Abstract
The programmed self-assembly of patterned aperiodic molecular structures is a major challenge in nanotechnology and has numerous potential applications for nanofabrication of complex structures and useful devices. Here we report the construction of an aperiodic patterned DNA lattice (barcode lattice) by a self-assembly process of directed nucleation of DNA tiles around a scaffold DNA strand. The input DNA scaffold strand, constructed by ligation of shorter synthetic oligonucleotides, provides layers of the DNA lattice with barcode patterning information represented by the presence or absence of DNA hairpin loops protruding out of the lattice plane. Self-assembly of multiple DNA tiles around the scaffold strand was shown to result in a patterned lattice containing barcode information of 01101. We have also demonstrated the reprogramming of the system to another patterning. An inverted barcode pattern of 10010 was achieved by modifying the scaffold strands and one of the strands composing each tile. A ribbon lattice, consisting of repetitions of the barcode pattern with expected periodicity, was also constructed by the addition of sticky ends. The patterning of both classes of lattices was clearly observable via atomic force microscopy. These results represent a step toward implementation of a visual readout system capable of converting information encoded on a 1D DNA strand into a 2D form readable by advanced microscopic techniques. A functioning visual output method would not only increase the readout speed of DNA-based computers, but may also find use in other sequence identification techniques such as mutation or allele mapping.
The field of nanotechnology holds tremendous promise. If the molecular and supramolecular world can be controlled at will, then it may be possible to achieve vastly better performance for computers and memories, and it might open up a host of other applications in materials science, medicine, and biology. Because of this promise, numerous research teams have embarked on the development of various detailed aspects of nanotechnology, such as the use of physically strong and electrically active fullerene materials (1), and organic molecules that have electrical switching properties (2). The construction of molecular-scale structures is one of the key challenges facing science and technology in the 21st century. There are two distinct approaches to the fabrication of nanomaterials: top-down methods and bottom-up approaches. Top-down methods, exemplified by e-beam lithography, may be limited by their serial nature, whereas bottom-up methods using self-assembly are by nature highly parallel. Although self-assembly methods are well known and have been long used by chemists, they conventionally result in structures with limited complexity (e.g., regular, periodic patterning with a small number of programmed association rules), and most current methods do not allow the self-assembly to be readily reprogrammable.
In recent years, DNA has been advanced as a useful material for constructing periodically patterned structures (3–9), nanomechanical devices (10–13), and molecular computing systems (14–19). DNA also has been designed to direct the assembly of other functional molecules by the use of appropriate attachment chemistries (20–26). The diversity of materials with known DNA attachment chemistries considerably enhances the attractiveness of DNA nanostructures, which can be used to form superstructures on which other materials may be assembled.
Self-assembling DNA tiling lattices represent a versatile system for nanoscale construction (27, 28). The methodology of DNA lattice self-assembly begins with the chemical synthesis of single-stranded DNA molecules, which self-assemble into DNA branched motif building blocks known as tiles. DNA tiles can carry sticky ends that preferentially match the sticky ends of other particular DNA tiles, thereby facilitating the further assembly into tiling lattices. Self-assembled 2D DNA tiling lattices composed of tens of thousands of tiles have been demonstrated by the Seeman, Winfree, and Reif groups and visualized by molecular imaging techniques such as atomic force microscopy (AFM) (6–9). Recent experiments by the Seeman and Reif groups have demonstrated computation (logical exclusive or) via molecular assembly of DNA tiles (15, 17).
Previous DNA self-assembly systems provided either small computational assemblies or large lattices with simple periodic patterning. There remains the further challenge of programming the self-assembly of patterned aperiodic DNA lattices with increasing size and increasing pattern complexity. This holds the potential of allowing us to build templates, on which molecular electronics and robotics components can be positioned with precision and specificity. A fully programmable aperiodic DNA tiling system will also find use as a visual readout method that will increase the ease and speed of output from DNA-based computers.
There are at least three major strategies for the formation of patterned DNA tiling lattice self-assemblies:
Unmediated algorithmic self-assembly is the most general method for 2D pattern formation. It entails the use of a small set of DNA tiles that self-assemble in a predictable manner. Unmediated algorithmic self-assembly is Turing-universal (29) and is thus theoretically capable of creating complex structures from simple rules. This method has the advantage of being very general and not requiring an input DNA strand encoding a pattern; instead, the 2D pattern is generated de novo based on the neighbor relations encoded within the DNA tile set. Unmediated algorithmic self-assembly of complex patterns requires delicate control of physical phenomena, which include nucleation rates, crystal growth rates, spontaneous nucleation, and error rates in solutions containing many distinct DNA tile types.
