequences of the P1/P2 and P4/P5 arm-type sites. (C) Detection of a second mIHF-independent SC. SCs were formed on ice as in Fig. 1B by using attP, varying amounts of Int-L5 (Left; from left to right: 0, 0.24, 0.72, 0.024, 0.072, 0.24, 0.72, and 2.4 pmol Int-L5; these lanes contain 0.12 pmol attB per reaction) and varying amounts of attB (Right; from left to right: 0.06, 0.12, and 0.6 pmol attB per reaction; these lanes contain 0.24 pmol Int-L5 per reaction), in the presence or absence of mIHF as indicated. (D) P1/P2 sites prefer to bridge with attB. Radiolabeled DNA fragments containing only one pair of arm-type sites (P1/P2, 497 bp; P4/P5, 318 bp; P6/P7, 876 bp) and no core sites were mixed and incubated on ice in the presence or absence of 0.12 pmol of attB DNA (as indicated) and varying amounts of Int-L5 (−, no Int-L5 added; left to right: 0.024, 0.072, 0.24, 0.72, and 2.4 pmol Int-L5 per reaction) in the absence of mIHF under conditions identical to complex formation reactions. The positions of the P1/P2-attB complex, the P4/P5-attB complex, and the free P1/P2, P4/P5, and P6/P7 DNA fragments are indicated. (E) Effect of P1/P2-core intersite spacing on recombination and complex formation. Reactions were performed either on ice or at room temperature (as indicated) by using attP DNA fragments ranging from 451 bp to 458 bp, depending on the size of the insertion. Results of reactions with attP insertions of +4 bp, +6 bp, and +11 bp are shown, and reactions with insertions of +13 bp, +15 bp, +17 bp, and +21 bp were performed but are not shown. attP DNAs with insertions of +13 bp, +15 bp, and +17 bp did not recombine; however, those with insertions of +21 bp did, and none interfered with complex formation (data not shown). (F) Proposed structures of protein–DNA complexes. SC2 is postulated to contain Int-mediated bridges between attB and the P1/P2 arm-type sites. SC1 contains these same bridges but also contains Int-mediated, mIHF-stabilized intramolecular bridges between the attP core and the P4/P5 sites. Because the spacing changes shown in E do not affect complex 1 formation, we suggest that it is unfolded or open, such that the helical phasing of the P1/P2 sites and the core is not important. However, because nonintegral DNA insertions inhibit recombination, we propose that it is necessary for SC1 to fold into a more compact or closed structure in order for strand exchange to occur. The mIHF host factor is shown as light gray balls situated between the attP core and P4 in complex 1 and also between P2 and the core in the putative active complex. DNase I footprinting suggests that there may also be a unit of mIHF bound just to the left of the core in SC1 (7).
This interpretation is supported by the behavior of substrates in which either the P1/P2 or P4/P5 arm-type sites are the only Int-binding sites present (Fig. 2D). Two key properties of these sites are revealed. First, the P1/P2 sites form electrophoretically stable complexes with attB DNA at Int-L5 concentrations at which P4/P5 sites do not; the P4/P5 sites do form a complex with attB DNA, but it is observed only at higher Int-L5 concentrations, similar to those that promote SC3 formation with attP DNA (Fig. 2C). The complex also seems to be electrophoretically unstable (Fig. 2D). The P1/P2 sites thus have a significant preference over P4/P5 for forming intermolecular bridges with attB DNA. Secondly, P1/P2∷attB complexes are seen at Int-L5 concentrations at which no complexes are observed in the absence of attB DNA (Fig. 2D); presumably, either the P1/P2–Int interaction is weaker without attB, or the complexes are formed but are not stable during electrophoresis. This observation is reminiscent of the previous finding (5) that the P1/P2 sites of attP are unoccupied by Int-L5 in the absence of attB when mIHF is present, and when the attP core-type sites are bridged to P4/P5 (and thus unable to bridge to P1/P2). Both observations could be explained by a model in which the binding affinity of Int-L5 to arm-type sites is enhanced when the C-terminal domain is bound simultaneously to core-type sites.
The Role of SC1.