Another approach is sequential stepwise assembly, similar to strategies used frequently in synthetic chemistry. It begins with separate assembly of a small number of distinct molecular building blocks (MBBs). These MBBs are then combined to construct larger assemblies in a stepwise fashion, possibly with an initial building block attached to a solid support, to enable removal of excess reactants after each step. In the context of DNA lattices, these MBBs may be DNA tiles, as described above. Using a stepwise approach, the number of distinct MBBs can be greatly reduced by repeatedly reusing a small number of MBB types in a controlled sequential assembly scheme, which introduces specific MBBs at successive times into a unit such as a fluid chamber. Such sequential assembly might allow for the synthesis of complex molecular structures, while extensively reusing a small number of MBB types. A potential drawback is the time-consuming nature of such an externally mediated stepwise assembly process.
Directed nucleation assembly is the method used here for self-assembly of complex patterned lattice. It uses a preassembled input DNA strand that encodes the required pattern information; other oligonucleotides then assemble into specified tiles around this input scaffold strand, forming the desired 1D or 2D pattern of tiles. The directed nucleation assembly technique was previously prototyped (30); here we demonstrate the use of a scaffold strand that encodes pattern information.
We describe the self-assembly of a DNA lattice, using directed nucleation assembly with an information-carrying input strand. A DNA barcode lattice with an aperiodic pattern encoding five bits of information (01101) was formed by assembling double crossover (DX) DNA tiles (4) around scaffold DNA strands. The barcode lattice displays banding patterns dictated by the sequence of bit values programmed on the input strand. To demonstrate the programmability of this technique, we modified and resynthesized the scaffold strand and also modified one of the strands composing each tile to create DNA lattice displaying the pattern 10010, which is inverted compared with the original barcode pattern. The success of this design may be extended to form more complicated patterns. Furthermore, error rates should be reduced, because tiles can only assemble to form lattice in the presence of scaffold strands, and fully half of the internal tile-to-tile associations are hard-wired through the scaffold strands.
Materials and Methods
Molecular Design and Synthesis. The sequences of the DNA tiles were designed by using the program sequin (31) and are listed in Figs. 6–8, which are published as supporting information on the PNAS web site, www.pnas.org. All DNA strands in this study were synthesized commercially by Integrated DNA Technologies (Coralville, IA; www.idtdna.com). DNA strands were purified by denaturing gel electrophoresis; bands were cut out of 12–20% denaturing gels. They were then eluted in a solution containing 500 mM ammonium acetate, 10 mM magnesium acetate, and 1 mM EDTA.
Formation of Scaffold Strand by Ligation of Shorter DNA. To assemble the barcode lattice, a scaffold strand was constructed by ligation. For the scaffold strand representing barcode 01101, three oligonucleotides, each containing 109 bases, were synthesized, purified, and brought together by annealing them with two 18-mer linker strands. The three 109-mers were then ligated to form the scaffold DNA strand. To construct the scaffold strand representing barcode 10010, one 109-mer and two 79-mer oligonucleotides were annealed with linker strands and ligated. Ligation reactions were performed in 1× ligation buffer (Amersham Pharmacia). All strands were mixed stoichiometrically at a concentration of 2 μM, and the solution was heated to 90°C for 7 min and cooled slowly to room temperature (1–2 h). Thirty units of T4 DNA ligase (Amersham Pharmacia) was added, and the reaction was allowed to proceed at 16°C for 16 h. The reaction was stopped by heating the sample at 95°C for 20 min and then drying it. Thereafter the full-length scaffold strand was purified by denaturing gel electrophoresis.
Formation of Hydrogen-Bonded Complexes. Complexes were formed by mixing a stoichiometric quantity of each strand (750 nM), as estimated by OD260, in 1× TAE/Mg (40 mM Tris·HCl, pH 8.0/20 mM acetic acid/2 mM EDTA/12.5 mM magnesium acetate). This solution was cooled slowly from 90°C to room temperature in a 1-liter water bath over the course of 4 h or longer.
AFM Imaging. A 5-μl sample drop was spotted on freshly cleaved mica (Ted Pella, Inc., Redding, CA) and left to adsorb to the surface for 3 min. Then, 30 μl of 1× TAE/Mg buffer was placed onto the mica, and another 30 μl of buffer was pipetted onto the AFM tip. Imaging was performed under 1× TAE/Mg in a fluid cell on a Multimode NanoScope IIIa, using NP-S tips (Veeco, Woodbury, NY).
Results
Design of the Barcode DNA Lattices. Demonstrations of large-scale 2D DNA self-assemblies using DX and triple crossover tiles had been performed, using unmediated self-assembly techniques (6, 7). Here we constructed aperiodic patterned DNA lattices (barcode lattice) by a directed nucleation self-assembly process using DNA DX tiles around a scaffold DNA strand. The scaffold strand provides barcode patterning information, which is represented by the presence (bit value = 1) or absence (bit value = 0) of DNA hairpin loops protruding from the DNA lattice. To demonstrate the programmability of this technique, we designed two types of DNA barcode lattice, one with barcode pattern 01101, the other with its inverse barcode pattern 10010. The banding patterns of the barcode lattices were dictated by the sequence of bit values programmed on their input strands.
The DX tile (6, 31) consists of two double-helical domains joined twice by strand exchange at two crossover points. Another way to think of the DX complex is that it is formed from two four-arm branched-junction complexes that have been ligated twice at adjacent arms. The upper tile shown in Fig. 1a is a simple DX complex (DAE), and the lower tile is a DX complex (DAE + 2J) with two hairpin loops protruding from the central helix of the DAE tile, with one loop projecting on each side of the tile plane. Regardless of which side faces down during sample preparation, there will be loops protruding up out of the lattice plane. These hairpin loops serve as topographic markers, which are observable by AFM. The DX tiles are stable and well behaved (32, 33), both in solution and when analyzed on nondenaturing polyacrylamide gels.
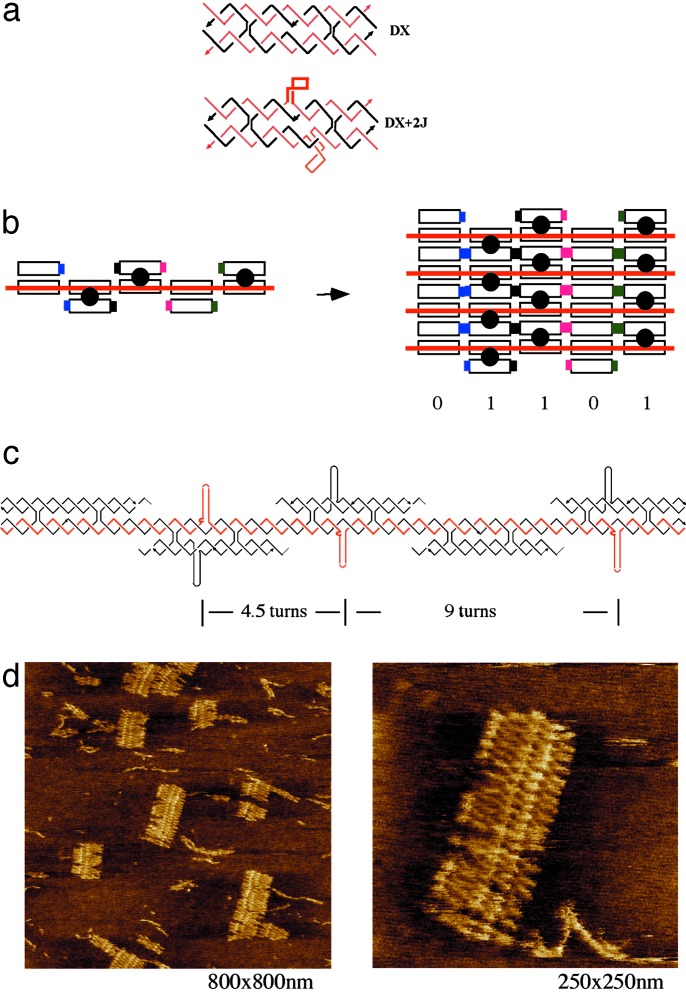
Fig. 1.
Self-assembly of 01101 barcode lattice around scaffold DNA strand. (a) (Upper) DAE tile, one type of antiparallel DNA DX tile. The tile drawing shows the five strands (three black and two red). The two red strands are continuous strands going through the tile in opposite directions (arrowheads mark 3′ ends). There are two crossover points connecting the two domains. There are two helical turns between the two crossover points. (Lower) DAE + 2J tile. This tile type has two hairpin loops protruding out of the central helix region of the DAE complex; one loop (thick line) is coming out of the plane and the other (thinner line) into the plane. The hairpin loops serve as topographic markers in AFM imaging of the lattices. (b) Schematic of self-assembly of barcode lattice layers based on DAE tiles around a scaffold strand. (Left) A five-tile crenellated horizontal layer is shown with an input scaffold strand running through the layer (red). The scaffold strand is required for the tiles to assemble. (Right) A lattice of four layers is illustrated (note that sticky ends are still available on the upper and lower layers for addition of more layers). The sticky ends are represented by different colored pads matching one other. The barcode information (01101) is represented by either the presence (designated 1) or the absence (designated 0) of a stem loop (shown as a black circle) protruding out of the tile plane. (c) Strand structure of one barcode layer. This layer represents barcode information of 01101. The red strand is the scaffold strand required for the tile assembly. The distance between adjacent hairpin loops is indicated by the number of helical turns. (d) AFM visualization of DNA barcode lattice (01101). The scale of each image is indicated in its lower right corner. Up to 24 layers of DNA have been self-assembled; the desired stripe pattern is clearly visible. Each layer contains five DX tiles and is ≈75 nm wide. The distance between the two closer adjacent stripes is ≈16 nm. The distance between the two further adjacent stripes is ≈31 nm. See Fig. 9, which is published as supporting information on the PNAS web site, for a large-area scan AFM image.