The experiments described above are consistent with a model in which the attP arm-type sites play specialized roles, with P4/P5 forming intramolecular bridges with the attP core, and with P1/P2 interacting with attB via an intermolecular bridge. The former interactions are observed in the intasome, and the latter are seen in SC2. It seems likely that both of these sets of interactions are present in SC1, and this likelihood is supported by in situ DNase I footprinting (7). However, SC1 accumulates during a time course of the reaction and does not behave as a simple obligatory intermediate within which strand exchange occurs (7). Although the reason for this behavior is not clear, we note that there are at least two types of higher-order structures that could support these protein–DNA interactions: a compact closed structure held together by a tetramer of Int (similar to the Cre synaptic tetramer; ref. 15) or an open structure lacking these interactions (see Fig. 2). To discern between these types of structures, we constructed a series of mutant attP substrates in which the spacing between the P1/P2 sites and the attP core was altered and assessed the effects on recombination and complex formation (Fig. 2E). We observed that this spacing is critical for recombination, and insertion of either one (Fig. 2E) or two (data not shown) integral DNA turns supports recombination, whereas nonintegral numbers of turns do not. However, none of the insertions interfere with formation of any of the complexes, including SC1. In view of the relatively short distance between the core and the P2 site (≈95 bp), it seems unlikely that a closed structure could tolerate insertions of half a helical turn of DNA, and these data are more consistent with an open structure as shown in Fig. 2F.
Recombinogenic Potential of Protein–DNA Complexes.
Although linear attP and attB substrates can undergo recombination in vitro, it is far from clear what role the various protein–DNA complexes play in the recombinational pathway. To examine this role, we have tried to evaluate the recombinational potential of the complexes by first separating them by native gel electrophoresis and then performing recombination within the gel matrix (“in gello”). We reasoned that those complexes that are electrophoretically stable would remain intact within the acrylamide matrix and might undergo recombination under appropriate conditions. Recombinant products could then be identified by a second dimension of electrophoresis in the presence of SDS. The major limitation in interpreting such events is that complexes could dissociate and reassemble into alternative structures during the recombination reaction.
Initially, we evaluated the recombinogenic potential of the intasome (Fig. 3A). First, Int-L5 and mIHF were incubated with attP DNA, and the intasome complexes were separated from free attP DNA by native gel electrophoresis (similar to the third lane in Fig. 1B). A vertical slice was then excised from the gel and incubated in a reaction buffer containing a 45-bp attB DNA substrate, and the products were identified (Fig. 3A, attB reaction). Although recombinant products were observed, in gello recombination was stimulated by the addition of mIHF (attB/mIHF reaction; Fig. 2A), indicating that the intasome does not have all of the mIHF required for recombination. Addition of Int-L5 provides little stimulation of recombination (attB/Int reaction), although some Int-L5 does seem to diffuse into the gel matrix (products are generated from free attP DNA when Int-L5, mIHF, and attB are provided; Fig. 3A). Control experiments demonstrated that neither Int-L5 nor mIHF migrates as free protein at the positions of any of the protein–DNA complexes (T. Huang and G.F.H., unpublished observations).
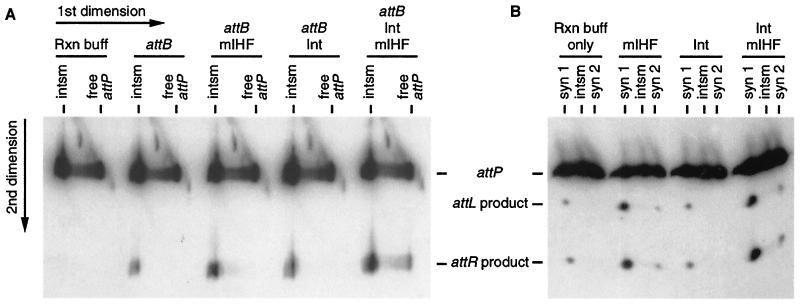
Figure 3.