Simple periodic 2D lattices composed of these DX tiles have been described (6). The barcode lattice described here was designed based on the DAE and DAE + 2J tiles. Fig. 1b illustrates the strategy of the first barcode lattice (01101) assembly around a scaffold strand. A crenellated DAE and DAE + 2J layer contains a scaffold input strand that goes through the layer and is required for tile assembly. Lattice assembly propagates up and down, by means of sticky-end associations joining adjacent layers to one another. Fig. 1c shows the strand structure of the repeating barcode layer for the 01101 barcode lattice. The red strand is the scaffold strand required for assembly of the DX tile layer. The barcode information of 01101 is represented either by the presence of stem loops (designated 1) or their absence (designated 0). The DNA sequences used in the experiment can be found in Figs. 6–8.
Self-Assembly of DNA Barcode Lattice Around Input Scaffold Strands. Both nanotechnological and computational applications of DNA self-assembly depend, in part, on the ability to assemble and visualize DNA building blocks in higher-order structures. DNA lattices with more complex patterns are desired for purposes of templating nanoelectronic components, circuits, and functional nanodevices. Using directed nucleation, we have constructed two types of barcode lattice displaying aperiodic banding patterns. Two AFM images of the self-assembled barcode lattice are shown in Fig. 1d. In this design, each input layer of the DNA barcode contains five DX tiles, corresponding to 22.5 helical turns (≈76.5 nm) of DNA. In good agreement with our design, the measured width of the barcode lattice is ≈75 nm. The designed distance between the two closer hairpin loops is 4.5 helical turns of DNA (15.3 nm), and the measured distance is ≈16 nm; further, the designed distance between the two further adjacent hairpin loops is nine helical turns (30.6 nm), and the measured distance is ≈31 nm. Both measurements are in good agreement with our design. The zoomed-in view of the barcode lattice in Fig. 1d reveals that a typical fragment of lattice contained up to ≈24 layers assembled together.
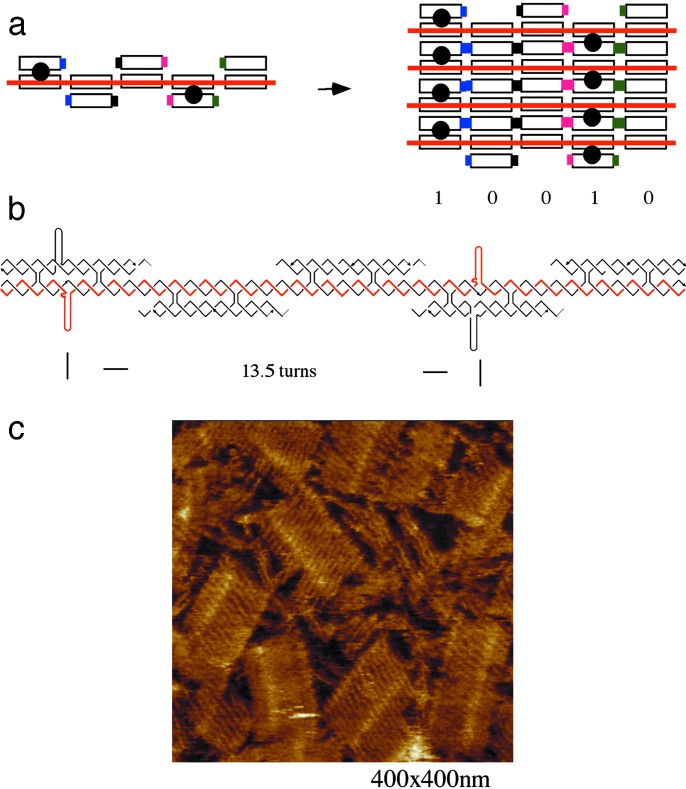
To demonstrate the programmability of the directed nucleation technique, we synthesized a modified scaffold strand along with one strand from each tile. Compared with the original barcode lattice, the resulting self-assembly displayed the inverted pattern (10010); Fig. 2a illustrates a schematic of the structure for this design. Two of the five DX tiles are DAE + 2J-containing pairs of hairpin loops directed out of the plane of the tile, and the self-assembly of the tiles around the input scaffold strand (red) produces an aperiodic pattern of 10010 in the array. Fig. 2b illustrates the strand structure of this design, and an AFM image of the self-assembled barcode lattice is shown in Fig. 2c. As with the first barcode lattice, each assembled layer contains five DX tiles, corresponding to 22.5 helical turns (≈76.5 nm) of DNA, while the measured width of the barcode lattice is ≈76 nm. The designed distance between the two hairpin loops is 13.5 turns of DNA (≈45.9 nm), while the measured distance is ≈47 nm. Thus, both measurements agree well with our design dimensions.
Fig. 2.
Inverse pattern barcode lattice (10010). (a) Schematic drawing of the barcode lattice representing the bit sequence 10010, which is the inverse of the first barcode of 01101. A single layer is shown (Left), and a four-layer lattice fragment is given (Right). (b) Strand structure of one barcode layer that represents barcode information of 01101. The red strand is the scaffold strand required for the tile assembly. The distance between adjacent hairpins is indicated in helical turns. (c) An AFM image at scale of 400 × 400 nm. Each layer contains five DX tiles, ≈75 nm wide, and the distance between the two stripes (designated 1) is ≈45 nm, as expected. See Fig. 9 for a large-area scan AFM image.
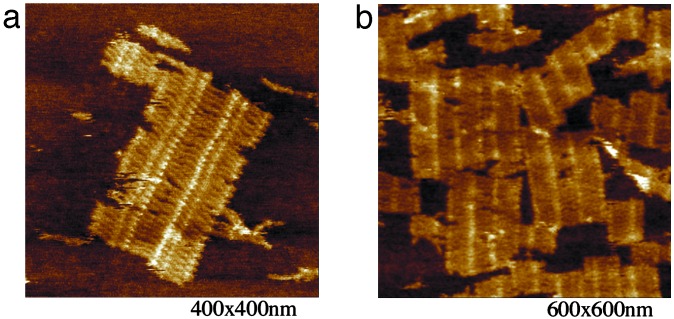
Observation of Blunt-End Helix Stacking Between DNA Tile Lattices. It is well known that base-stacking interactions contribute to the stability of DNA double-helices. Base-stacking interactions are caused by intramolecular forces between the aromatic rings of adjacent base pairs in DNA double-helices. When stacking occurs between bases located on separate helices, it is referred to as helix stacking. In the crystal structure of a Holliday junction complex (34), pairs of the four helical arms stack onto one another helix end to helix end; this results in two continuous strands and two crossover strands. Our AFM results provide clear evidence that helix stacking is a considerable factor in DNA tile self-assembly. The two barcode lattices in our design both have blunt-end helical arms on the two ends of each tile layer. In addition to the barcode lattice we observed in Figs. 1d and 2c, there are distributions of larger lattice pieces corresponding to alignment of two or more lattice fragments, with their helices almost perfectly aligned end to end. Fig. 3a shows three blocks of the first barcode lattice aligned to each other with opposite orientation (i.e., 01101-10110-01101 from left to right). Fig. 3b shows segments of the second barcode lattice aligned to each other to form a larger piece of lattice. Even without sticky-end associations, the lattice units preferred to align with one another end to end. In these larger lattices, the barcode lattice can align with neighboring fragments either in the same or opposite orientation. These results suggest that helix-stacking interactions participate in the process of the DNA lattice self-assembly to a greater extent than previously suspected, and that these interactions should not be ignored in future studies.
Fig. 3.
AFM images showing blunt-end helix stacking of tile assemblies. The scale is indicated below each image. (a) AFM image showing the alignment of two pieces of the first barcode lattice (01101) oriented in opposite directions. (b) AFM image showing the alignment of fragments of the second barcode lattice (10010) to form larger pieces of lattice.
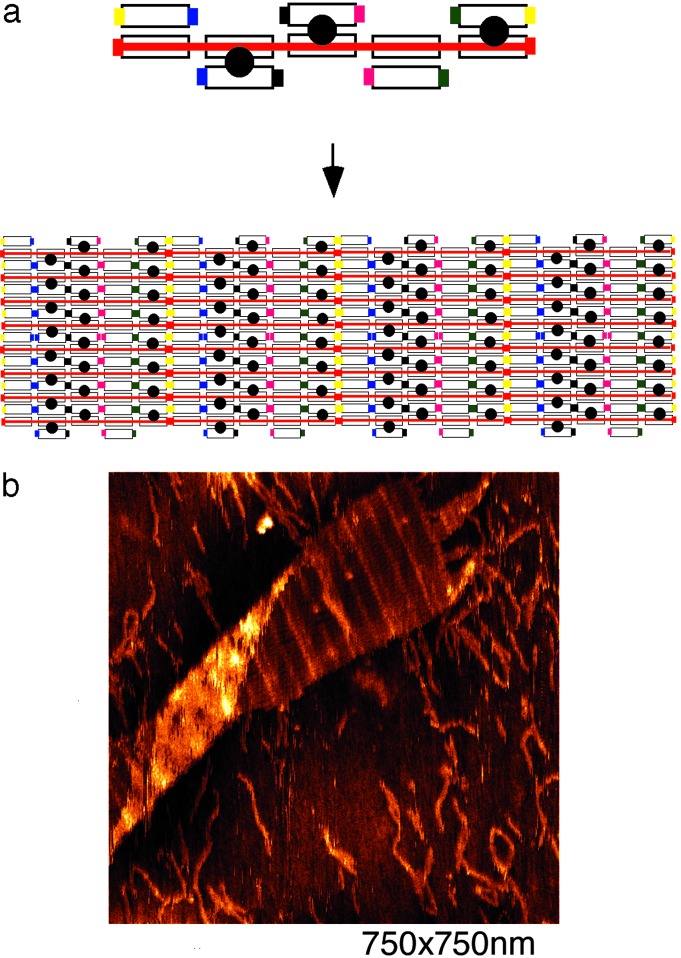
Self-Assembly of a Ribbon Lattice from Repeating DNA Barcode Units. We have extended the self-assembly of the barcode lattice further, by adding two pairs of complementary sticky ends to each end of the layers, joining the units into long ribbons. To demonstrate this, we have used the first barcode lattice. Fig. 4a shows a schematic of the five-tile layers with attached sticky ends and how they should link together to form long ribbons of repeating five-bit patterns. Fig. 4b is an AFM image that shows that the single-unit barcode lattice was successfully concatenated to form ribbon lattice displaying repeating patterns of barcode information (i.e., repeating of 10110 from right to left). Again, measured distances between features in the AFM images accord closely with the design. The objective of this experiment was to demonstrate that single-unit barcode lattice can serve as a distinguishable pixel packet for visual output within the context of a larger lattice. To generate a more useful overall pattern, barcode lattices containing more information encoded in longer scaffold strands, and distinct units that associate in specified order, can be designed.