Activation of recombination in gello. (A) In gello recombination of intasomes. For the first dimension, five reactions similar to the one shown in Fig. 1B lane 3 were run on a native gel to separate intasomes from free attP DNA (labeled at the P1 end), and the lanes were excised. Each gel slice was soaked in reaction (Rxn) buffer either alone or with attB, mIHF, and/or Int-L5 (as indicated). The five gel slices were laid on top of a polyacrylamide gel containing SDS for the second dimension. The last lane (attB/Int/mIHF), in which free attP DNA yields products, demonstrates that attB DNA, mIHF, and Int-L5 do diffuse into the gel slice. (B) In gello recombination of SCs. The intasome, SC1, and SC2 were formed on ice as in Fig. 1B lane 4. The regions of four lanes containing the intasome and both SCs were excised and soaked in reaction buffer (without attB) either alone or with mIHF and/or Int-L5 (as indicated). Recombination products were then identified as in A. For B, complexes were formed by using an attP DNA radiolabeled on both ends, such that both products are visualized. For both A and B, the positions of the intasome and SCs as they ran in the first dimension are labeled on the top, and positions of the DNAs in the second dimension are indicated at the side.
Using similar methods, we find that SC1—but not SC2—can undergo recombination in the absence of any additional proteins. When a gel slice containing all of the complexes is incubated at room temperature, products are generated from only SC1 and not from any other complexes (Fig. 3B), and this generation occurs in a time-dependent manner (data not shown). However, the ability of SC1 to undergo recombination is stimulated substantially by the inclusion of mIHF in the reaction buffer; some recombination is also seen from SC2 under these conditions (Fig. 3B). This experiment suggests that the poor level of recombination in SC1 results at least in part from a less-than-full complement of mIHF, either because of a deficiency of mIHF binding in solution (i.e., before electrophoresis) or because of its loss during subsequent incubation of the gel slice. Little further stimulation of recombination is seen when Int-L5 is also provided.
Assembly Pathways for Integrative Recombination.
These in gello experiments are consistent with the idea that both the intasome and SC1 are intermediates in recombination, although dissociation and reassembly into alternative structures during the in gello incubation cannot be ruled out. SC2 is also able to undergo some recombination when mIHF is present, although it is not easy to see how SC2 could be on the same assembly pathway as the intasome. To investigate this process further, we examined the formation of complexes as a function of incubation time, both on ice and at room temperature (Fig. 4A). Although gel electrophoresis is an admittedly somewhat crude assay for this kinetic experiment, the results suggest that SC2 is formed early in the reaction and accumulates to a high level but then diminishes as recombination proceeds (at room temperature). Thus, SC2 seems to be an early intermediate, in part explaining the relatively poor conversion to products in the in gello experiment; SC1 and the intasome do not appear until later times. The interpretation of this experiment is complicated by the reversibility of many of the steps and not knowing whether the complexes observed after gel electrophoresis match those that exist during the reaction both in quantity and composition. Nevertheless, these observations are consistent with early synapsis representing a productive assembly pathway.
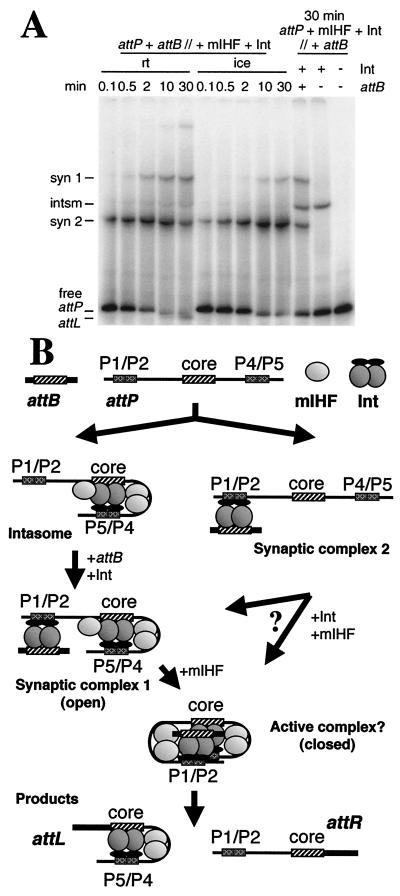
Figure 4.
Pathways for integrative recombination. (A) Recombination reactions were performed by using attP DNA radiolabeled on both ends, mixing all DNA and protein components concurrently at the temperature indicated with no preincubation (attP + attB//+ mIHF + Int) and with incubation at the temperature indicated for 0.1 to 30 min. As a control, the intasome was preformed on ice by preincubation of attP, Int-L5 (as indicated), and mIHF, and attB was added later (as indicated) and incubated on ice for 30 min (attP + mIHF + Int//+ attB). At the 10- and 30-min room temperature time points, a doublet can be distinguished near the free attP position, which is the formation of the attL product complex after strand exchange (7). (B) Multiple assembly pathways. At the top are shown the reaction components attP, attB, Int, and mIHF. On the left is shown the synapsis late pathway in which Int and mIHF first form an intasome complex that then captures attB DNA to generate SC1. Further addition of mIHF promotes folding of this open structure into a postulated compact but active complex in which strand exchange occurs to release the products, free attR DNA, and an attL complex. On the right is shown the alternative synapsis early pathway, in which the initial complex formed is SC2, followed by assembly into complex 1 or perhaps directly into the closed active complex. The only one of these complexes not identified by native gel electrophoresis is the putative active closed complex, which is postulated to form slowly (such that complex 1 accumulates) but is rapidly converted into products.
Discussion
The experiments described above provide significant insights into how higher-order macromolecular structures are assembled for integrative recombination of mycobacteriophage L5. A model that is consistent with these observations is shown in Fig. 4B. A principal feature of this model is that there are multiple assembly pathways available. In one pathway, the intasome is an obligatory intermediate, as indicated by the ability of the intasome to capture attB DNA in the in gello recombination experiments. In the alternative pathway, SC2 is an early intermediate, as supported by the kinetic experiment shown in Fig. 4A. Although we cannot rule out the possibility that there is only a single assembly pathway that requires dissociation and reassembly of either the intasome or SC2 (and all of the assembly steps are presumably reversible), there is no a priori reason to make this assumption, because the protein–DNA interactions required for intasome and SC2 formation do not seem to be mutually exclusive.
The intasome pathway for assembly is not dissimilar to that reported for lambda recombination, in which an attP intasome captures naked attB DNA (17). However, we note that the arrangement of the arm-type sites in attP and their occupancy by Int-λ is quite different from the arrangement of L5. In particular, in the lambda intasome, arm-type sites on both sides of the attP core are occupied, whereas in the L5 intasome, only those to the right of the core are bound by Int-L5. A consequence of this arrangement is that the structure of the L5 attP intasome is virtually identical to that of the predicted L5 attL intasome; indeed, attL is released from the recombination reaction as this protein–DNA complex (7). This process is distinct from the lambda system where the attP and attL intasomes are quite different (20).
It is unclear whether—as in the lambda pathway (17)—the L5 intasome captures attB as naked DNA. The in gello experiments suggest that the intasome may indeed contain all four required Int-L5 protomers, because recombination is observed without further addition of Int-L5 and the presence of Int-L5 does not stimulate the reaction substantially (Fig. 3A). Alternatively, it is possible that the intasome contains subrecombinogenic amounts of Int-L5, and the Int-L5 protomers required for capture of attB by the P1/P2 sites are cannibalized from other intasomes. The two-protomer intasome is attractive from the perspective of the excision reaction. If there are two protomers in the attP intasome, there are likely to be two in the attL intasome as well, which could then assemble with an attR intasome that also contains two protomers of Int-L5. Preliminary experiments show that the L5 excisionase (21) does promote formation of an attR-intasome complex, although—as with the attP and attL complexes—the stoichiometries are not yet known.
The possibility of an early step involving synapsis is a significant departure from the lambda assembly pathway. The primary support for this conclusion is from the kinetic experiment (Fig. 4A), although the ability of the P1/P2 sites to form intermolecular bridges with attB in the absence of any other interactions is strongly supported by the data shown in Fig. 2. The observation that P1/P2 sites do this bridging preferentially over P4/P5 is curious, because this preference must reside in the subtle differences between the sequences of the sites (or their sequence contexts) and cannot be explained by the action of mIHF or DNA bending (which influences intasome formation). The specialized roles of the arm-type sites in integration is also reflected in in vivo observations where removal of the P1/P2 pair of sites reduces integration to below detectable levels—presumably because of the inability to capture attB—whereas loss of the P4/P5 pair reduces integration only to about 1% (6), reflecting their role in higher-order assembly and activation of recombination.