Fig. 4.
Ribbon lattice formed from repeating DNA barcode units. (a) Schematic drawing showing the concatenation of the single-unit aperiodic barcode units into a repeating pattern. Two more pairs of sticky ends were included on the ends of the tile layer (compare with Fig. 1b). The sticky ends are represented by colored pads (yellow and red). (b) An AFM image showing the ribbon lattice displaying a repeating pattern of the barcode information. Distances between stripes are as expected.
Discussion
One major goal of DNA nanotechnology includes the growth of larger lattices and the assembly of more complex periodic and aperiodic structures. We found that self-assembly of DNA tiles around a defined scaffold strand is an effective means of pursuing these goals. In our experiments, we used ligation to generate the scaffold strand. To extend this technique to generate more complex patterns, one needs to construct longer scaffold strands. Unfortunately, ligation offers fairly low product yields; however, there are alternative strategies that can increase scaffold strand production: (i) segmental assembly via primerless PCR can be adapted to scaffold strand synthesis (35); (ii) PCR can be used to amplify very long scaffold strands produced by ligation; and (iii) in the long run, larger quantities of DNA can be produced by the use of phagemid cloning vectors and bacterial cultures, with the DNA tile information encoded in the singlestrand copy of the cloned product.
The barcode lattice as described in Figs. 1 and 2 has fairly well-defined edges, which provide promising templates for studying the binding of other molecules, such as nanocrystals, other nucleic acids, or proteins, to the DNA lattice. The surface features of the barcode lattice are easily modified, by varying the barcode information encoded in the scaffold strand. We have shown that this technique is programmable by constructing barcode lattices with various patterns. Their use will improve detection of the interaction of the DNA lattice with other components beyond that possible in a large 2D periodic DNA tile lattice (6–9). Protein–protein or protein–DNA interaction can be studied by controlling the distance between binding sites.
The helix-stacking effect between blunt-end DNA double-helices described above suggests that its importance in self-assembly of DNA tiles, lattice, and higher-order superstructures should not be neglected. Sticky ends provide specificity to helix–helix associations, but the importance of base-stacking energetics in these interactions (even devoid of single-stranded DNA base-pairing) is revealed by the prevalence of stacking between lattice pieces. A special DNA unit lattice with defined dimensions could be designed to study helix stacking between DNA double-helices with different base pair contents. To avoid helix stacking in the self-assembly of DNA tile lattices, capping loops [unpaired poly(T) sequences] could be placed on the ends of some or all portions of the barcode lattice helices.
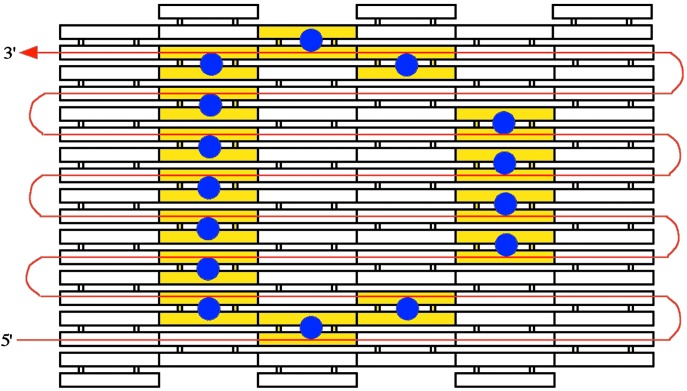
The scaffold strands described here encode 1D information. However, 2D patterns might also be constructed by the self-assembly of tiles around a DNA scaffold strand, as illustrated in Fig. 5, where the DNA sequence information to generate a letter D is encoded in the scaffold strand. Once the rest of the tiles assemble around this scaffold strand, the tiles (yellow) containing protruding stem loops or other topographical markers (blue dots) will form the 2D DNA lattice with the desired D pattern. In this approach, the scaffold strand winds through row by row of the 2D DNA lattice, so that a single strand participates in every layer of the lattice.