The specific fates of the intasome and SC2 are unclear, although the in gello experiments suggest that SC1 may indeed be a recombinational intermediate, because it can undergo recombination (assuming that it does not have to dissociate first). However, when taken together, the observations that the P1/P2-core intersite spacing is important for recombination but not for SC1 formation (Fig. 2E), that in gello recombination by SC1 is stimulated substantially by mIHF (Fig. 3B), and that SC1 facilitates strand exchange rather poorly (Fig. 3B; ref. 7) suggest that SC1 is an open complex (see Fig. 2) that requires mIHF for bending the DNA between P1/P2 and the core to form a compact closed complex within which recombination can then occur. The reason why this putative closed complex is not observed could be accounted for by assuming that its passage through recombination or reversion to the open configuration is rapid. The transition from open to closed complex may thus represent the limiting step in the recombination reaction with linear DNA substrates, and it may be this step that is facilitated by DNA supercoiling (19).
Acknowledgments
We thank J. Lewis for comments on the manuscript. C.E.A.P. was supported in part by a Mellon Predoctoral Fellowship from the University of Pittsburgh. This work was supported by National Institutes of Health Grant GM49647.
Abbreviations
- attP
phage attachment site
- attB
bacterial attachment site
- attL
left attachment junction
- attR
right attachment junction
- Int-L5
L5 integrase
- mIHF
mycobacterial integration host factor
- SC
synaptic complex
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140014297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140014297
References
- 1.Lee M H, Pascopella L, Jacobs W R, Jr, Hatfull G F. Proc Natl Acad Sci USA. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee M H, Hatfull G F. J Bacteriol. 1993;175:6838–6841. doi: 10.1128/jb.175.21.6836-6841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hatfull G F. News. 1994;60:255–260. [Google Scholar]
- 4.Snapper S, Lugosi L, Jekkel A, Melton R, Keiser T, Bloom B R, Jacobs W R., Jr Proc Natl Acad Sci USA. 1988;85:6987–6991. doi: 10.1073/pnas.85.18.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedulla M L, Lee M H, Lever D C, Hatfull G F. Proc Natl Acad Sci USA. 1996;93:15411–15416. doi: 10.1073/pnas.93.26.15411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peña C E A, Lee M H, Pedulla M L, Hatfull G F. J Mol Biol. 1997;266:76–92. doi: 10.1006/jmbi.1996.0774. [DOI] [PubMed] [Google Scholar]
- 7.Peña C E A, Kahlenberg J M, Hatfull G F. J Bacteriol. 1999;181:454–461. doi: 10.1128/jb.181.2.454-461.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi Y, Nash H A. Cold Spring Harbor Symp Quant Biol. 1979;43:1099–1109. doi: 10.1101/sqb.1979.043.01.122. [DOI] [PubMed] [Google Scholar]
- 9.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V L, Peirson L S, III, et al. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nunes-Düby S E, Kwon H J, Tirumalai R S, Ellenberger T, Landy A. Nucleic Acids Res. 1998;26:391–406. doi: 10.1093/nar/26.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moitoso de Vargas L, Pargellis C A, Hasan N, Bushman E, Landy A. Cell. 1988;54:923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- 12.Landy A. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 13.Tirumalai R S, Kwon H J, Cardente E H, Ellenberger T, Landy A. J Mol Biol. 1998;279:513–527. doi: 10.1006/jmbi.1998.1786. [DOI] [PubMed] [Google Scholar]
- 14.Hickman A B, Waninger S, Scocca J J, Dyda F. Cell. 1997;89:227–237. doi: 10.1016/s0092-8674(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 15.Guo F, Gopaul D N, van Duyne G D. Nature (London) 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 16.Nash H A. In: Escherichia coli and Salmonella cellular and molecular biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2363–2376. [Google Scholar]
- 17.Richet E, Abcarian P, Nash H A. Cell. 1988;52:9–17. doi: 10.1016/0092-8674(88)90526-0. [DOI] [PubMed] [Google Scholar]
- 18.Peña C E A, Stoner J E, Hatfull G F. J Bacteriol. 1996;17:5533–5536. doi: 10.1128/jb.178.18.5533-5536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peña C E A, Kahlenberg J M, Hatfull G F. Nucleic Acids Res. 1998;26:4012–4018. doi: 10.1093/nar/26.17.4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Landy A. Science. 1992;256:198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis J A, Hatfull G F. Mol Microbiol. 2000;35:350–360. doi: 10.1046/j.1365-2958.2000.01695.x. [DOI] [PubMed] [Google Scholar]