Fig. 5.
Schematic drawing of a proposed 2D pattern using a directed nucleation assembly technique. In red is an input single-stranded DNA scaffold strand that encodes a 2D pattern (each odd row traversing from left to right, and each even row traversing right to left). Specific DNA tiles self-assemble around each segment of this input strand. The tiles then (or perhaps concurrently) self-assemble into a 2D tiling lattice with the predetermined pattern.
Another approach for constructing a 2D pattern is to form the pattern in distinct stages row by row, with the assembly of each row dictated by a distinct scaffold strand (sequential stepwise assembly). For example, the process may begin with the self-assembly of a single initial row of tiles around an initial scaffold strand, providing a row of patterning in a way similar to that shown in Figs. 1 and 2. This scaffold strand also would have a tail section designed to provide solid support to a surface. After washing, the strands composing tiles of another layer and a further scaffold strand would be introduced into the solution for self-assembly into a further row of tiles, providing a second row of patterning. The tiles of these two rows can be designed so as to bind the current row and align it to the previous one. This process would be repeated in stages, each time providing for the self-assembly of an additional row of patterned tiles. With careful design of the tiles, a small constant number of tile types can be shown to suffice to construct any number of distinct rows. While this alternative approach is more labor intensive, it may be feasible for assembling moderately sized 2D patterned DNA lattices.
DNA self-assembly is expected to be useful in various applications in nanofabrication and for arrangement of molecular components. An illustrative example of a more complex target pattern is to create the layout for a demultiplexed random access memory (RAM) circuit, which could potentially serve as a template for molecular electronics (36). A RAM circuit essentially consists of a periodic lattice of memory elements coupled to a means to specifically address any given element for writing or reading a logical bit. Directed nucleation self-assembly may also permit the fabrication of other patterns useful for nanoelectronic and nanoscale objects for other fields of technology.
Supplementary Material
Acknowledgments
We thank Prof. Erik Winfree (California Institute of Technology, Pasadena) for providing access to a Multimode Nanoscope IIIa AFM, Drs. Paul Rothemund and Nick Papadakis for useful advice on the operation of the AFM, and Arnold E. Reif for helpful editorial suggestions. This work was supported by National Science Foundation Grants EIA-00-86015 (to J.H.R.), EIA-0218376 (to T.H.L.), and EIA-0218359 (to H.Y.) and Defense Advanced Research Projects Agency/Air Force Office of Scientific Research Grant F30602-01-2-0561 (to J.H.R.).
Abbreviations: AFM, atomic force microscopy; MBB, molecular building block; DX, double crossover.
References
- 1.Bernholc, J., Brenner, D., Nardelli, M. B., Meunier, V. & Roland, C. (2002) Annu. Rev. Mater. Res. 32 347-375. [Google Scholar]
- 2.Joachim, C., Gimzewski, J. K. & Aviram, A. (2000) Nature 408 541-548. [DOI] [PubMed] [Google Scholar]
- 3.Seeman, N. C. (2003) Nature 421 427-431. [DOI] [PubMed] [Google Scholar]
- 4.LaBean, T. H. (2003) in Computational Biology and Genome Informatics, eds. Wang, J. T. L., Wu, C. H. & Wang, P. P. (World Scientific, River Edge, NJ), pp. 35-38.
- 5.Reif, J. H. (2002) Comput. Sci. Eng. Mag.: Special Issue on Bio-Computation 4 32-41. [Google Scholar]
- 6.Winfree, E., Liu, F., Wenzler, L. A. & Seeman, N. C. (1998) Nature 394 539-544. [DOI] [PubMed] [Google Scholar]
- 7.LaBean, T. H., Yan, H., Kopatsch, J., Liu, F., Winfree, E., Reif, J. H. & Seeman, N. C. (2000) J. Am. Chem. Soc. 122 1848-1860. [Google Scholar]
- 8.Mao, C., Sun, W. & Seeman, N. C. (1999) J. Am. Chem. Soc. 121 5437-5443. [Google Scholar]
- 9.Sha, R., Liu, F., Millar, D. P. & Seeman, N. C. (2000) Chem. Biol. 7 743-749. [DOI] [PubMed] [Google Scholar]
- 10.Mao, C., Sun, W., Shen, Z. & Seeman, N. C. (1999) Nature 397 144-146. [DOI] [PubMed] [Google Scholar]
- 11.Yurke, B., Turberfield, A. J., Mills, A. P., Simmel, F. C. & Neumann, J. L. (2000) Nature 406 605-608. [DOI] [PubMed] [Google Scholar]
- 12.Yan, H., Zhang, X., Shen, Z. & Seeman, N. C. (2002) Nature 415 62-65. [DOI] [PubMed] [Google Scholar]
- 13.Li, J. J. & Tan, W. (2002) Nano Lett. 2 315-318. [Google Scholar]
- 14.Adleman, L. M. (1994) Science 266 1021-1024. [DOI] [PubMed] [Google Scholar]
- 15.Liu, Q., Wang, L., Frutos, A. G., Condon, A. E., Corn, R. M. & Smith, L. M. (2000) Nature 403 175-179. [DOI] [PubMed] [Google Scholar]
- 16.Mao, C., LaBean, T. H., Reif, J. H. & Seeman, N. C. (2000) Nature 407 493-496. [DOI] [PubMed] [Google Scholar]
- 17.Yan, H., Feng, L., LaBean, T. H. & Reif, J. H. (2003) DNA Computing, Ninth International Workshop, eds. Chen, J. & Reif, J. (Springer, Berlin), in press.
- 18.Benenson, Y., Paz-Elizur, T., Adar, R., Keinan, E., Livneh, A. & Shapiro, E. (2001) Nature 414 430-434. [DOI] [PubMed] [Google Scholar]
- 19.Braich, R. S., Chelyapov, N., Johnson, C., Rothemund, P. W. K. & Adleman, L. (2002) Science 296 499-502. [DOI] [PubMed] [Google Scholar]
- 20.Mirkin, C. A. (2000) Inorg. Chem. 39 2258-2272. [DOI] [PubMed] [Google Scholar]
- 21.Zanchet, D., Wolfgang, C. M. M., Parak, J., Gerion, D. & Alivisatos, A. P. (2001) Nano Lett. 1 32-35. [Google Scholar]
- 22.Elghanian, R., Storhoff, J. J., Mucic, R. C., Letsinger, R. L. & Mirkin, C. A. (1997) Science 277 1078-1081. [DOI] [PubMed] [Google Scholar]
- 23.Loweth, C. J., Caldwell, W. B., Peng, X. G., Alivisatos, A. P. & Schultz, P. G. (1999) Angew. Chem. Int. Ed. 38 1808-1812. [DOI] [PubMed] [Google Scholar]
- 24.Xiao, S., Liu, F., Rosen, A., Hainfeld, J., Seeman, N. C., Musier-Forsynth, K. M. & Kiehl, R. A. (2002) J. Nanoparticle Res. 4 313-317. [Google Scholar]
- 25.Niemeyer, C. M., Burger, W. & Peplies, J. (1998) Angew. Chem. Int. Ed. 37 2265-2268. [DOI] [PubMed] [Google Scholar]
- 26.Braun, E., Eichen, Y., Sivan, U. & Ben-Yoseph, G. (1998) Nature 391 775-778. [DOI] [PubMed] [Google Scholar]
- 27.Seeman, N. C. (1999) Trends Biotechnol. 17 437-443. [DOI] [PubMed] [Google Scholar]
- 28.Seeman, N. C. (2001) Nano Lett. 1 22-26. [Google Scholar]
- 29.Winfree, E. (1995) in DIMACS Series in Discrete Mathematics and Theoretical Computer Science: Proceedings of the First DIMACS Workshop on DNA-Based Computers, eds. Lipton, R. & Baum, E. B. (Am. Math. Soc., Providence, RI), Vol. 27, pp. 199-221. [Google Scholar]
- 30.LaBean, T. H., Winfree, E. & Reif, J. H. (1999) in DIMACS Series in Discrete Mathematics and Theoretical Computer Science: Proceedings of the Fifth DIMACS Workshop on DNA-Based Computers, eds. Winfree, E. & Gifford, D. K. (Am. Math. Soc., Providence, RI), pp. 123-140.
- 31.Seeman, N C. (1990) J. Biomol. Struct. Dyn. 8 573-581. [DOI] [PubMed] [Google Scholar]
- 32.Fu, T.-J. & Seeman, N. C. (1993) Biochemistry 32 3211-3220. [DOI] [PubMed] [Google Scholar]
- 33.Li, X., Yang, X., Qi, J. & Seeman, N. C. (1996) J. Am. Chem. Soc. 118 6131-6140. [Google Scholar]
- 34.Ortiz-Lombardia, M., Gonzalez, A., Eritja, R., Aymami, J., Azorin, F. & Coll, M. (1999) Nat. Struct. Biol. 6 913-917. [DOI] [PubMed] [Google Scholar]
- 35.Stemmer, W. P. C. (1994) Proc. Natl. Acad. Sci. USA 91 10747-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothemund, P. W. K. (2001) Ph.D. thesis (Univ. of Southern California, Los Angeles).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.